Abstract
During Bacillus subtilis sporulation, the SpoIIIE DNA translocase moves a trapped chromosome across the sporulation septum into the forespore. The direction of DNA translocation is controlled by the specific assembly of SpoIIIE in the mother cell and subsequent export of DNA into the forespore. We present evidence that the MinCD heterodimer, which spatially regulates cell division during vegetative growth, serves as a forespore-specific inhibitor of SpoIIIE assembly. The deletion of minCD increases the ability of forespore-expressed SpoIIIE to assemble and translocate DNA, and causes otherwise wild-type cells to reverse the direction of DNA transfer, producing anucleate forespores. We propose that two distinct mechanisms ensure the specific assembly of SpoIIIE in the mother cell, the partitioning of more SpoIIIE molecules into the larger mother cell by asymmetric cell division and the MinCD-dependent repression of SpoIIIE assembly in the forespore. Our results suggest that the ability of MinCD to sense positional information is utilized during sporulation to regulate protein assembly differentially on the two faces of the sporulation septum.
Keywords: DNA segregation/MinCD/polarity/ protein assembly/sporulation
Introduction
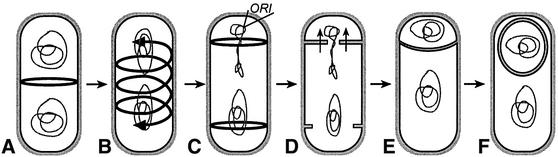
The generation of asymmetry is essential for development in both eukaryotes and prokaryotes, since it allows the products of a single cell division event to adopt dissimilar developmental programs. The sporulating bacterium Bacillus subtilis provides a simple developmental system to investigate the generation of asymmetry. A key step in sporulation is the asymmetrically positioned cell division event that gives rise to two progeny of differing size and developmental fate: a smaller forespore, which ultimately becomes the spore, and a larger mother cell, which eventually lyses after contributing to spore development (Piggot and Losick, 2002). Two dramatic alterations in cellular architecture prepare the cell for polar division: reorganization of the chromosomes into an elongated structure known as the axial filament (Ryter, 1965) and relocalization of the cell division machinery from midcell to the poles (Levin and Losick, 1996). Recent studies indicate that axial filament formation results from the anchoring of both chromosomes to the cell poles (Thomaides et al., 2001) and the asymmetric partitioning of the future forespore chromosome, with the origin-proximal 30% condensed near one cell pole (Pogliano et al., 2002). Relocalization of the cell division machinery occurs when the medial ring of the key cell division protein FtsZ is converted into a spiral intermediate through which it relocalizes to two rings, one near each cell pole (Ben-Yahuda and Losick, 2002) (Figure 1B and C). This event requires elevated expression of both FtsZ and the SpoIIE phosphatase, a bifunctional protein required for both polar division and activation of the first cell-specific transcription factor, σF. Although the two polar division sites are formed concurrently, they are activated sequentially, producing a single forespore (Lewis et al., 1994; Pogliano et al., 1999). Cell-specific gene expression commences first in this smaller cell and subsequently in the larger mother cell, which produces three proteins responsible for repressing division at the second polar division site (Pogliano et al., 1999; Eichenberger et al., 2001). Mutants lacking mother cell-specific gene expression therefore divide at both polar sites, producing abortively disporic sporangia that contain two forespores and an anucleate mother cell (Setlow et al., 1991). Thus, during B.subtilis sporulation, the establishment of asymmetry requires a dramatic reorganization of cellular architecture, while the maintenance of asymmetry requires cell-specific gene expression.
Fig. 1. The sporulation pathway of B.subtilis. (A) Vegetative B.subtilis cells divide at midcell, assembling medial rings of the cell division protein FtsZ (rings) and separately condensing their chromosomes prior to cytokinesis. (B) At the onset of sporulation, production of the SpoIIE phosphatase and higher levels of FtsZ allow FtsZ to form a spiral intermediate, and (C) relocalize to two rings, one near each cell pole. At the same time, the axial filament forms, with the chromosomes anchored to the cell poles and the future forespore chromosome partitioned to condense the origin-proximal 30% near the cell pole. (D) Division occurs at one FtsZ ring, thereby trapping the forespore chromosome in the septum, from which it is cleared by the SpoIIIE DNA translocase (arrows). Three proteins produced in the mother cell inhibit division at the second division site, thereby ensuring the maintenance of asymmetry. (E) During the phagocytosis-like process of engulfment, the mother cell membrane migrates around the forespore, (F) which ultimately is fully enclosed in the mother cell cytoplasm.
In addition to its asymmetric position, the sporulation septum differs from a vegetative septum as it bisects the forespore chromosome, thereby trapping the origin-proximal 30% of the chromosome in the forespore. The remaining 70% of the trapped chromosome is translocated subsequently across the sporulation septum by SpoIIIE (Wu and Errington, 1994; Wu et al., 1995), which assembles into a focus at the septal midpoint and probably serves as a DNA channel (Wu and Errington, 1997; Bath et al., 2000; Errington et al., 2001). This post-septational chromosome segregation causes transient genetic asymmetry, since the forespore lacks the bulk of its genetic complement prior to the completion of chromosome translocation. This genetic asymmetry facilitates activation of the first cell-specific transcription factor (σF), by allowing the proteolytic depletion of the anti-σF factor SpoIIAB from the forespore during the period in which its gene remains trapped in the mother cell (Dworkin and Losick, 2001). Genetic asymmetry also impacts activation of mother cell-specific transcription, since the gene encoding the forespore-expressed signaling protein necessary for activation of mother cell-specific gene expression (spoIIR) must be located in the chromosomal domain initially trapped in the forespore so that it can be expressed immediately after polar septation, thereby activating mother cell-specific transcription before the production of abortively disporic sporangia (Khvorova et al., 2000; Zupancic et al., 2001).
