Abstract
We have identified genes regulated by starvation and sugar signals in Drosophila larvae using whole-genome microarrays. Based on expression profiles in the two nutrient conditions, they were organized into different categories that reflect distinct physiological pathways mediating sugar and fat metabolism, and cell growth. In the category of genes regulated in sugar-fed, but not in starved, animals, there is an upregulation of genes encoding key enzymes of the fat biosynthesis pathway and a downregulation of genes encoding lipases. The highest and earliest activated gene upon sugar ingestion is sugarbabe, a zinc finger protein that is induced in the gut and the fat body. Identification of potential targets using microarrays suggests that sugarbabe functions to repress genes involved in dietary fat breakdown and absorption. The current analysis provides a basis for studying the genetic mechanisms underlying nutrient signalling.
Keywords: fat/feeding/microarrays/starvation/sugar
Introduction
The ability of organisms to adapt to fluctuating food conditions is critical for their survival. For most animals, the bulk diet consists of carbohydrates, fats and proteins, and classic biochemical studies have led to an elucidation of the major enzymatic steps in the metabolism of these nutrients (Lehninger, 1970; Stryer, 1975). The interplay of different organ systems in coordinating cellular metabolism under various physiological conditions has also been extensively analysed (Ganong, 1991). However, the genetic and molecular mechanisms underlying these processes are far less understood. Recent studies in vertebrates have identified a number of molecules that regulate nutrient signalling and metabolic activity. These include transcription factors that control a battery of genes involved in metabolism, such as ADD1/SREBP1, PPARs, C/EBPs (Spiegelman and Flier, 1996; Flier and Hollenberg, 1999), a forkhead/winged helix factor FOXC2 (Cederberg et al., 2001) and a transcriptional coactivator PCG-1, which regulates gluconeogenesis (Herzig et al., 2001; Yoon et al., 2001), as well as hormones such as leptin, which controls fat homeostasis and feeding behaviour (Friedman and Halaas, 1998). Malfunctioning of physiological pathways underlying nutrient signalling and energy homeostasis can have major consequences for human health, and the modern society is facing ever increasing cases of physiological disturbances such as eating disorders, diabetes and obesity. As the dietary requirement for sugars, fats and amino acids is essentially universal, many aspects of the basic logic of nutrient signalling should be conserved. The finding that both Drosophila and Caenorhabditis elegans possess components of insulin signalling supports this view (Lehner, 1999; Brogiolo et al., 2001; Gems and Partridge, 2001).
As part of our analysis of Drosophila larval feeding behaviour, we previously identified lipase 3 (lip3) and phosphoenolpyruvate carboxykinase (pepck) as being upregulated upon starvation (Zinke et al., 1999). Upon addition of sugar, this upregulation was completely suppressed for lip3, but not for pepck. These results demonstrated that different nutrient conditions can have very specific effects on gene expression patterns in Drosophila larvae. We have now used Affymetrix microarrays to identify genes regulated by starvation and by sugar in order to study the mechanisms underlying nutrient signalling. Based on the pattern of response to different nutrient conditions and on existing knowledge of metabolic pathways, we could categorize the identified genes into groups that reflect distinct physiological functions. We have further characterized a zinc finger transcription factor that is one of the earliest and highest upregulated genes upon sugar ingestion. Identification of potential target genes indicates that this transcription factor functions to repress genes involved in dietary fat breakdown and absorption.
Results
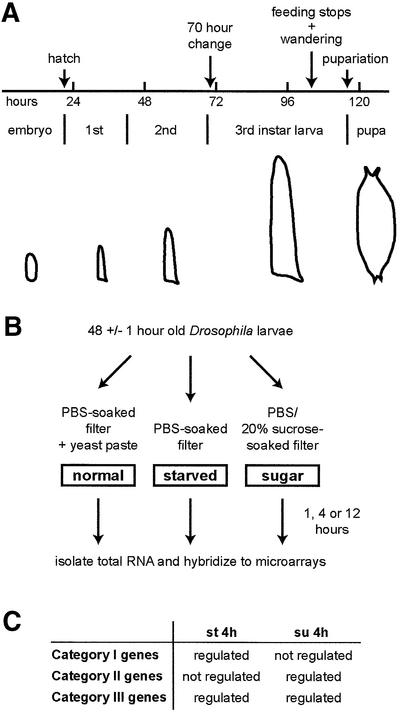
Drosophila larvae are continuous feeders and show large growth in a relatively short time period. About 5 days after egg laying (AEL), they stop feeding, leave the food to enter the wandering stage and pupariate shortly thereafter (Figure 1A). Within this normal developmental progression, there are several notable variations that become apparent under different environmental conditions. One intriguing observation was made by Beadle et al. (1938). When larvae are starved before 70 h AEL, they die within several days, whereas if they are starved after this time point, they do not grow, but still survive and differentiate to give rise to small adult flies. The authors concluded that some ‘organizational change occurs in larvae at about 70 h’ and termed this the ‘70 h change’ (Beadle et al., 1938). This survival after the 70 h change period is independent of whether the larvae are starved or placed on sugar; however, before the 70 h, larvae placed in sugar live for much longer than those under starvation conditions (over a week as compared with ∼2 days; see also Britton and Edgar, 1998; Zinke et al., 1999). Clearly, there is a difference in the metabolic programme that becomes activated across this point upon change in nutrient status. As the period before 70 h is critical for survival, we decided to perform the experiments prior to this point. For each time and nutrient condition, two chips were used with each chip being hybridized to the samples collected independently (Figure 1B).
Fig. 1. Larval growth pattern and experimental outline. (A) Drosophila larvae pupariate 5 days AEL under standard conditions, i.e. 25°C, non-crowding. They undergo two molts and grow rapidly in size. The 70 h point is not a morphologically or behaviourally identifiable stage, in contrast to pupariation, wandering behaviour or feeding stoppage (see text for details). (B) Larvae grown on yeast paste on apple juice agar plates (47–49 h AEL) were washed and transferred to appropriate nutrient conditions. After 1, 4 and 12 h, total RNA was isolated and hybridized to microarrays. (C) Three categories were formed based on the expression profile of genes after 4 h treatment in starvation (st 4h) and sugar (su 4h) conditions (see text for details).
Categorization of nutrient-dependent genes
We first determined what genes are up- or downregulated in animals that had been starved or sugar fed for 4 h. Instead of listing the regulated genes from just one nutrient condition, we categorized the genes based on their behaviour in the two conditions (Figure 1C): genes that are regulated in starved, but not in sugar-fed, larvae (category I); genes that are not regulated in starved, but are in sugar-fed, larvae (category II); and genes that are regulated in both starved and sugar-fed larvae (category III). We decided to use this double cut-off approach because it allowed a better insight into the putative functions of the regulated genes.
