Abstract
Tripeptidyl peptidase II (TPP II) is an exopeptidase of the subtilisin type of serine proteases that is thought to act downstream of the proteasome in the ubiquitin–proteasome pathway. Recently, a key role in a pathway parallel to the ubiquitin–proteasome pathway has been ascribed to TPP II, which forms a giant protease complex in mammalian cells. Here, we report the 900-fold purification of TPP II from Drosophila eggs and demonstrate via cryo-electron microscopy that TPP II from Drosophila melanogaster also forms a giant protease complex. The presented three-dimensional reconstruction of the 57 × 27 nm TPP II complex at 3.3 nm resolution reveals that the 150 kDa subunits form a superstructure composed of two segmented and twisted strands. Each strand is 12.5 nm in width and composed of 11 segments that enclose a central channel.
Keywords: cryo-electron microscopy/three-dimensional reconstruction/tripeptidyl peptidase II
Introduction
The obvious necessity for compartmentalization of critical processes in the crowded cellular environment has led to the evolution of a number of large oligomeric protein complexes that bury their active sites in a secluded compartment. The paradigm of a self-compartmentalizing protease is the 20S proteasome, which, in eukaryotes, is capped bilaterally with the 19S regulatory complex to form the 26S proteasome (Baumeister et al., 1998; Voges et al., 1999). This 2.4 MDa complex is the most downstream element of the ubiquitin–proteasome pathway, which controls diverse vital cellular functions (for reviews, see Coux et al., 1996; Zwickl et al., 2001; Glickman and Ciechanover, 2002). However, the potential of T-cell lymphoma cells to adapt to otherwise lethal concentrations of covalent inhibitors of the proteasome has led to an emerging consensus that there might be a parallel pathway allowing the cell to react more flexibly in coping with its diverse physiological tasks, such as proteolytic house-keeping, major histocompatibility (MHC) class I-associated antigen presentation, apoptosis and the stress response (Glas et al., 1998). A key role in this alternative pathway is ascribed to tripeptidyl peptidase II (TPP II), which is upregulated in response to inhibition of the proteasome (Geier et al., 1999; Wang et al., 2000). In fact, endopeptidolytic activity has been demonstrated for this protease (Geier et al., 1999), although TPP II is primarily an exopeptidase of the subtilisin type of serine proteases and is otherwise thought to act downstream of the proteasome in the ubiquitin– proteasome pathway (Tomkinson, 1999), analogous to the role of tricorn protease in prokaryotes (Tamura et al., 1998). In addition to this soluble complex, a membrane-bound form of TPP II acting as a cholecystokinin-inactivating peptidase exists in rat cerebral cortex (Rose et al., 1996).
Thus far, TPP II has only been found in eukaryotes. The mammalian variant has a molecular subunit mass of 138 kDa (Tomkinson, 1998), whereas the insect, worm, plant and fungal form carry an additional insert in the C-terminal region and have a molecular mass of ∼150 kDa (Renn et al., 1998; DDBJ/EMBL/GenBank accession Nos U23176, AL080282 and AL117210, respectively). The closest structural, albeit not functional homolog of TPP II is the archaeal protease pyrolysin, a membrane-bound endopeptidase from Pyrococcus furiosus (Voorhorst et al., 1996).
The majority of proteases have molecular masses in the range of 20–70 kDa while the self-compartmentalizing proteases are often oligomers of 500–1000 kDa and some of them (e.g. tricorn; Walz et al., 1997) assemble further in vivo to form giant homo-oligomeric superstructures. TPP II, which is estimated to have a molecular mass of 5–9 MDa, falls into this category (Geier et al., 1999).