A more subtle asymmetry is necessary for the completion of chromosome segregation into the forespore, a step that requires SpoIIIE to act in a vectoral manner, to specifically move the trapped chromosome into the forespore. Recently, we have provided evidence that the direction of DNA transfer is ensured by the specific assembly of SpoIIIE on the mother cell face of the septum, where it acts as a DNA exporter (Sharp and Pogliano, 2002b). Our results demonstrate that SpoIIIE exports DNA from the cell in which it is synthesized, and suggest the existence of a regulator of SpoIIIE assembly capable of discriminating between the forespore and mother cell faces of the sporulation septum to ensure the correct polarity of SpoIIIE assembly and DNA transfer (Sharp and Pogliano, 2002b).
Little is known about how positional information is encoded or sensed in bacterial cells. Two conserved proteins with well-established roles in spatial regulation are MinC and MinD (hereafter MinCD), which act in concert to restrict division to the cell midpoint during vegetative growth (de Boer et al., 1989; Levin et al., 1992; Varley and Stewart, 1992; Marston et al., 1998; Marston and Errington, 1999). These proteins prevent assembly of FtsZ at non-medial positions, thereby inhibiting polar division; in their absence, division takes place near the cell poles, producing small anucleate cells (minicells) and longer multinucleate filaments. In Escherichia coli, MinCD oscillate from pole to pole (Hu and Lutkenhaus, 1999; Raskin and de Boer, 1999a,b), while in B.subtilis, MinCD appear to localize stably to both cell poles (Marston et al., 1998; Marston and Errington, 1999). The ability of MinCD to spatially regulate cell division and to disrupt FtsZ polymerization (Hu et al., 1999) prompted us to explore the possibility that they also regulate SpoIIIE assembly during sporulation. Our results suggest that MinCD contribute to the forespore-specific inhibition of SpoIIIE assembly, and thereby play a key role in establishing cellular polarity during sporulation.
Results
Effects of MinCD on SpoIIIE localization
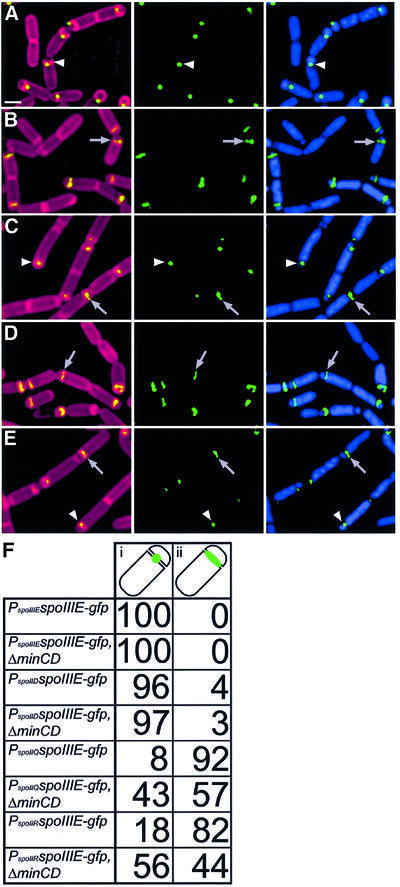
To determine whether MinCD regulates SpoIIIE assembly, we first tested the effects of a minCD deletion on the localization of SpoIIIE–green fluorescent protein (GFP). When expressed from its native promoter, SpoIIIE–GFP localized as a focus at the middle of the sporulation septum (Figure 2A, arrowhead), where it probably forms a channel through which the forespore chromosome is translocated (Wu and Errington, 1997). Subsequently, this protein focus moved around the forespore to the cell pole, where it participated in the final step of engulfment, membrane fusion (Sharp and Pogliano, 1999). However, when expressed in the forespore from the spoIIQ promoter, SpoIIIE–GFP failed to assemble foci in most sporangia, instead localizing as a line along the polar septum (Figure 2B, arrow) (Sharp and Pogliano, 2002b). In wild-type cells expressing PspoIIQ-spoIIIE, only 8% of sporangia with flat polar septa show normal focus assembly (Figure 2B, arrow); however, when minCD are deleted, this frequency rises to 43% (Figure 2C, arrowhead, and F). Similar results were obtained when SpoIIIE was expressed in the forespore at lower levels from the spoIIR promoter, where SpoIIIE–GFP foci were seen in 18% of wild-type sporangia (Figure 2D, arrow) compared with 56% of ΔminCD mutant sporangia (Figure 2E, arrowhead, and F). The absence of MinCD had no effect on focus assembly when spoIIIE was expressed from either its native promoter or the mother cell-specific spoIID promoter (data not shown; scored in Figure 2F). These results suggest that MinCD are necessary for the forespore-specific inhibition of SpoIIIE DNA translocase assembly.
Fig. 2. Effect of minCD deletion on SpoIIIE–GFP localization. Cells were harvested at t1.5 and stained as described in Materials and methods. Images show Mitotracker red-stained membranes (red), DAPI-stained DNA (blue) and SpoIIIE–GFP (green). (A) SpoIIIE–GFP expressed from its native promoter efficiently assembles into foci at the septal midpoint (arrowhead; strain KP629). Scale bar = 1 µm. (B and C) SpoIIIE–GFP expressed at high levels in the forespore (PspoIIQ-spoIIIE-gfp) in (B) a wild-type (KP630) or (C) a ΔminCD strain (KP657), the latter of which allows additional foci to assemble. Arrowheads indicate foci of SpoIIIE–GFP at the septal midpoint, while arrows indicate lines of SpoIIIE–GFP along the septum. (D and E) Localization of SpoIIIE–GFP expressed at low levels in the forespore (from PspoIIR-spoIIIE-gfp) in (D) a wild-type (KP632) or (E) a ΔminCD strain (KP659). Again, more foci assemble in the ΔminCD strain. (F) Percentage of sporangia with the fusions indicated showing SpoIIIE–GFP localization as either (i) a focus or (ii) a line along a septum that appeared complete. Only sporangia with flat polar septa were scored, to ensure that only the early stages of SpoIIIE–GFP localization and production were scored. A total of 120–200 sporangia were scored for each strain.