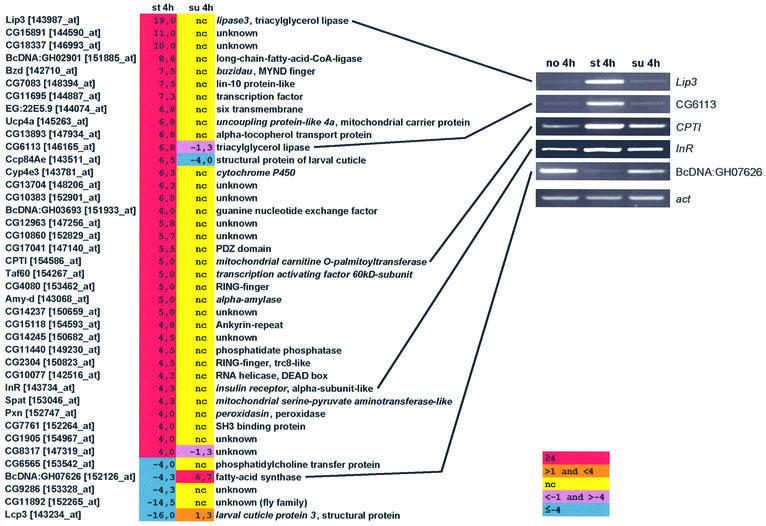
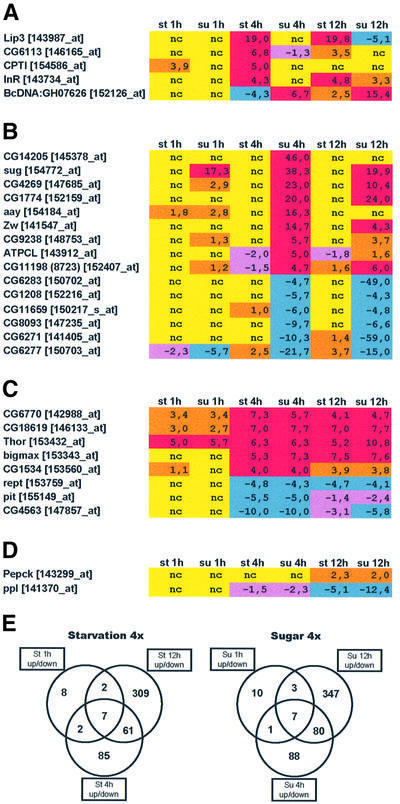
(I) Regulated under starvation but not in sugar. The first category comprised genes whose expression is affected upon starvation, but not in sugar condition, i.e. the effect of starvation can be attributed to lack of sugar. The following criteria were applied: we listed all genes regulated upon starvation, setting the fold cut-off value at 4.0-fold. From this list, we took those genes whose expression in sugar was unchanged (we defined this as fold change value ≤1.0 or ≥–1.0; see also Supplementary data available at The EMBO Journal Online), or regulated in the opposite direction (Figure 2). Genes whose expression was only partially affected by sugar were compiled into the ‘complex regulated’ group and were not considered further in the study. About 35% of the starvation upregulated genes made it into category I; of the starvation downregulated genes, <10% made this list (see Supplementary table S1). The genes fulfilling this criteria are listed in Figure 2. On top of the upregulated list is lip3, a gene that we previously showed by northern analysis as being upregulated upon starvation and completely suppressed in sugar (Zinke et al., 1999).
Fig. 2. List of genes that are regulated upon starvation, but not in sugar conditions. Thus, the regulation upon starvation is completely sugar dependent. The treatment was for 4 h. Tabular listing of genes and their fold regulation. Gene names are from FlyBase annotations (Flybase, 1999); the numbers in brackets are Affymetrix probe set names. RT–PCR for selected genes is shown on the right. nc, no change; fold change value of ≤1.0 or ≥–1.0.
RT–PCR analysis further confirmed the findings for lip3 and for several other selected genes (Figure 2). Many of the upregulated genes are clearly implicated to function in fat catabolism (Lehninger, 1970; Stryer, 1975), including BcDNA:GH02901 (fatty acid CoA ligase; activates fatty acids for β-oxidation), CG6113 (lysosomal lipase/cholesterol esterase) and CPTI (mitochondrial carnitine palmitoyltransferase, which transports fatty acid CoAs into mitochondria for β-oxidation). There are others that are likely to be involved in energy homeostasis. These include amy-d (α-amylase), insulin receptor and Ucp4a (uncoupling protein-like 4a); some members of the mitochondrial carrier proteins in mammals have been implicated in regulating insulin secretion (Polonsky and Semenkovich, 2001). There is also a downregulation of the gene encoding fatty acid synthase, which makes fatty acids from malonyl CoA (we note that we did not observe its upregulation in sugar with RT–PCR, as is shown from the microarray). The fatty acid synthase gene is also known to be downregulated under starvation in mammals (Sul and Wang, 1998).
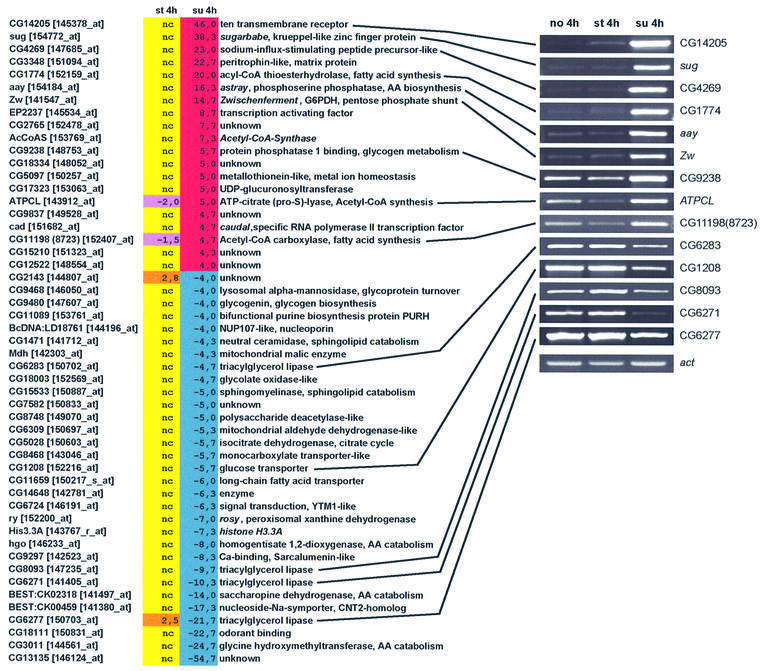
(II) Regulated in sugar but not under starvation. While comparing the starvation and the sugar data, we noticed an unexpected pattern: most of the highest regulated genes on sugar condition were not affected at all upon starvation (see Supplementary table S1). Thus, using the same strict criteria as above, we listed all genes regulated by sugar (cut-off of 4.0-fold), but not by starvation (or regulated in the opposite direction). About 30% of the genes up regulated by sugar made it into category II; the percent of downregulated genes making it to this list was about the same (Figure 3; Supplementary table S1).
Fig. 3. List of genes that are regulated in sugar condition, but not under starvation. Tabular listing of genes and their fold regulation, and selected RT–PCR analysis. We only included those that are regulated with sugar and that showed no change under starvation. Again, we included those genes whose regulation went in the opposite manner to sugar, as for example ATPCL and the lipase CG6277. See Figure 2 for colour code.
A most informative feature of this category is that among the eight identifiable enzymes found in the upregulated group, five are predicted to have a clear role in fatty acid synthesis (Lehninger, 1970; Stryer, 1975). These include acetyl CoA carboxylase (rate-limiting enzyme in fatty acid biosynthesis), acyl CoA thioesterhydrolase (decreases fat breakdown in mitochondria; Poupon et al., 1999), ATP-citrate lyase (produces cytoplasmic acetyl CoA for fatty acid synthesis), Zwischenferment (Zw; encodes glucose 6 phosphate dehydrogenase, which generates NADPH reducing power needed for fatty acid synthesis) and acetyl CoA synthase (makes acetyl CoA de novo from acetate and CoA). There is also a downregulation of four triacylglycerol lipases (CG6271, CG6277, CG6283 and CG8093). Interestingly, the two highest upregulated genes in this category encode a 10-transmembrane receptor (CG14205) and a putative zinc finger transcription factor (CG3850, which we have termed sugarbabe, or sug; see below).