The structural characterization of TPP II has not proceeded much since its discovery almost 20 years ago. Electron microscopic investigations of the mammalian complex have shown that TPP II forms a ‘double bow’ oligomer (50 × 20 nm) (Macpherson et al., 1987; Harris and Tomkinson, 1990). In the only report of a two-dimensional image analysis of TPP II, the occurrence of ovoid and dumb-bell-shaped particles comprising eight segments each was taken to reflect conformational changes (Geier et al., 1999). Significant progress in the structural characterization of this giant assembly is required to understand its facultative adoption of proteasomal functions, its regulation and its reaction mechanism, as well as its specificity. However, the extraordinary size of the TPP II complex, its lability and its propensity to form even larger aggregates (Harris and Tomkinson, 1990; Renn et al., 1998; Tomkinson, 2000) appear to be very severe impediments for three-dimensional crystallization. In contrast, single-particle electron microscopy is the method of choice for studying protein complexes of this type. Therefore, we have chosen this approach to determine the structure of TPP II from Drosophila melanogaster. We have purified the protein from Drosophila eggs and propose a model of the three-dimensional architecture of the complex at 3.3 nm resolution.
Results
Protein characterization
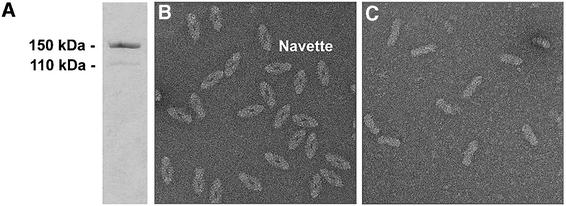
Drosophila TPP II was purified ∼900-fold by chromatography on Superose 6, Q-Sepharose HP and phenyl-Sepharose HP. Whereas the purified complex is remarkably stable under the buffer conditions used, it is relatively labile during the purification procedure. When sodium chloride is used instead of ammonium sulfate, the total activity drops drastically in the second chromatographic step. Although the activity largely recovers in the third step, both large fragments and aggregates of the supercomplex are observed in addition to the intact complex in the electron microscope. However, addition of up to 1 M sodium chloride to purified TPP II does not impair its activity and structural integrity (data not shown). TPP II is also quite sensitive to nicking by endogenous proteases. The specific activities at the different stages of the purification are displayed in Table I. As shown in Figure 1A, the apparent Mr of purified TPP II is ∼150 kDa on SDS–gels, which corresponds well to the value of 149 kDa deduced from the nucleotide sequence. A weak additional band at ∼110 kDa is a result of proteolytic nicking which may not be prevented completely. A proteolytic fragment of similar size was observed with recombinant Drosophila TPP II (Renn et al., 1998).
Table I. Specific H-AAF-AMC-cleaving activity measured in the presence of bestatin at 30°C during the course of protein purification.
| Stage of purification | Activity (nmol/h/mg) |
|---|---|
| Crude extract | 58 |
| Superose 6 | 889 |
| Q-Sepharose HP | 11 734 |
| Phenyl-Sepharose | 48 890 |
Fig. 1. Purification of TPP II from D.melanogaster. (A) SDS–gel. (B) TPP II particles purified in the presence of DTT (1 mM, initial chromatographic step; 10 mM, final step). (C) TPP II particles purified in the absence of DTT in the final chromatographic step.
TPP II complexes prepared in the presence of dithiothreitol (DTT) are shown in Figure 1B. Whereas the particles exhibit their characteristic appearance in electron micrographs under reducing conditions and are uniform in size and shape, this is not the case in the absence of reducing agent (Figure 1C). The number of particles observed per unit area is also significantly smaller. However, the specific activity was identical in both samples. In both the presence and absence of DTT, the activity remained stable for 6 weeks at 4°C and the morphology of the particles in both samples did not change at all. Thus, the preservation of the 17 thiol groups per monomer is essential for the native structure of TPP II, but less so for its peptidase activity. So far, there seems to have been a consensus in the literature that a high glycerol concentration (30%) is required to maintain the integrity of the complex. However, our data indicate that Drosophila TPP II is quite stable at low glycerol concentration (5%), at least when stored in the presence of ∼200 mM ammonium sulfate.