Deletion of MinCD improves the efficiency of reverse chromosome translocation
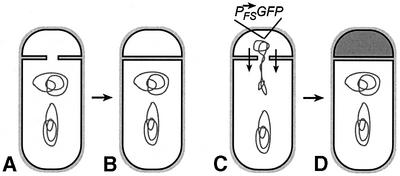
If the reduced assembly of foci by forespore-produced SpoIIIE explains its reduced ability to support chromosome translocation (Sharp and Pogliano, 2002b), then the ΔminCD mutation should improve the ability of forespore-produced SpoIIIE to translocate DNA. In order to test this prediction, we needed to discriminate between anucleate minicells produced during growth of ΔminCD strains and anucleate forespores resulting from reverse chromosome translocation. This was accomplished by fusing either gfp or spoIIIE-gfp to the forespore-specific spoIIQ promoter. These GFP reporters were integrated in the origin-proximal region of the chromosome, which is located in the forespore immediately after division and prior to chromosome translocation. Anucleate cells containing GFP fluorescence were forespores that must have contained DNA, activated forespore gene expression (Figure 3C) and have undergone reverse chromosome translocation (Figure 3D). In contrast, anucleate cells lacking GFP must have never contained DNA or activated forespore-specific gene expression (Figure 3A and B), and therefore are either minicells or anucleate forespores resulting from the defective anchoring of the chromosome to the cell pole (which has been reported to occur in certain minicell-producing mutants; Mulder et al., 1990; Äkerlund et al., 1992; Thomaides et al., 2001).
Fig. 3. Method to discriminate anucleate forespores resulting from reverse chromosome translocation from minicells. (A and B) During growth, minCD mutants produce small anucleate cells resulting from sporulation-independent polar division. These anucleate cells can be discriminated from anucleate forespores resulting from reverse chromosome translocation by using a forespore-expressed gfp fusion gene located in the origin-proximal chromosome domain. Forespores that never contained a chromosome, such as those which result from defective polar anchoring of the chromosome (Mulder et al., 1990; Äkerlund et al., 1992; Thomaides et al., 2001), will appear identical to minicells in this assay. (C and D) Anucleate forespores containing GFP fluorescence (indicated by shading in D) must have once contained a chromosome, activated forespore-specific gene expression and subsequently translocated the forespore chromosome into the mother cell.
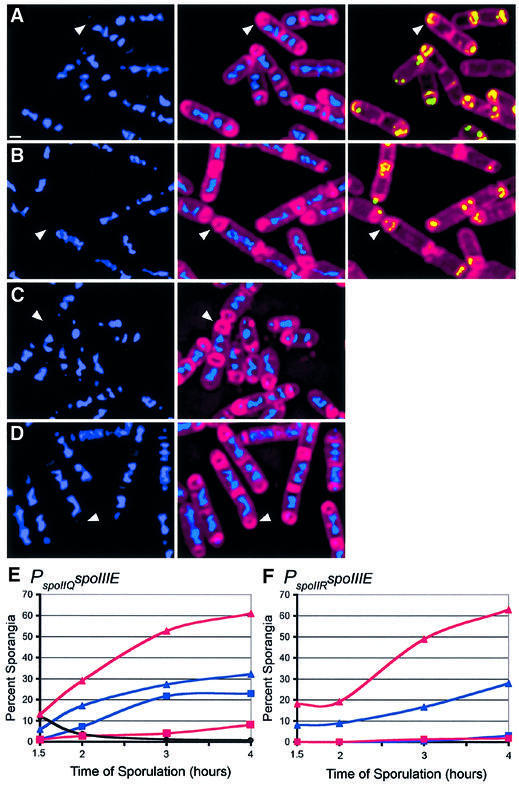
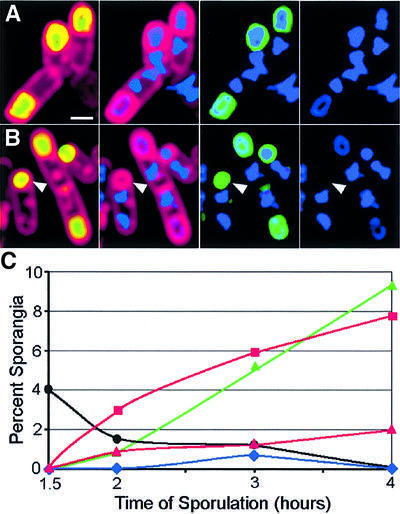
Deletion of minCD increased the efficiency with which SpoIIIE produced by the forespore-specific spoIIQ promoter catalyzed reverse chromosome translocation, from 32% in the wild type to 61% in ΔminCD 4 h after the onset of sporulation (t4) (Figure 4A, B and E). We also noted that in ΔminCD strains, the frequency of anucleate cells lacking GFP fluorescence (minicells) declined from 10% at t1.5 to <0.5% at t4 (Figures 4E and 5C). Therefore, by later times of sporulation, the contribution of minicells to the total population of anucleate cells was negligible. This observation allowed us to also test the effect of the ΔminCD mutation on reverse chromosome translocation when SpoIIIE–GFP is expressed from the weaker spoIIR promoter, although we could not readily detect the low level of GFP fluorescence produced by this fusion after the chemical fixation employed to ensure uniform permeability to 4′,6-diamidino-2-phenylindole (DAPI). The ΔminCD mutation also increased the number of anucleate cells produced by PspoIIR-spoIIIE, from 28% in the wild type to 63% in ΔminCD at t4 (Figure 4C, D and F). These results suggest that the additional SpoIIIE–GFP foci assembled in ΔminCD forespores are active and capable of promoting reverse chromosome translocation.