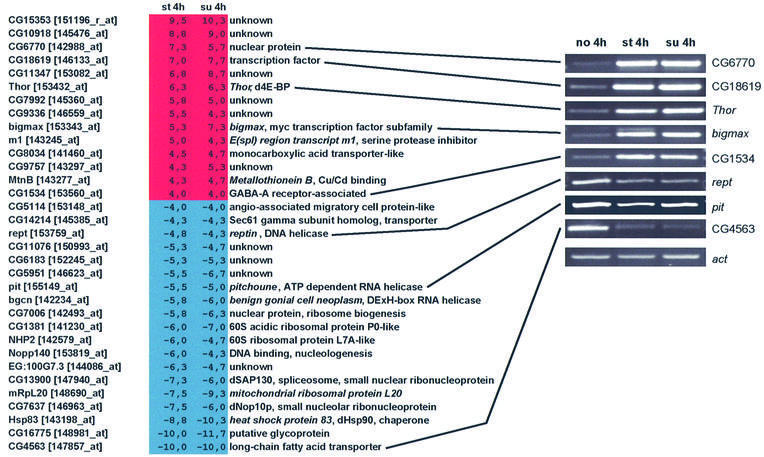
(III) Regulated both under starvation and in sugar. The third category comprised genes that are affected under starvation and where sugar does not alter this effect (Figure 4). The reasoning behind this was that larval growth is completely stopped under either starvation or sugar conditions, and we hoped to identify genes that control cell growth, with a special interest in those that might be regulated by dietary amino acids. Here we included those genes in which the fold change value in sugar did not differ by >2.0, up or down, in comparison to the starvation value (cut-off of 4.0). A particularly informative gene in this category is Thor, which encodes a homologue of mammalian PHAS1/4E-BP. This gene is upregulated to a similar extent under starvation and in sugar condition, consistent with the view that it is responding to a cell growth signal and not a sugar-dependent signal. Interestingly, this factor is involved in translational control and is a target of mTOR (mammalian target of rapamycin), which in turn is thought to be regulated by amino acids (reviewed in Schmelzle and Hall, 2000).
Fig. 4. List of genes that are regulated upon starvation and in sugar condition. Tabular listing of genes and their fold regulation, and selected RT–PCR analysis. We took the regulated genes from starvation data and listed those genes that did not differ from this by >2-fold in sugar. See Figure 2 for colour code.
The fact that many of the downregulated genes encode RNA helicases or ribosomal proteins further supports the view that this category identifies genes involved in protein biosynthesis and cell growth. There are also three putative transcription factors (CG6770, CG18619 and bigmax) high on the group of upregulated genes. CG6770 shows homology to mammalian P8 transcriptional regulator, which is implicated in pancreatic growth and regeneration (Mallo et al., 1997), while CG18619 has high homology to human cAMP responsive element-binding protein-like 2 (CREBL2; 73% amino acid identity; Hoornaert et al., 1998). The activation of these transcription factors might be required to stop cell growth in the face of nutrient deprivation.
Early response to nutrient signals
The above microarray data were obtained after 4 h nutrient treatment. Next, we performed the same experiments with different time periods, namely 1 and 12 h treatments, in order to distinguish early from late regulated genes (Figure 5). The lists shown in Figure 5A–C are limited to those genes for which RT–PCR was performed at the 4 h time point (for data on all genes, see Supplementary table S1) or by in situ hybridization (see below). In the category of genes regulated under starvation, but not in sugar, one of the earliest upregulated genes is CPTI (Figure 5A). The activity of this enzyme was shown to be very sensitive to inhibition by malonyl CoA, a key intermediate in fatty acid synthesis (Jackson et al., 1999). In the category of genes regulated in sugar, but not under starvation, the highest and earliest upregulated gene is sug, a putative transcription factor, whereas one of the earliest downregulated genes is CG6277, a triacylglycerol lipase (Figure 5B). Note that this lipase, along with three other lipases in this category (CG8093, CG6283 and CG6287), has distinct kinetics of repression. In the category of genes regulated under starvation and in sugar, one of the highest and earliest upregulated genes is the putative translation regulator Thor (Figure 5C). The two transcription factor-encoding genes CG6770 and CG18619 are also upregulated within 1 h of nutrient deprivation, supporting the notion that they act very early on in the signalling pathway.
Fig. 5. Time course of gene regulation: 1, 4 and 12 h after appropriate nutrient condition. The panels show genes tested with RT–PCR at 4 h or in situ hybridization. (A) Category I genes: regulated upon starvation, but not in sugar (sorted by regulation in st 4h). (B) Category II genes: regulated in sugar, but not upon starvation (sorted by regulation in su 4h). (C) Category III genes: regulated upon starvation and in sugar (sorted by regulation in st 4h). (D) Regulation of pepck and ppl: note the late regulation of these genes (see text for details). (E) Venn diagrams showing the increase in number of genes regulated (cut-off of 4.0-fold) with increasing times (1, 4 and 12 h) of starvation (left) or sugar conditions (right). See Figure 2 for colour code. st 1h, starved for 1 h; su 1h, sugar fed for 1 h; st 4h, starved for 4 h; su 4h, sugar fed for 4 h; st 12h, starved for 12 h; su 12h, sugar fed for 12 h.
These data help further characterize the different classes of nutrient-dependent genes into early and late responding. In this regard, we note that the gene pumpless (ppl), which encodes an amino acid catabolic enzyme, was shown by northern analysis to be downregulated after 24 h starvation, whereas the pepck gene was upregulated (Zinke et al., 1999). In the current microarray analysis, ppl does not make the 4-fold cut-off for the 4 h starvation period, but its downregulation can be seen at 12 h (Figure 5D); in the case of pepck, there is no change at 4 h starvation, but there is a slight upregulation at 12 h. As pepck is the rate-limiting step in gluconeogenesis, it may be that this gene becomes upregulated only after longer periods of starvation, when the stored energy source is first used up. Venn diagrams of the number of genes regulated at 1, 4 and 12 h (Figure 5E) show that the number of regulated genes increases as time of starvation or sugar treatment increases, as expected.
Genes regulated by sugar but not by starvation: a role in sugar to fat conversion
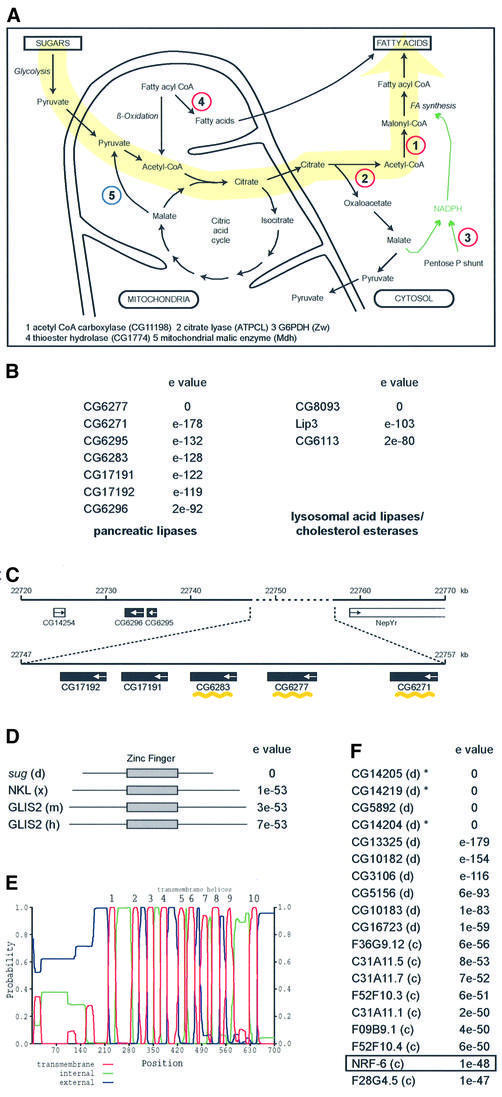
From the three categories of nutrient-dependent genes, we decided to investigate in more detail the category II genes, i.e. those that are regulated in sugar, but not under star vation. This was because many of these genes could be placed within a defined metabolic pathway, namely one that converts carbohydrates to fats (Figure 6A). Consistent with this view, there are four lipase genes that become downregulated, which makes physiological sense since the organism would want to decrease fat breakdown when accelerating its fatty acid synthesis programme. One of the lipases, CG8093, is very similar to the two lipases that become upregulated upon starvation (lip3 and CG6113; Figure 6B). These lipases belong to the vertebrate lysosomal acid lipase/cholesterol esterase family and mutations in a human homologue lipase A can lead to Wolman disease and cholesteryl ester storage disease (Anderson et al., 1994). The other three belong to the secreted pancreatic lipases, which also have an important role in fat metabolism (Lowe, 1997; Figure 6B); these break down dietary fats into forms that can be absorbed by the gut. As mentioned earlier, these lipases have different kinetics of downregulation, and it is interesting that three are arranged tandemly amongst a larger lipase cluster (Figure 6C). Also downregulated is a long chain fatty acid transporter (CG11659; Figures 3 and 5B). Based on these observations, we were especially intrigued by the two highest upregulated genes in this category: sug and CG14205. The former encodes a C2H2 zinc finger protein with homology to genes in Xenopus, mouse and human (Figure 6D; Lamar et al., 2001), whereas the latter encodes a 10-transmembrane protein that belongs to a highly related family found in both flies and worms (Figure 6E and F). One of the genes in the family is nrf-6, which was identified in a screen for fluoxetine (Prozac) resistance (Choy and Thomas, 1999). These observations suggested that the two genes may act in a regulatory cascade that responds to sugar signals.