Electron microscopy
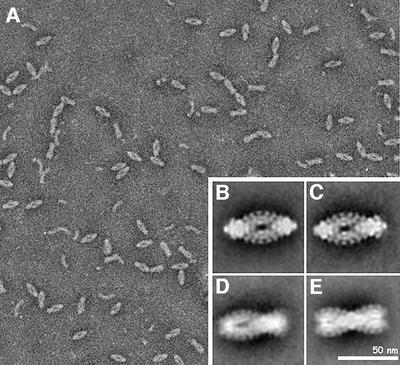
On a supporting carbon film, negatively stained TPP II molecules adopt a preferred orientation, resulting in a pointed oval shape (navette shape; Figure 2A). In addition to this navette view, dumb-bell views exhibiting a central constriction and views with a constriction ‘off-center’ can also be observed. Furthermore, bows are visible, which correspond to partially dissociated complexes. Accordingly, characteristic class averages of TPP II negatively stained in methylamine tungstate classify into groups representing different localizations of their constriction (Figure 2B–E).
Fig. 2. Purified TPP II particles negatively stained in methylamine tungstate. (A) Overview. (B–E) Class averages. (B) Navette view, (C and D) views with off-center constrictions and (E) dumb-bell view.
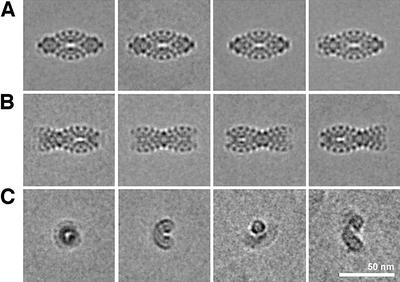
Whereas TPP II particles in negative stain on carbon film adopt only a limited number of orientations, the situation in non-supported ice layers is different: while ‘side views’ still dominate (Figure 3A and B), a small subset of particles represent end-on views, such as a circular average, a kidney-shaped average, a bow around a triangle and a boomerang-shaped view (Figure 3C). The navette view and the dumb-bell view shown in Figure 3A, first panel and Figure 3B, second panel, respectively, exhibit a high degree of symmetry and are the most elongated views. Therefore, both appear to be oriented as true side views without a rotation around their transverse axis. In the most symmetrical navette view, the convex sides of the bows are lined with five globular densities, suggesting that each strand is composed of an odd number of segments. In the dumb-bell view, a large globular density on each side of the constriction marks the center. Here, the convex sides of the bows emerging left and right from the constriction are lined with five globular densities each, suggesting that the number of segments in the strands is 11 in total.
Fig. 3. Class averages of TPP II particles embedded in vitreous ice. (A) Several views of the TPP II particle in navette orientation. (B) Dumb-bell views and views showing different off-center locations of the constriction. (C) Class averages showing more compact rather than elongated views.
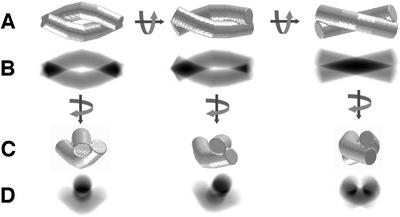
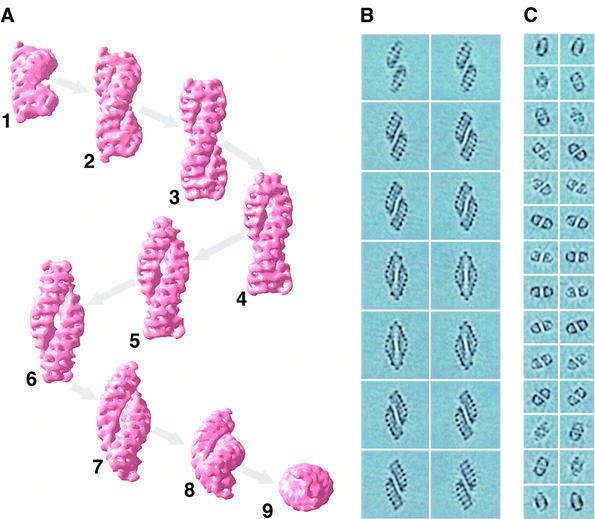
The presence of dumb-bell views, navette views and, occasionally, bows strongly suggests that TPP II is composed of two strands twisted around each other. This was proposed previously by Harris and Tomkinson (1990), whereas Geier et al. (1999) interpreted these different views as a consequence of conformational changes. In order to verify that this double bow architecture agrees with the views we had obtained from ice-embedded TPP II particles, we created a model composed of two solid single bows. Figure 4A and B explains how the navette view can be transformed into a dumb-bell view by rotating the model around its longitudinal axis and how the projec tions change during this transition. Rotation of the model around its transverse axis produces end-on views (Figure 4C), and the corresponding projections (Figure 4D) resemble the class averages of ice-embedded TPP II particles shown in Figure 3C. After we had confirmed that the twisted double bow is most probably the basic architecture of TPP II, we used the double bow model as a reference for projection matching (EMAN; Ludtke et al., 1999).