Fig. 4. Effect of the minCD deletion on chromosome translocation mediated by forespore-expressed SpoIIIE. (A–D) Samples from t3 were fixed and lysozyme treated to ensure uniform permeability to fluorescent stains, then the membranes were stained with FM 4-64 (red) and the DNA stained with DAPI (blue) as described (Materials and methods). Arrowheads indicate sporangia that have undergone reverse translocation. Sporangia expressing spoIIIE at high levels in the forespore (PspoIIQ-spoIIIE-gfp) in (A) a wild-type (KP630) or (B) a ΔminCD mutant background (KP657). SpoIIIE–GFP (green) fluorescence is also shown for this strain, to distinguish between minicells and anucleate forespores. Sporangia expressing spoIIIE at low levels in the forespore (PspoIIR-spoIIIE-gfp) in (C) wild-type (KP632) or (D) ΔminCD mutant background (KP659). GFP fluorescence is not shown, since it was not possible to detect SpoIIIE–GFP fluorescence reproducibly after fixation, due to the low levels of expression from the spoIIR promoter. The scale bar in (A) = 1 µm. (E and F) Chromosome translocation kinetics when spoIIIE was expressed at (E) high (PspoIIQ-spoIIIE-gfp) or (F) low (PspoIIR-spoIIIE-gfp) levels in the forespore. Chromosome translocation was scored and is presented as the percentage of sporangia showing forward (squares) and reverse (triangles) translocation in ΔminCD mutant (red) or wild-type (blue) backgrounds. The black line in (E) indicates minicell frequency in ΔminCD PspoIIQ-spoIIIE-gfp. At least 200 sporangia were scored for each strain.
Fig. 5. Anucleate forespores are produced by the ΔminCD mutant. Micrographs of (A) wild-type (KP646) and (B) ΔminCD sporangia (KP651) at t3 fixed and stained (as described in Materials and methods), showing FM 4-64-stained membranes (red), DAPI-stained DNA (blue) and forespore-expressed GFP (green; PspoIIQ-gfp). The arrowhead in (B) indicates an anucleate forespore. The scale bar in (A) = 1 µm. (C) Scoring of minicell (black circles) and anucleate forespore production. In the ΔminCD mutant, the number of minicells declined during sporulation (black circles), whereas the frequency of anucleate forespores increased (red squares). When minCD was expressed in the mother cell of the ΔminCD mutant (strain KP653), the frequency of anucleate forespores was unchanged (green triangles). Expression of minCD in the forespore of the ΔminCD mutant reduced anucleate forespore production (red triangles; strain KP652), whereas introduction of a spoIIIE mutation abolished anucleate forespore production in the ΔminCD mutant (blue diamonds; strain KP655). At least 250 sporangia were scored for each strain.
When SpoIIIE is expressed in the forespore at high levels (from PspoIIQ-spoIIIE), some forward DNA translocation is observed, an effect likely to be due to the escape of SpoIIIE into the mother cell, where it assembles into a functional DNA channel more rapidly than in the forespore (Sharp and Pogliano, 2002b). Deletion of minCD significantly reduced this forward chromosome translocation (from 23% to 8%; Figure 4E), suggesting that the ΔminCD mutation reduces the kinetic disadvantage of SpoIIIE assembly in the forespore, allowing forespore-expressed SpoIIIE to assemble into a functional channel before it can escape into the mother cell.
Normally, production of SpoIIIE in the mother cell closely mimics the wild-type situation, supporting efficient forward translocation and no reverse translocation. However, the deletion of minCD allowed mother cell-expressed SpoIIIE to support some reverse translocation (4% of 292 sporangia scored at t4), which was never observed in cells with wild-type minCD (Sharp and Pogliano, 2002b). This suggests that in the absence of MinCD, mother cell-expressed SpoIIIE was able to enter the forespore and assemble into an active DNA transporter, which then exported the forespore chromosome. Together, these results suggest that the ΔminCD mutation eliminates a forespore-specific inhibitor of reverse chromosome translocation.
MinCD regulates the polarity of SpoIIIE produced prior to division
SpoIIIE normally is produced prior to septation, rather than after division as in the preceding experiments. If MinCD normally regulate the polarity of SpoIIIE DNA translocase assembly, then a ΔminCD strain with a wild-type copy of spoIIIE should occasionally produce anucleate forespores resulting from the reversal of SpoIIIE polarity. Again, we used forespore-expressed GFP to distinguish minicells from anucleate forespores produced by reverse chromosome translocation (Figure 3), and observed neither anucleate forespores nor minicells in wild type (>2000 sporangia examined). However, ΔminCD sporangia produced 8% anucleate forespores (Figure 5B, arrowhead) and 0.5% minicells by t4 (Figure 5B and C). A spoIIIE minCD double mutant produced the same frequency of minicells as the ΔminCD single mutant, but no anucleate forespores (Figure 5C), confirming that anucleate forespores result from a SpoIIIE-mediated DNA translocation event. Thus, in an otherwise wild-type strain, the absence of MinCD can lead to a reversal in the direction that SpoIIIE translocates DNA.
To confirm that MinCD acts specifically in the forespore to prevent reverse chromosome segregation, we tested the effect of mother cell- and forespore-produced MinCD on the polarity of chromosome translocation in the ΔminCD mutant. When minCD was expressed in the mother cell of a ΔminCD mutant, there was no effect on the frequency of reverse chromosome translocation (Figure 5C). However, when minCD was expressed in the forespore of the ΔminCD mutant, the frequency of reverse chromosome translocation dropped from 8% to <2% at t4 (Figure 5C). Thus, MinCD is required specifically in the forespore to prevent reverse chromosome translocation. The failure to completely abolish reverse chromosome translocation when minCD was expressed in the forespore could result from the delay in minCD expression relative to spoIIIE expression and localization: in this experiment, SpoIIIE assembles during polar septation, while MinCD is produced after polar septation. Therefore, concentrations of MinCD might be limiting in some forespores, allowing some reverse translocation. However, because forespore-expressed MinCD inhibits reverse translocation to a significant extent, it is possible that MinCD can disassemble pre-existing SpoIIIE complexes, similar to its proposed role in disassembling FtsZ polymers (Hu et al., 1999).