Fig. 6. Candidate genes involved in sugar to fatty acid conversion. (A) Schematic view of a metabolic pathway leading from sugar breakdown to fat synthesis. Genes represented by red circles are upregulated in sugar and those in blue are downregulated. (B) Lipase gene families with members regulated in starved or sugar-fed animals. CG6277, CG6271, CG6283 and CG8093 are all downregulated in sugar-fed larvae, but unaffected in starved larvae; lip3 and CG6113 are upregulated in starved larvae and unaffected in sugar-fed larvae. Blast searches were performed with CG6277 and CG8093 using NCBI Blastp. (C) Tandemly located genes in a lipase cluster (3R; chromosomal locus 97D15-E1). Black boxes indicate lipase genes, the arrows indicate direction of transcription. The three downregulated lipases are noted by a yellow wavy line. White boxes denote the nearest non-lipase genes at this locus. The numbers in kilobases (kb) correspond to those defined in Flybase. (D) Homology of sug to other transcription factor genes. The closest ones belong to Gli zinc finger trancription factors of Xenopus (x), mouse (m), human (h). The zinc finger domain is shown in a grey box; sug protein has five C2H2 motifs (sequence data not shown). (E) Hydropathy plot of CG14205, showing the 10 transmembrane domains; from graphical output of TMHMM (transmembrane helix region prediction program) of Sonnhammer et al. (1998). (F) Homology of CG14205 to other Drosophila (d) and C.elegans (c) genes. All are blasts with NCBI. The asterisks indicate that the three genes are tandemly located. The nrf-6 gene of C.elegans, which was identified as being required for fluoxetine resistance, is boxed.
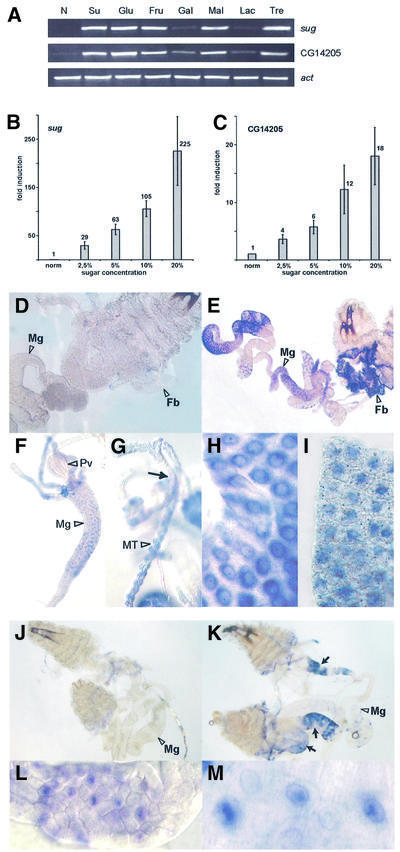
To investigate this, we first checked the response of sug and CG14205 to different sugars. The disaccharides maltose and trehalose, as well as the monosaccharides glucose and fructose, upregulated both genes to about the same extent as sucrose after 4 h treatment (Figure 7A). These results suggested that the induction of sug and CG14205 gene expression is dependent on metabolic flux through glycolysis. By contrast, the disaccharide lactose and the monosaccharide galactose showed little or no effect, suggesting that these may not be preferred sugar sources for Drosophila larvae. We then monitored the expression levels in response to varying sucrose concentration (Figure 7B and C). There is a concentration-dependent increase in the expression with increasing sugar concentration, emphasizing the importance of dietary sugar for sug and CG14205 regulation.
Fig. 7. Sugar-dependent activation of sug and CG14205. (A) Activation of CG14205 and sug by different sugars as shown by RT–PCR analysis. Normal food condition (N), sucrose (Su), glucose (Glu), fructose (Fru), galactose (Gal), maltose (Mal), lactose (Lac) and trehalose (Tre). The concentration of all the sugar solutions is 20%. Note that galactose and lactose have a much less activating effect on the two genes than other sugars tested. (B) Concentration-dependent activation of sug by sugar by real-time PCR analysis. (C) Concentration-dependent activation of CG14205 by sugar by real-time PCR analysis. (D–I) In situ hybridization with sug probe. (D) Larvae grown on normal food; no staining is detectable (Mg, midgut; Fb, fat body). (E) Larvae grown on sugar; note staining in fat body and parts of the midgut. (F) Anterior part of the midgut; the proventriculus (Pv) is not stained. (G) Staining in Malpighian tubules (MT); the arrow indicates the point where MT extends from the gut. (H) Close-up of midgut cells. (I) Close-up of fat body. (J–M) In situ hybridization with CG14205 probe. (J) Larvae grown on normal food; no staining is detectable (Mg, midgut). (K) Larvae grown on sugar; note staining in three parts of the gut (arrows). (L and M) Close-ups of gut cells. Note that CG14205 is highly induced in specific parts of the gut but not in the fat body.
We then asked in which tissues the genes sug and CG14205 are expressed. Therefore, we performed in situ hybridizations on larvae grown in normal food and in sugar (Figure 7D–M). sug showed no expression in the larval tissues taken from animals grown in yeast paste (Figure 7D). However, the gene became specifically expressed in the gut, the fat body and the Malphigian tubules of sugar-fed larvae (Figure 7E–I). There was also speckled cytoplasmic expression in the fat body that we did not observe in the gut, but the significance of this is unknown. CG14205 also showed no expression in normal-fed larvae (Figure 7J), but sugar-fed larvae showed high expression in three specific regions of the gut (Figure 7K–M). No expression was detected in the fat body. As the gut, the fat body and the Malphigian tubules play key roles in Drosophila metabolism, these results fit well with the view that the genes sug and CG14205 are involved in nutrient signalling.