Discussion
During B.subtilis sporulation, the SpoIIIE DNA translocase must act in a vectoral manner across the sporulation septum specifically to move the trapped forespore chromosome into, rather than out of, the forespore. Previously, we provided evidence that SpoIIIE acts as a DNA exporter whose vectoral activity is conferred by its differential assembly in the two cells of the sporangium (Sharp and Pogliano, 2002b). Specifically, we found that mother cell-expressed, but not forespore-expressed, SpoIIIE localizes to the septal midpoint and efficiently catalyzes chromosome translocation into the forespore. Because SpoIIIE is synthesized prior to polar septation and is present in both cells of the sporangium, these results suggest the existence of a mechanism to regulate the assembly of a functional SpoIIIE DNA translocase differentially on each side of the sporulation septum.
Here we demonstrate that the absence of MinCD allows forespore-expressed SpoIIIE to assemble a focus at the septal midpoint and catalyze reverse chromosome translocation with higher efficiency. MinCD are well-conserved proteins responsible for inhibiting cell division at polar sites in E.coli and B.subtilis, thereby restricting cell division to the cell midpoint (reviewed by Margolin, 2001; Rothfield et al., 2001). Our results suggest that MinCD play an unanticipated second role during sporulation, repressing SpoIIIE assembly in the forespore to ensure that the DNA translocase assembles with the appropriate polarity in the septum. In keeping with this hypothesis, inactivation of minCD in an otherwise wild-type background leads to the production of anucleate forespores in a SpoIIIE-dependent manner.
It therefore appears that the ability of MinCD to sense positional information is utilized during sporulation to specifically inhibit assembly of the SpoIIIE DNA translocase on the forespore face of the sporulation septum. MinC has been shown to disassemble FtsZ filaments in vitro (Hu et al., 1999), which, together with its MinD-dependent localization to the cell poles (Hu and Lutkenhaus, 1999; Marston and Errington, 1999; Raskin and de Boer, 1999a), suggests that MinCD restrict vegetative division to midcell by disassembling FtsZ rings at the cell poles. MinCD might play a similar role in regulating SpoIIIE assembly, acting in the forespore to disassemble SpoIIIE foci. Alternatively, they might interact with another factor, which itself modulates SpoIIIE assembly; further experiments are required to resolve this question. However, our results clearly demonstrate that MinCD affect SpoIIIE localization, and suggest that these proteins are able to contribute to the polarity with which an individual protein assembles within the septum.
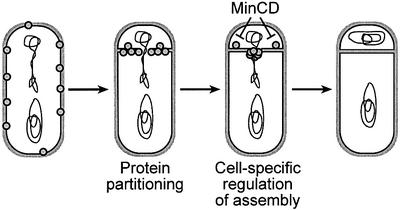
We propose that two processes act in concert to ensure the mother cell-specific assembly of the SpoIIIE DNA translocase (Figure 6). The first mechanism involves differences in abundance of SpoIIIE in the two cells of the sporangium (Sharp and Pogliano, 2002b). Prior to polar septation, SpoIIIE is distributed uniformly in the cell membrane; however, as a consequence of asymmetric septation, more SpoIIIE molecules are partitioned into the mother cell than the forespore. Indeed, after division, the mother cell contains ∼8-fold more SpoIIIE than the forespore (Sharp and Pogliano, 2002b). SpoIIIE is subsequently recruited to the nascent septum from this pool of free protein, forming a ring at the site of membrane invagination (Sharp and Pogliano, 2002a). We propose that the larger pool size of SpoIIIE in the mother cell leads to a higher concentration of SpoIIIE on the mother cell face of the growing sporulation septum, thereby providing a kinetic advantage to complex assembly in the mother cell. The contribution of protein partitioning to the polarity of SpoIIIE assembly can be overcome by producing SpoIIIE only after polar septation, in which case forespore-expressed SpoIIIE is able to support reverse chromosome translocation, although at a reduced efficiency relative to forward chromosome translocation.
Fig. 6. Model for the role of MinCD in cell-specific SpoIIIE assembly. We propose that the polarity of SpoIIIE assembly is controlled by two events: (i) the asymmetric partitioning of SpoIIIE by polar septation, which results in the mother cell containing ∼8-fold more SpoIIIE than the forespore (Sharp and Pogliano, 2002b); and (ii) the MinCD-dependent inhibition of SpoIIIE assembly in the forespore. Together, these two pathways serve to ensure that SpoIIIE assembles specifically on the forespore face of the septum, thereby ensuring the correct polarity of DNA translocation.
Secondly, we propose that MinCD-dependent repression of SpoIIIE assembly in the forespore serves to ensure that any SpoIIIE molecules partitioned into the forespore during polar division are unable to assemble into an active DNA translocase. Our observation that inactivation of minCD in an otherwise wild-type strain allows just 8% reverse chromosome translocation suggests that in most sporangia the increased SpoIIIE pool size in the mother cell achieved by protein partitioning suffices to ensure the correct polarity of SpoIIIE assembly. However, eliminating MinCD-dependent repression of translocase assembly more than doubles the level of reverse chromosome translocation observed when we eliminate the advantage of pool size by expressing SpoIIIE only in the forespore. Under these circumstances, 60% of sporangia show reverse chromosome translocation, while most of the remaining 40% show untranslocated chromosomes.
The fact that we have not yet been able to produce 100% anucleate forespores suggests the existence of additional regulatory mechanisms to ensure accurate chromosome partitioning during sporulation. One candidate for such a factor is DivIVA, which interacts with MinCD to regulate cell division during vegetative growth (Edwards and Errington, 1997; Marston et al., 1998). Interestingly, DivIVA has also been proposed to anchor the forespore chromosome to the cell pole prior to asymmetric division, since divIVA minD double mutants produce more anucleate cells than minD mutants during sporulation (Thomaides et al., 2001). However, during cell division, DivIVA is thought to regulate polar division by recruiting the MinCD division inhibitors to the cell poles, where they inhibit FtsZ assembly (Marston et al., 1998; Marston and Errington, 1999). It therefore seems likely that if DivIVA affects SpoIIIE assembly, it would do so by controlling the localization and function of MinCD.
Another regulatory mechanism that might contribute to accurate chromosome partitioning is the inherent polarity of the DNA substrate, which might also favor forward over reverse chromosome translocation. Recently, evidence has been provided suggesting that the direction in which the E.coli homolog of SpoIIIE, FtsK, moves DNA is determined by the polarity of the DNA substrate itself (Corre and Louarn, 2002). Similarly, it is possible that specific asymmetric sequence motifs are located on the region of the chromosome trapped in the invaginating septum, and that these contribute to the polarity of DNA transfer. A second mechanism by which chromosomal polarity might contribute to the direction of DNA transfer is provided by the observation that most genes in the bacterial chromosome are arranged so that transcription proceeds away from the origin of replication, an arrangement that probably reduces collisions between RNA and DNA polymerase. Similarly, during forward chromosome translocation, most RNA polymerase molecules move along DNA in the same direction as SpoIIIE, whereas during reverse chromosome translocation, RNA polymerase and SpoIIIE would move in opposite directions, perhaps resulting in collisions between RNA polymerase and SpoIIIE that might slow the rate of DNA translocation.
At each key step of B.subtilis sporulation, overlapping regulatory mechanisms act in concert to ensure the appropriate outcome. This is true of the switch from medial to asymmetric cell division, which requires the production of the SpoIIE phosphatase and increased levels of FtsZ (Ben-Yahuda and Losick, 2002). It is also true of activation of the first cell-specific transcription factor, σF, which requires both specific protein localization (Arigoni et al., 1995; Duncan et al., 1995) and transient genetic asymmetry, which allows the specific depletion of the anti-σF factor from the forespore (Dworkin and Losick, 2001). Here we present evidence that the polarity of DNA transfer is controlled similarly by multiple mechanisms including protein partitioning, cell-specific regulation of protein assembly and an as yet unidentified third mechanism. It seems likely that these overlapping regulatory mechanisms serve to convey robustness to sporulation, ensuring that a viable spore is produced even in a changing and uncertain environment.
Materials and methods
Strains, growth conditions and genetic manipulation
Strains were induced to sporulate by the resuspension method (Sterlini and Mandelstam, 1969). All strains (Table I) are derivatives of the wild-type strain PY79 (Youngman et al., 1984) and were produced using standard methods (Hoch, 1991). Disruptions of amyE were achieved by integrating the specified plasmid construct by a double recombination event. All plasmids used for amyE integration were derivatives of pDG1662 (Guerout-Fleury et al., 1996) with a variety of modifications as described in Sharp and Pogliano (2002b), and those summarized below. All PCR amplification was done using the Roche Expand High Fidelity PCR Kit. Restriction digests were performed with the specified restriction enzymes (New England Biolabs), ligations used T4 DNA ligase (New England Biolabs) and all cloning was performed in E.coli DH5α.
Table I. Bacillus subtilis strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| PY79 | Wild type | Youngman et al. (1984) |
| KP92 | spoIIIE36 | Wu and Errington (1994) |
| KP141 | ΔspoIIIE::spc | Pogliano et al. (1997) |
| KP629 | ΔspoIIIE::spc, amyE::PspoIIIEspoIIIE-gfpΩcat | Sharp and Pogliano (2002b) |
| KP630 | ΔspoIIIE::spc, amyE::PspoIIQspoIIIE-gfpΩcat | Sharp and Pogliano (2002b) |
| KP631 | ΔspoIIIE::spc, amyE::PspoIIDspoIIIE-gfpΩcat | Sharp and Pogliano (2002b) |
| KP632 | ΔspoIIIE::spc, amyE::PspoIIRspoIIIE-gfpΩcat | Sharp and Pogliano (2002b) |
| KP646 | amyE::PspoIIQ-gfpΩcat | This study |
| KP651 | amyE::PspoIIQ-gfpΩtet, ΔminCD::erm | This study |
| KP652 | amyE::PspoIIQ-gfpΩtet, ΔminCD::erm, sacA::PspoIIQminCDΩcat | This study |
| KP653 | amyE::PspoIIQ-gfpΩtet, ΔminCD::erm, sacA::PspoIIDminCDΩcat | This study |
| KP654 | ΔspoIIIE::spc, amyE::PspoIIQ-gfpΩtet, ΔminCD::erm | This study |
| KP655 | spoIIIE36, amyE::PspoIIQ-gfpΩtet, ΔminCD::erm | This study |
| KP656 | ΔspoIIIE::spc, amyE::PspoIIIEspoIIIE-gfpΩcat, ΔminCD::erm | This study |
| KP657 | ΔspoIIIE::spc, amyE::PspoIIQspoIIIE-gfpΩcat, ΔminCD::erm | This study |
| KP658 | ΔspoIIIE::spc, amyE::PspoIIDspoIIIE-gfpΩcat, ΔminCD::erm | This study |
| KP659 | ΔspoIIIE::spc, amyE::PspoIIRspoIIIE-gfpΩcat, ΔminCD::erm | This study |
The first modification to pDG1662 involved inserting a promoterless super bright variant of gfp to yield pMDS12 (Cormack et al., 1996). The gfp gene was amplified using the primers GACTGAGAATTCGGAT CCAAGCTTACTAGTAGTAAAGGAGAAGAACTTTTCACTG and CTGACTAGATCTCTATTACGGCCGTTTGTATAGTTCATCCA TGCCATGTG (template homology shown in bold), digested with EcoRI and BglII and ligated into pDG1662 digested with EcoRI and BamHI. We next constructed pMDS13, pMDS14, pMDS16 and pMDS78, all derivatives of pMDS12, with the spoIIQ, spoIID, spoIIIE and spoIIR promoters and translational start sites, respectively, cloned upstream of gfp. The spoIIQ promoter and translational start site was amplified using the primers GACTGAAGATCTGCTAGCGCCAT AAGTGAGCGGATGCCAAG and CTGACTAAGCTTGGATCCG TTTTCTTCCTCTCTCATTGTTTCATC, the spoIID promoter and translational start site with GACTGAAGATCTGCTAGCGTTGA TTTAGCAAACTATATCAACGG and CTGATCACTAGTGGATC CTGCGAATTGTTTCATATTCAGCTGC, the spoIIIE promoter and translational start site with GACTGAAGATCTGCTAGCAACGTA AACCGATGATCATCC and CTGACTAAGCTTGGATCCTTTCT TTGCCACACTCATCACCTTAC, and the spoIIR promoter and translational start site with GACTAGATCTGCTTTCTTTGTTGC GGCCATACC and CTGAAAGCTTGGATCCTACTGTTTTTTTC ATCGGTCCCCAC. Each promoter fragment was digested with BglII and HindIII and ligated into pMDS12 digested with BamHI and HindIII. Finally, the spoIIIE genes were inserted between the promoters and gfp. The full-length spoIIIE gene was amplified using the primers CTGACTGGATCCCGAAAATCAAGAAAAAAACAGGCGAAA and GACTGAACTAGTAGAAGAGACCTCATCATATTTCT CTT, digested with SpeI and BamHI and ligated into pMDS13, pMDS14, pMDS16 and pMDS78 digested with the same enzymes to yield pMDS24, pMDS27, pMDS33 and pMDS80, respectively.