Potential targets of the transcription factor sug
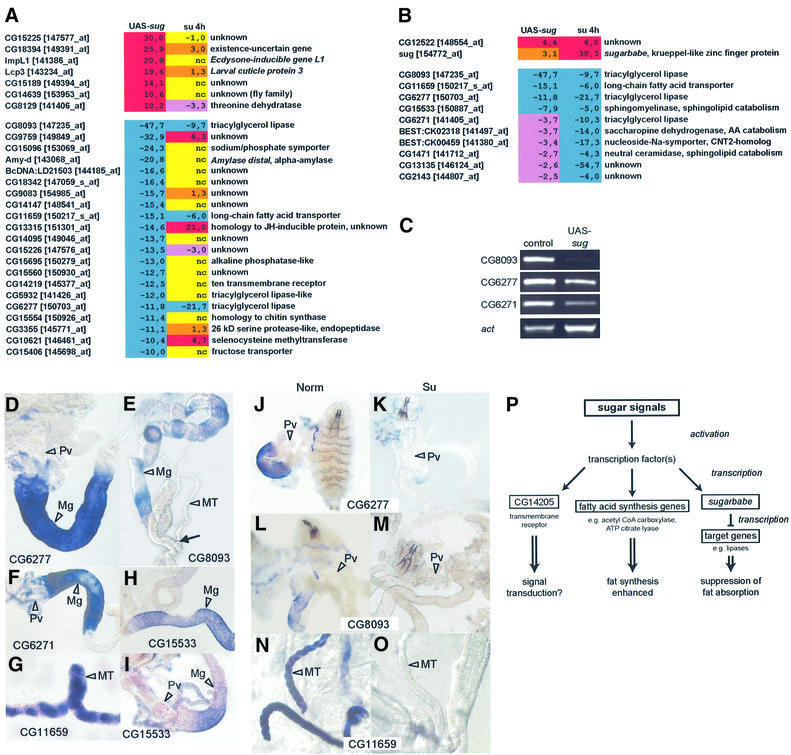
The expression profile of sug stands out because it is the highest and earliest upregulated gene in sugar-fed larvae (see Figure 5B; Supplementary table S1). Real-time RT–PCR analysis confirmed the microarray data (sug was upregulated some 25-fold after 1 h sugar treatment, whereas CG14205 was upregulated only ∼2-fold; data not shown). Furthermore, it became expressed in the gut, the fat body and Malpighian tubules, suggesting that the gene may be involved in metabolic control. In this context, it may, for example, activate those fat synthesis genes that are upregulated in sugar, or repress those that are downregulated, or both. It could also function by first activating potential signalling receptors such as CG14205. To distinguish between these possibilities, we wanted to identify putative targets of sug through microarrays. We therefore used the heat shock Gal4-UAS system to overexpress sug in the larvae under normal food conditions in order to determine what genes were up- or downregulated. Heat shock-Gal4/UAS-sug-harbouring larvae growing on normal food were heat shocked for 40 min at second instar stage. We note that a single 40 min heat shock resulted in a 1 day delay of pupariation and increased male lethality (data not shown). RNA was isolated 6 h after heat shock and hybridized to GeneChips. From the resulting data, we looked for genes that showed a similar expession profile to sugar-fed larvae. Therefore, next to each regulated gene, we also listed its corresponding expression in 4 h sugar-fed condition (Figure 8A). We were immediately struck by the fact that among the highest regulated genes (cut-off of 10.0-fold), none corresponded to any upregulated genes, whereas three matched the downregulated genes: CG8093 (lysosomal lipase/cholesterol esterase), CG11659 (long chain fatty acid transporter) and CG6277 (pancreatic lipase). We then used a double cut-off strategy to group the potential sug-regulated genes: we listed all genes regulated upon heat-shock sug with a cut-off of 2.0-fold and then asked which of these were in the category of genes regulated by sugar, but not under starvation (Figure 8B). Of the upregulated genes upon sug overexpression, none of the fat synthesis genes, such as acetyl CoA carboxylase, was on the list (there was only one gene, besides sug, which was on this list). By contrast, 10 genes were among the downregulated, including an enzyme in sphingolipid catabolism (sphingomyelinase, CG15533) and another pancreatic lipase (CG6271).
Fig. 8. Identification of potential sug target genes by microarrays. (A) RNA was isolated 6 h after the end of heat shock treatment and hybridized to Affymetrix GeneChip arrays. A list is given of all genes that are strongly regulated (cut-off of 10.0-fold) in larvae overexpressing sug (UAS-sug; left column). Next to each is the fold change of the corresponding gene observed in 4 h sugar-fed larvae (right column). Note that two lipases (CG8093 and CG6277) and a fatty acid transporter (CG11659) show a similar pattern (downregulated) in both cases. See Figure 2 for colour code. (B) All genes up- or downregulated by >2.0-fold upon sug overexpression (left column), and which are also similarly regulated in sugar-fed larvae (category II genes of Figure 3; right column). Note the downregulation of genes encoding lipases and other genes involved in fat catabolism. (C) RT–PCR of three lipase genes in larvae overexpressing sug. (D–I) In situ hybridization of selected potential sug target genes. (D) CG6277, pancreatic lipase (Pv, proventriculus; Mg, midgut). (E) CG8093, lysosomal lipase/cholesterol esterase (MT, Malpighian tubules; the arrow indicates the point where MT extends from the gut). (F) CG6271, pancreatic lipase. (G) CG11659, long chain fatty acid transporter. (H and I) CG15533, sphingomyelinase. (J–O) In situ hybridization of CG6277, CG8093 and CG11659 in normal (Norm, left column) and sugar (Su, right column) conditions. Pv, proventriculus; MT, Malpighian tubules. Note that sugar suppresses expression of all three genes in the larval tissues. (P) A model for the activation of genes by sugar signals. See text for details.
Many genes regulated by sug overexpression show no change in sugar-fed condition or show even an opposite regulation (Figure 8A). However, it should be kept in mind that the sug overexpression study was carried out in normal food conditions, and it is likely that different combinations of nutrient signals can lead to different results. In this context, we find it intriguing that the gene CG14219 is downregulated upon sug overexpression, but is unaffected in sugar-fed larvae (Figure 8A). CG14219 encodes a 10-transmembrane receptor and shows very high homology to CG14205 (the two genes are also tandemly located in the genome; see Figure 6F), which is one of the highest upregulated genes in sugar-fed larvae (see Figure 3). CG14205, in turn, is not regulated at all in sug overexpression. Thus, like the lipases, highly homologous genes can have very different regulatory behaviour in response to nutrient signals.
RT–PCR analysis confirms repression of the three lipase genes (CG8093, CG6277 and CG6271) upon sug overexpression (Figure 8C). In order to see where these lipases, as well as several other genes potentially involved in fat metabolism, are expressed and to see whether they overlap the domain of expression of sug, we performed in situ hybridizations on normal-fed larvae. The pancreatic lipase genes (CG6271 and CG6277) are both expressed in the anterior portion of the midgut, just posterior to the proventriculus (Figure 8D and F); the lysosomal lipase/cholesterol esterase CG8093 and sphingomyelinase CG15533 are also expressed in the gut (Figure 8E, H and I), whereas the fatty acid transporter CG11659 is expressed in the Malphigian tubules (Figure 8G). All these are sites where sug is also expressed (see Figure 7E–I). We then took the three highest regulated genes, CG6277, CG8093 and CG11659, and tested whether their expression in these tissues is indeed downregulated upon sugar treatment. They are all repressed in the appropriate tissues upon sugar treatment (Figure 8J–O). Taken together, these results support the view that these genes involved in fat metabolism are putative transcriptional targets of the zinc finger protein sug.
Discussion
We have used microarrays to identify genes in Drosophila larvae that are regulated by nutrient signals. In experiments of this type, there are various ways to organize the data. We opted to use a double cut-off approach, in which we compared the changes in gene expression level in starved and sugar-fed animals after a specific time period. We then organized the genes into different categories based on the similarity of their expression profile in the two nutrient conditions. This approach has been quite effective in highlighting genes that are involved in specific aspects of sugar-dependent and -independent metabolic processes.