Cell-specific expression of minCD was accomplished by fusing the entire coding sequence of minCD to either the spoIID or spoIIQ promoters and translational start sites and integrating the hybrid genes at the sacA locus using derivatives of pRM52 (a gift from R.Middleton and A.Hofmeister). The sacA gene is located within the chromosomal domain trapped in the forespore by polar septation. The coding sequence of minCD was amplified using the primers CAGTGGATCCAAAA AGCAATATG and GTCACGGCCGTTAAGTCTTACTCCGAAAA ATG.
Microscopy
Cells were grown to the desired stage of sporulation, harvested and processed in two different ways. When examining GFP localization, 0.5 ml of live cells were mixed with 5 µl of Mitotracker Red at 10 µg/ml and 10 µl of DAPI at 5 µg/ml (Molecular Probes), and concentrated 10-fold as described in Sharp and Pogliano (2002b). After staining, 3 µl of cells were applied to a slide, immobilized with poly-l-lysine-treated coverslips and visualized with an Applied Precision optical sectioning microscope. To evaluate chromosome translocation efficiency, cells were fixed (Pogliano et al., 1995), permeabilized with 0.4 mg/ml lysozyme and stained with 0.2 µg/ml DAPI and 2 µg/ml FM 4-64 (Molecular Probes). Images were acquired and deconvolved using DeltaVision v2.10 software (Applied Precision); typically, three optical sections were taken for each sample (Sharp and Pogliano, 2002b). Figures were assembled using Photoshop v5.0 software (Adobe).
Acknowledgments
Acknowledgements
We thank Eric Becker, Linda Liu and Aileen Rubio for their helpful comments on this manuscript, and Rebecca Middleton and Antje Hofmeister for providing plasmids. This work was supported by the National Science Foundation (NSF 0135955) and the National Institutes of Health (GM57045).
References
- Äkerlund T., Bernander,R. and Nordström,K. (1992) Cell division in Escherichia coli minB mutants. Mol. Microbiol., 6, 2073–2083. [DOI] [PubMed] [Google Scholar]
- Arigoni F., Pogliano,K., Webb,C., Stragier,P. and Losick,R. (1995) Localization of protein implicated in establishment of cell type to sites of asymmetric division. Science, 270, 637–640. [DOI] [PubMed] [Google Scholar]
- Bath J., Wu,L.J., Errington,J. and Robinson,C. (2000) Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell–prespore division septum. Science, 290, 995–997. [DOI] [PubMed] [Google Scholar]
- Ben-Yahuda S. and Losick,R. (2002) Asymmetric cell division in B.subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell, 109, 257–266. [DOI] [PubMed] [Google Scholar]
- Cormack B.P., Valdivia,R.H. and Falkow,S. (1996) FACS-optimized mutants of the green fluorescent protein (GFP). Gene, 173, 33–38. [DOI] [PubMed] [Google Scholar]
- Corre J. and Louarn,J.-M. (2002) Evidence from terminal recombination gradients that FtsK uses replichore polarity to control chromosome terminus positioning at division in Escherichia coli. J. Bacteriol., 184, 3801–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P.A., Crossley,R.E. and Rothfield,L.I. (1989) A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E.coli. Cell, 56, 641–649. [DOI] [PubMed] [Google Scholar]
- Duncan L., Alper,S., Arigoni,F., Losick,R. and Stragier,P. (1995) Activation of cell-specific transcription by a serine phosphatase at the site of asymmetric division. Science, 270, 641–644. [DOI] [PubMed] [Google Scholar]
- Dworkin J. and Losick,R. (2001) Differential gene expression governed by chromosomal spatial asymmetry. Cell, 107, 339–346. [DOI] [PubMed] [Google Scholar]
- Edwards D.H. and Errington,J. (1997) The Bacillus subtilis DivIVA protein targets to the division septum and controls the site-specificity of cell division. Mol. Microbiol., 24, 905–915. [DOI] [PubMed] [Google Scholar]
- Eichenberger P., Fawcett,P. and Losick,R. (2001) A three-protein inhibitor of polar septation during sporulation in Bacillus subtilis. Mol. Microbiol., 42, 1147–1162. [DOI] [PubMed] [Google Scholar]
- Errington J., Bath,J. and Wu,L.-J. (2001) DNA transport in bacteria. Nat. Rev. Mol. Cell. Biol., 2, 538–545. [DOI] [PubMed] [Google Scholar]
- Guerout-Fleury A.M., Fransdsen,N. and Stragier,P. (1996) Plasmids for ectopic integration in Bacillus subtilis. Gene, 180, 57–61. [DOI] [PubMed] [Google Scholar]
- Hoch J.A. (1991) Genetic analysis in Bacillus subtilis. Methods Enzymol., 204, 305–320. [DOI] [PubMed] [Google Scholar]
- Hu Z. and Lutkenhaus,J. (1999) Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol., 34, 82–90. [DOI] [PubMed] [Google Scholar]
- Hu Z., Mukherjee,A., Pichoff,S. and Lutkenhaus,J. (1999) The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl Acad. Sci. USA, 96, 14819–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A., Chary,V.K., Hilbert,D.W. and Piggot,P.J. (2000) The chromosomal location of the Bacillus subtilis sporulation gene spoIIR is important for its function. J. Bacteriol., 182, 4425–4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin P.A. and Losick,R. (1996) Transcription factor SpoOA switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev., 10, 478–488. [DOI] [PubMed] [Google Scholar]
- Levin P.A., Margolis,P.S., Setlow,P., Losick,R. and Sun,D. (1992) Identification of Bacillus subtilis genes for septum placement and shape determination. J. Bacteriol., 174, 6717–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P.J., Partridge,S.R. and Errington,J. (1994) σ factors, asymmetry and the determination of cell fate in Bacillus subtilis. Proc. Natl Acad. Sci. USA, 91, 3849–3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W. (2001) Spatial regulation of cytokinesis in bacteria. Curr. Opin. Microbiol., 4, 647–652. [DOI] [PubMed] [Google Scholar]