Starvation response and lipases
One response to nutrient deprivation is to break down fat, and this is consistent with two of the lipase genes, lip3 and CG6113, being upregulated upon starvation. However, other lipases, e.g. CG6277, CG6283, CG6271 and CG8093, are not affected at all under starvation, but become downregulated in sugar conditions. The two lipase genes upregulated under starvation both belong to the lysosomal acid lipase/cholesterol esterase family. One of the lipases that is unaffected under starvation, but is downregulated under sugar, also belongs to this lipase family; however, the other three clustered lipases that show the same regulatory pattern belong to a different family, namely the secreted pancreatic lipases. Therefore, the regulatory behaviour of the different lipase genes is not dependent on what type of lipase they encode. Furthermore, the four downregulated lipases have diverse expression kinetics, depending on the time period the larvae spend on sugar, and it is interesting to note that the repressive action of the zinc finger gene sug on the four lipases is also quite diverse. These results point to a finely tuned homeostasis system that coordinates the activities of the various lipases in pace with changing physiological demands. The Drosophila genome contains over 60 genes encoding lipases (Rubin et al., 2000), and clearly, the specific physiological role and transcriptional regulation of the lipases form an interesting issue on their own.
Genes involved in converting carbohydrate to fat
In animals, there is an efficient mechanism for converting carbohydrate into fat (Lehninger, 1970; Stryer, 1975). Many of the genes that are upregulated in sugar, but remain unchanged under starvation, are clearly implicated in fatty acid synthesis, including acetyl CoA carboxylase and ATP citrate lyase. The transcription of their mammalian homologues is also regulated by glucose signals (Newgard and McGarry, 1995). Thus, in the presence of high sugar, even in absence of other nutrients, there also appears to be a conversion of sugar to fat in Drosophila. This is consistent with the observation that lipids are still present in the fat body of larvae grown on sugar, whereas it is completely broken down upon starvation (Zinke et al., 1999). The transcriptional regulation of genes encoding these enzymes must lie at the end of a signalling cascade initiated by sugar-dependent signals. The two highest upregulated genes in this category, the transcription factor sug and the 10-transmembrane receptor CG14205, are good candidates for operating in this cascade. CG14205 is part of a large family whose members show very high homology to each other. There is also a similar family of C.elegans genes. Interestingly, the defining member of this family, nrf-6, was identified in a screen for mutants resistant to fluoxetine. This anti-depressant drug, commonly known as Prozac, works as a selective serotonin uptake inhibitor. The nrf-6 gene in C.elegans is expressed in the gut (Choy and Thomas, 1999), the same tissue where CG14205 is also expressed in Drosophila. These proteins are unlike any other known protein, and we could not find any homologous sequences outside of Drosophila and C.elegans. At this point, we can only guess at the function of CG14205, but one possibility is that it acts as a receptor for invertebrate-specific neuropeptides or hormones.
sug has highest homology to C2H2 Gli-like zinc finger transcription factors (Lamar et al., 2001) and is highly induced in sugar-fed larvae in the fat body and parts of the midgut. The transcriptional activation of this gene probably represents one of the first transcriptional responses when Drosophila larvae are shifted from normal to sugar conditions. Overexpression of sug results in dramatic repression of genes involved in fat catabolism. Potential direct targets of sug include the lipases CG8093, CG6277 and CG6271, all of which are also downregulated in sugar-fed larvae. In fact, the pancreatic lipases (to which family CG6277 and CG6271 belong) are the targets of orlistatin (Xenical), a drug used in weight loss therapy (Hill et al., 1999). Based on these observations, we propose a model (Figure 8P) in which an as yet unknown transcription factor(s) is activated by a sugar signal. This factor(s) then activates sug, as well as a set of genes involved in fatty acid synthesis, such as acetyl CoA carboxylase and ATP citrate lyase. sug then represses a set of lipase genes and other genes involved in fat catabolism. As sug is the earliest induced, the goal behind this regulatory mechanism seems to be to activate a repressor that suppresses fat breakdown prior to the activation of the fatty acid synthesis pathway. An open issue at this point is the identification of the transcriptional activator of sug (as well as of CG14205 and the fatty acid synthesis genes) and the corresponding signalling pathway leading to its activation. In this context, we have found five tightly clustered binding sites for C/EBP proteins in the upstream regulatory region of sug (our unpublished observation). As the C/EBP proteins have been shown to be important for regulating various metabolic genes in vertebrates (Roesler, 2000), Drosophila C/EBP homologues may be involved in sug activation.
Nutrient-dependent cell growth genes: regulation by amino acid supply?
Growth is stopped regardless of whether the larvae are placed under starvation conditions or on sugar. Thus, genes that become regulated upon starvation and show similar regulation in sugar are good candidates for genes involved in cell growth. Protein biosynthesis drives cell growth, and this category has several components that may be regulated by amino acid-dependent signals. One candidate is Thor, since its mammalian homologue Phas1/4E-BP is involved in controlling translation initiation. As PhasI/4E-BP is involved in inhibiting translation by binding to the translation initiation factor 4E, the upregulation of Thor is expected to inhibit translation and growth. The response of Thor to starvation or to sugar is fast, with transcription being upregulated within 1 h. Thor also plays a role in the immune response, and it is interesting that Thor becomes upregulated upon bacterial infection as well (Bernal and Kimbrell, 2000). It may be that one of the earliest responses to halt growth is the upregulation of Thor. One of the regulators of the vertebrate Thor homologue, Phas/4E-BP, is mTOR (target of rapamycin), which in turn is thought to be regulated by amino acids (Hara et al., 1998; Schmelzle and Hall, 2000). Furthermore, mutants in the Drosophila homologue dTOR show several features of normal larvae starved for amino acids (Oldham et al., 2000; Zhang et al., 2000). Another informative gene is pitchoune (pit), which is downregulated upon starvation and is required for larval growth (Zaffran et al., 1998). It is a potential target of dMyc, which regulates cellular growth (Johnston et al., 1999). pit encodes a RNA helicase and is thought to be involved in protein biosynthesis (Zaffran et al., 1998). It is noteworthy that many (>40%) of the other downregulated genes in this category are also RNA helicases or components required for ribosomal function and protein biosynthesis. We have also identified three putative transcription factors that may be involved in controlling cell growth in response to nutrient signals. CG6770 has homology to a rat and human P8 homologue, a gene implicated in pancreatic growth and regeneration (Mallo et al., 1997), while CG18619 has high homology to a human cAMP-responsive element binding protein-like 2 protein (Hoornaert et al., 1998), and the third, bigmax, is a member of the max family of proteins (Gallant et al., 1996). Whether these transcription factors are regulated in response to amino acid shortage remains to be determined.
Nutrient signalling and feeding behaviour
Altering nutrient conditions brings about distinct feeding responses in most animals, and numerous compounds have varied effects on appetite (Blundell, 1991; Rosenbaum et al., 1997). Therefore, one question is how specific components of diet, such as sugars, fats and amino acids, differentially alter feeding behaviour. For Drosophila, placing larvae on filter paper soaked with either saline alone or sugar results in immediate dispersal. If a clump of yeast is added, larvae will quickly move toward this source; when the yeast supply is depleted, they will again disperse. Clearly, sugar is not sufficient to quench this food-seeking or ‘hunger’ response in the larvae. The critical difference in the two conditions is that the animals live much longer on sugar than under starvation. Thus, the goal of the change in the metabolic programme upon sugar seems to be to buy time: the larvae stop their growth and start storing the available sugar into fat, while at the same time dispersing to increase their chance of finding better food. Although we do not know whether any of the nutrient-controlled genes identified here affect feeding behaviour, there are some indications that they may. For example, inhibitors of fatty acid synthase activity have been shown to reduce feeding in mouse, probably by a malonyl CoA-dependent signal that acts centrally via neuropeptides and neurotransmitters (Loftus et al., 2000). In addition, mice lacking acetyl CoA carboxylase 2 gene have reduced fat, but increased food intake (Abu-Elheiga et al., 2001). We also find it interesting that the trans membrane receptor CG14205 shows high homology to a C.elegans gene, which was identified in a screen for fluoxetine resistance, since fluoxetine has been shown to affect feeding behaviour in worms and mammals (Wurtmann and Wurtmann, 1977; Choy and Thomas, 1999; Sawin et al., 2000).
Our study also forms a basis for investigating the ‘70 h change’ first described by Beadle et al. (1938), in which larvae starved before 70 h of age die, whereas those starved after 70 h survive and give rise to small flies. Comparing the current data on nutrient-dependent gene expression, which was performed before the 70 h change, with those after this time point, may provide insights into the mechanisms underlying this change in metabolic programming during larval growth. Finally, as sugars, fats and proteins are universal dietary components, our analysis provides a starting point for a comparative genomics approach to elucidate conserved mechanisms in nutrient signalling in different organisms.
Materials and methods
Larval growth conditions
Normal food consisted of apple juice agar plates with yeast, and the larvae were raised at 25°C. Larvae were collected 47–49 h AEL, thoroughly washed with water and then placed on filter paper on a Petri dish soaked with either phosphate-buffered saline (PBS) or PBS and the appropriate concentration and type of sugar. For the control on normal food, larvae were subjected to the same washing procedure and placed on PBS-soaked filter paper containing a portion of yeast paste.
RNA purification
Fifty to 200 larvae were washed thoroughly with water, transferred to lysis buffer (supplied with RNA isolation kit) and homogenized (Ultra-Turrax T25basic) at full speed for 2 min. The lysates were then vortexed for 1 h at room temperature. Total RNA was isolated by using NucleoSpin RNA II or NucleoSpin RNA L kit (Macherey & Nagel; includes DNase I treatment). For real-time PCR, total RNA was cleaned up for a second time by using NucleoSpin RNA II (includes DNase I treatment).
Microarray analysis
The GeneChip hybridizations were carried out by a service provider licensed by Affymetrix (Affymetrix Service Supplier, Steinbeis-Transferzentrum Proteom-Analyse, Rostock, Germany). The protocol used was as follows. First strand synthesis was carried out by a T7-(dT)24 primer and SuperScript II reverse transcriptase (Gibco) using 50 µg of whole RNA samples. Second strand synthesis was performed according to the Superscript Choice System (Gibco). An in vitro transcription reaction was carried out (BioArray HighYield RNA Transcript Labeling Kit; Enzo) and the fragmented cRNA was hybridized to the Affymetrix Drosophila genome arrays (14 010 genes including controls). The washing procedure was carried out using the GeneChip Fluidics Station (Affymetrix) according to the manufacturer’s protocol. Staining was perfomed by R-phycoerythrin–streptavidin (Molecular Probes) followed by an antibody amplification procedure using a biotinylated anti-streptavidin antibody (Vector Laboratories). Scanning was carried out using the GeneArray Scanner (Hewlett Packard). The data were collected with software provided by Affymetrix and analysed in GeneSpring (Silicon Genetics) and in Microsoft Excel. We used the given fold changes for genes with a difference call increase (I) or decrease (D). According to the Affymetrix service, bad data in I or D were set as not determined (nd) and not taken into consideration. The values for each gene were averaged, and the categories were as described in the text. See Supplementary data for more details on data handling.
RT–PCR, real-time PCR and in situ hybridization analysis
For RT–PCR and real-time PCR, 1 µg of total RNA was used for first strand cDNA reaction using oligo d(T) primers (Amersham) and Superscript II reverse transcriptase (Gibco/Invitrogen) at 42°C for 50 min. PCR was performed on aliquots of this reaction in a total volume of 25 µl. Different cycles were used depending on the primers used (see Supplementary table S2).
For real-time PCR, the reaction consisted of cDNA first strand template (as for RT–PCR), primer mix (5 µM, different from RT–PCR primers, see Supplementary table S2) and SYBR Green PCR Master Mix (Applied Biosystems) in a total volume of 25 µl. Three reactions per template were performed in parallel. These were repeated with independently isolated RNA samples from different egg collections. The experiments were performed with ABI Prism 7000 SDS from Applied Biosystems. The values were normalized using actin 5C and analysed using Microsoft Excel, following the instructions provided by the manufacturer.
In situ hybridization was carried out with a single-stranded RNA probe using a modification of a standard procedure (Tautz and Pfeifle, 1989). Second instar larvae from a 12 h egg collection were used and were placed in sugar conditions for 12 h. After appropriate nutrient treatment, larvae were dissected, fixed with formaldehyde (1:10) and processed as for embryos. Glycine treatment was omitted.
Overexpression analysis
The sug cDNA was obtained from larvae with PCR, cloned into TOPO vector (Invitrogen) and then placed into p{UAST} vector. Eight independent lines were obtained, and two of these were used for analysis (UAS-sug1 and UAS-sug3). Heat shock was performed on larvae 44–48 h AEL old by placing the plates at 37°C for 40 min. For the control, heat shock-Gal4 was crossed with white flies. The microarray data were obtained with the UAS-sug-3 line.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Anne Marie Voie and Steve Cohen for helping us establish the fly injections, Christoph Melcher and Bettina Pankratz for working out the larval in situs, Simone Schindler for technical assistance, Anne Gaehler for microarray support and Dr Dirk Koczan for help in the Affymetrix data analysis. This work was supported by the Forschungszentrum Karlsruhe GmbH and DFG grants PA 787/1 and PA 787/2 to M.J.P.
References
- Abu-Elheiga L., Matzuk,M., Abo-Hasema,K. and Wakil,S. (2001) Continuous fatty acid oxidation and reduced fat storage in mice lacking acetyl-CoA carboxylase 2. Science, 291, 2613–2616. [DOI] [PubMed] [Google Scholar]
- Anderson R., Byrum,R., Coates,P. and Sando,G. (1994) Mutations at the lysosomal acid cholesteryl ester hydrolase gene locus in Wolman’s disease. Proc. Natl Acad. Sci. USA, 91, 2718–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle G., Tatum,E. and Clancy,C. (1938) Food level in relation to rate of development and eye pigmentation in Drosophila melanogaster. Biol. Bull., 75, 447–462. [Google Scholar]
- Bernal A. and Kimbrell,D. (2000) Drosophila Thor participates in host immune defense and connects a translational regulator with innate immunity. Proc. Natl Acad. Sci. USA, 97, 6019–6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell J. (1991) Pharmacological approaches to appetite suppression. Trends Pharm. Sci., 12, 147–157. [DOI] [PubMed] [Google Scholar]
- Britton J. and Edgar,B. (1998) Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development, 125, 2149–2158. [DOI] [PubMed] [Google Scholar]
- Brogiolo W., Stocker,H., Ikeya,T., Rintelen,F., Fernandez,R. and Hafen,E. (2001) An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol., 11, 213–221. [DOI] [PubMed] [Google Scholar]
- Cederberg A., Gronning,L., Ahren,B., Taken,K., Carlsson,P. and Enerback,S. (2001) FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia and diet-induced insulin resistance. Cell, 106, 563–573. [DOI] [PubMed] [Google Scholar]
- Choy R. and Thomas,J. (1999) Fluoxetine-resistant mutants in C.elegans define a novel family of transmembrane proteins. Mol. Cell, 4, 143–152. [DOI] [PubMed] [Google Scholar]
- Flier J. and Hollenberg,A. (1999) ADD-1 provides major new insight into the mechanism of insulin action. Proc. Natl Acad. Sci. USA, 96, 14191–14192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flybase (1999) The FlyBase database of the Drosophila genome projects and community literature. The FlyBase Consortium. Nucleic Acids Res., 27, 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J. and Halaas,J. (1998) Leptin and the regulation of body weight in mammals. Nature, 395, 763–770. [DOI] [PubMed] [Google Scholar]
- Gallant P., Shiio,Y., Cheng,P., Parkhurst,S. and Eisenman,R. (1996) Myc and Max homologs in Drosophila. Science, 274, 1523–1527. [DOI] [PubMed] [Google Scholar]
- Ganong W. (1991) Review of Medical Physiology. Appleton and Lange, Norwalk, CT.