- Marston A.L. and Errington,J. (1999) Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC. Mol. Microbiol., 33, 84–96. [DOI] [PubMed] [Google Scholar]
- Marston A.L., Thomaides,H.B., Edwards,D.H., Sharpe,M.E. and Errington,J. (1998) Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the midcell division site. Genes Dev., 12, 3419–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder E., El’Bouhali,M., Pas,E. and Woldringh,C.L. (1990) The Escherichia coli minB mutation resembles gyrB in defective nucleoid segregation and decreased negative supercoiling of plasmids. Mol. Gen. Genet., 221, 87–93. [DOI] [PubMed] [Google Scholar]
- Piggot P.J. and Losick,R. (2002) Sporulation genes and inter compartmental regulation. In Sonenshein,A.L., Hoch,J.A. and Losick,R. (eds), Bacillus subtilis and its Relatives: From Genes to Cells. American Society for Microbiology, Washington, DC, pp. 483–518.
- Pogliano J., Osborne,N., Sharp,M., Abanes-DeMello,A., Perez,A.R., Sun,Y.-L. and Pogliano,K. (1999) A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol. Microbiol., 31, 1149–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano J., Sharp,M.D. and Pogliano,K. (2002) Partitioning of chromosomal DNA during the establishment of cellular asymmetry in Bacillus subtilis. J. Bacteriol., 184, 1743–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano K., Harry,L. and Losick,R. (1995) Visualization of the subcellular location of sporulation proteins in Bacillus subtilis using immunofluorescence microscopy. Mol. Microbiol., 18, 459–470. [DOI] [PubMed] [Google Scholar]
- Pogliano K., Hofmeister,A.E.M. and Losick,R. (1997) Disappearance of the σE transcription factor from the forespore and the SpoIIE phosphatase from the mother cell contributes to the establishment of cell-specific gene expression during sporulation in Bacillus subtilis. J. Bacteriol., 179, 3331–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin D.M. and de Boer,P.A. (1999a) MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J. Bacteriol., 181, 6419–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin D.M. and de Boer,P.A. (1999b) Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl Acad. Sci. USA, 96, 4971–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfield L.I., Shih,Y.L. and King,G. (2001) Polar explorers: membrane proteins that determine division site placement. Cell, 106, 13–16. [DOI] [PubMed] [Google Scholar]
- Ryter A. (1965) Etude morphologie de la sporulation de Bacillus subtilis. Ann. Inst. Pasteur, 108, 40–60. [PubMed] [Google Scholar]
- Setlow B., Magill,N., Febbroriello,P., Nakhimousky,L., Koppel,D.E. and Setlow,P. (1991) Condensation of the forespore nucleoid early in sporulation of Bacillus species. J. Bacteriol., 173, 6270–6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp M.D. and Pogliano,K. (1999) An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc. Natl Acad. Sci. USA, 96, 14553–14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp M.D. and Pogliano,K. (2002a) The dynamic architecture of the Bacillus cell. In Sonenshein,A.L., Hoch,J.A. and Losick,R. (eds), Bacillus subtilis and its Relatives: From Genes to Cells. American Society for Microbiology, Washington, DC, pp. 15–20.
- Sharp M.D. and Pogliano,K. (2002b) Role of cell specific assembly of SpoIIIE in polarity of DNA transfer. Science, 295, 137–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterlini J.M. and Mandelstam,J. (1969) Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J., 113, 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomaides H.B., Freeman,M., Karoui,M.E. and Errington,J. (2001) Division site selection protein DivIVA of Bacillus subtilis has a second distinct function in chromosome segregation during sporulation. Genes Dev., 15, 1662–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley A.W. and Stewart,G.C. (1992) The divIVB region of the Bacillus subtilis chromosome encodes homologs of Escherichia coli septum placement (MinCD) and cell shape (MreBCD) determinants. J. Bacteriol., 174, 6729–6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.J. and Errington,J. (1994) Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science, 264, 572–575. [DOI] [PubMed] [Google Scholar]
- Wu L.J. and Errington,J. (1997) Septal localization of the SpoIIIE chromosome partitioning protein in Bacillus subtilis. EMBO J., 16, 2161–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.J., Lewis,P.J., Allmansberger,R., Hauser,P.M. and Errington,J. (1995) A conjugation-like mechanism for prespore chromosome partitioning during sporulation in B.subtilis. Genes Dev., 9, 1316–1326. [DOI] [PubMed] [Google Scholar]
- Youngman P., Perkins,J.B. and Sandman,K. (1984) New genetic methods, molecular cloning strategies and gene fusion techniques for Bacillus subtilis which take advantage of Tn917 insertional mutagenesis. In Hoch,J.A. and Ganesan,A.T. (eds), Genetics and Biotechnology of Bacilli. Academic Press, NY, pp. 103–111.
- Zupancic M.L., Tran,H. and Hofmeister,A.E. (2001) Chromosomal organization governs the timing of cell type-specific gene expression required for spore formation in Bacillus subtilis. Mol. Microbiol., 39, 1471–1481. [DOI] [PubMed] [Google Scholar]