- Gems D. and Partridge,L. (2001) Insulin/IGF signalling and ageing: seeing the bigger picture. Curr. Opin. Genet. Dev., 11, 287–292. [DOI] [PubMed] [Google Scholar]
- Hara K., Yonezawa,K., Weng,Q., Kozlowski,M., Belham,C. and Avruch,J. (1998) Amino acid sufficiency and mTOR regulate p70S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem., 273, 14484–14494. [DOI] [PubMed] [Google Scholar]
- Herzig S. et al. (2001) CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature, 413, 179–183. [DOI] [PubMed] [Google Scholar]
- Hill J., Hauptman,J., Anderson,J., Fujioka,K., O’Neil,P., Smith,D., Zavoral,J. and Aronne,L. (1999) Orlistat, a lipase inhibitor, for weight maintenance after conventional dieting: a 1-y study. Am. J. Clin. Nutr., 69, 1108–1116. [DOI] [PubMed] [Google Scholar]
- Hoornaert I., Marynen,P. and Baens,M. (1998) CREBL2, a novel transcript from the chromosome 12 region flanked by ETV6 and CDKN1B. Genomics, 51, 154–157. [DOI] [PubMed] [Google Scholar]
- Jackson V., Cameron,J., Zammit,V. and Price,N. (1999) Sequencing and functional expression of the malonyl CoA-sensitive carnitine palmitoyltransferase from Drosophila melanogaster. Biochem. J., 341, 483–489. [PMC free article] [PubMed] [Google Scholar]
- Johnston L., Prober,D., Edgar,B., Eisenman,R. and Gallant,P. (1999). Drosophila myc regulates cellular growth during development. Cell, 98, 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamar E., Kintner,C. and Goulding,M. (2001) Identification of NKL, a novel Gli-Krüppel zinc-finger protein that promotes neuronal differentiation. Development, 128, 1335–1346. [DOI] [PubMed] [Google Scholar]
- Lehner C. (1999) The beauty of small flies. Nat. Cell Biol., 1, E129–E130. [DOI] [PubMed] [Google Scholar]
- Lehninger A. (1970) Biochemistry. Worth, New York, NY.
- Loftus T., Joworsky,D., Frehywot,G., Townsend,C., Ronnett,G., Lane,M. and Kuhajda,F. (2000) Reduced food intake and body weight in mice treated with fatty acid synthase inhibitors. Science, 288, 2379–2381. [DOI] [PubMed] [Google Scholar]
- Lowe M. (1997) Molecular mechanisms of rat and human pancreatic triglyceride lipases. J. Nutr., 127, 549–557. [DOI] [PubMed] [Google Scholar]
- Mallo G., Fiedler,F., Calvo,E., Ortiz,E., Vasseur,S., Keim,V., Morisset,J. and Iovanna,J. (1997) Cloning and expression of the rat p8 cDNA, a new gene activated in pancreas during the acute phase of pancreatitis, pancreatic development and regeneration and which promotes cellular growth. J. Biol. Chem., 272, 32360–32369. [DOI] [PubMed] [Google Scholar]
- Newgard C. and McGarry,D. (1995) Metabolic coupling factors in pancreatic β-cell signal transduction. Annu. Rev. Biochem., 64, 689–719. [DOI] [PubMed] [Google Scholar]
- Oldham S., Montagne,J., Radimerski,T., Thomas,G. and Hafen,E. (2000) Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev., 14, 2689–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polonsky K. and Semenkovich,C. (2001) The pancreatic β cell heats up: UCP2 and insulin secretion in diabetes. Cell, 105, 705–707. [DOI] [PubMed] [Google Scholar]
- Poupon V., Begue,B., Gagnon,J., Dautry-Varsat,A., Cerf-Bensussan,N. and Benmerah,A. (1999) Molecular cloning and characterization of MT-ACT48, a novel mitochondrial acyl-CoA thioesterase. J. Biol. Chem., 274, 19188–19194. [DOI] [PubMed] [Google Scholar]
- Roesler W. (2000) What is a cAMP response unit? Mol. Cell. Endocrinol., 162, 1–7. [DOI] [PubMed] [Google Scholar]
- Rosenbaum M., Leibel,R. and Hirsch,J. (1997) Obesity. N. Engl. J. Med., 337, 396–407. [DOI] [PubMed] [Google Scholar]
- Rubin G. et al. (2000) Comparative genomics of the eukaryotes. Science, 287, 2204–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin E., Ranganathan,R. and Horvitz,H. (2000) C.elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron, 26, 619–631. [DOI] [PubMed] [Google Scholar]
- Schmelzle T. and Hall,M. (2000) TOR, a central controller of cell growth. Cell, 103, 253–262. [DOI] [PubMed] [Google Scholar]
- Sonnhammer E.L.L., von Heijne,G. and Krogh,A. (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. In Glasgow,J., Littlejohn,T., Major,F., Lathrop,R., Sankoff,D. and Sensen,C. (eds), Proceedings of the Sixth Inter national Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA, pp. 175–182. [PubMed]
- Spiegelman B. and Flier,J. (1996) Adipogenesis and obesity: rounding out the big picture. Cell, 87, 377–389. [DOI] [PubMed] [Google Scholar]
- Stryer L. (1975) Biochemistry. W.H.Freeman and Co., San Francisco, CA.
- Sul H. and Wang,D. (1998) Nutritional and hormonal regulation of enzymes in fat synthesis: studies of fatty acid synthase and mitochondrial glycerol-3-phosphate acyltransferase gene transcrip tion. Annu. Rev. Nutr., 18, 331–351. [DOI] [PubMed] [Google Scholar]
- Tautz D. and Pfeifle,C. (1989) A non-radioactive in situ hybridization method for the localization of specific RNAs reveals translational control of the segmentation gene hunchback. Chromosoma, 98, 81–85. [DOI] [PubMed] [Google Scholar]
- Wurtmann J. and Wurtmann,R. (1977) Fenfluramine and fluoxetine spare protein consumption while suppressing caloric intake by rats. Science, 198, 1178–1180. [DOI] [PubMed] [Google Scholar]
- Yoon F. et al. (2001) Control of hepatic gluconeogenesis through the transcriptional coactivator PCG-1. Nature, 413, 131–138. [DOI] [PubMed] [Google Scholar]
- Zaffran S., Chartier,A., Gallant,P., Astier,M., Arquier,N., Doherty,D., Gratecos,D. and Semeriva,M. (1998) A Drosophila RNA helicase gene, pitchoune, is required for cell growth and proliferation and is a potential target of dMyc. Development, 125, 3571–3584. [DOI] [PubMed] [Google Scholar]
- Zhang H., Stallock,J., Ng,J., Reinhard,C. and Neufeld,T. (2000) Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev., 14, 2712–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinke I., Kirchner,C., Chao,L., Tetzlaff,M. and Pankratz,M. (1999) Suppression of food intake and growth by amino acids in Drosophila: the role of pumpless, a fat body expressed gene with homology to vertebrate glycine cleavage system. Development, 126, 5275–5284. [DOI] [PubMed] [Google Scholar]