Fig. 4. Analysis of a synthetic model reveals that the twisted double strand concept is compatible with the views obtained of ice-embedded TPP II particles. (A) Surface representation of a double-strand model and rotation around its longitudinal axis. (B) Projections of the three- dimensional models shown in (A) in the direction perpendicular to the image plane. (C) A 75° rotation of the models shown in (A) around their transverse axis. (D) Projection of the three-dimensional models shown in (A) in the direction perpendicular to the image plane.
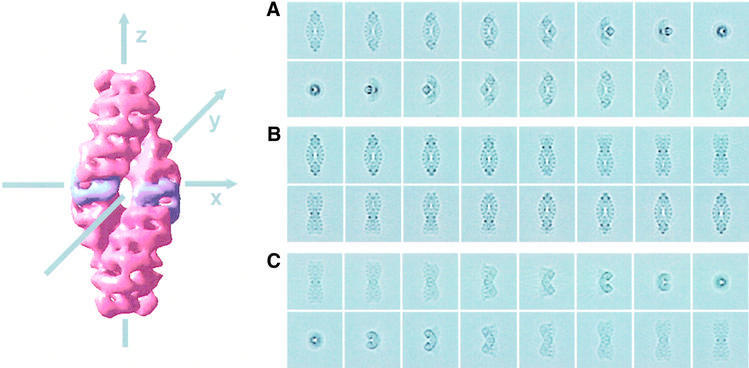
The reconstructed TPP II complex is shown in its navette orientation in Figure 5 together with projections through the reconstructed volume explaining the geometrical relationship between the averages of ice-embedded TPP II particles shown in Figure 3. Counting in either direction from the central segment (highlighted in the isosurface of Figure 5), five segments are visible, confirming that the number of segments in each strand is 11.
Fig. 5. Projections through the reconstructed TPP II volume. (A) Rotation of the TPP II complex around the x-axis (increment 12°) and orthogonal projection onto the image plane. (B) Rotation of the TPP II complex around the z-axis (increment 12°) and orthogonal projection onto the image plane. (C) Rotation of the TPP II complex around the y-axis (increment 12°) and projection in the direction of the x-axis. In the isosurface representation, the central segment of each strand is highlighted.
The surface views of TPP II in Figure 6A show different orientations of the complex. Precise Mr values of the complex currently are not available, and values reported in the literature range from Mr > 1 MDa (Renn et al., 1998) to Mr = 5–9 MDa (Geier et al., 1999). Therefore, we set the threshold for the isosurface to a value that best displays the same features that are seen in the projections and in the slices through the reconstructed volume. The dimensions of the complex are 57 nm along its longitudinal axis and 27 nm at its transverse axis. The complex is composed of two helical strands wound around each other; this winding is also obvious in x–y slices through the reconstructed volume (Figure 6C). The strands themselves are 12.5 nm in width and display a corrugated surface that clearly confirms the segmentation suggested by the two-dimensional data (Figure 6A and B). Interestingly, the cross-sectional area of a strand is not circular (Figure 6C), and the isosurface representation (Figure 6A) emphasizes the angular shape of the segments. The area where the single bows are jointed together at this chosen contour level comprises two segments of each strand at both ends of the twisted double strand. The slices through the reconstructed volume displayed in Figure 6B and C show that the centers of the strands are less dense than the outer boundary, indicating the presence of a central channel in the strands.
Fig. 6. Three-dimensional reconstruction of TPP II from D.melanogaster. (A) Surface representation of the complex. (1–3) Rotation of the TPP II reconstruction in tilted dumb-bell orientation around its transverse axis. (3–6) Rotation of the reconstruction in dumb-bell orientation around its longitudinal axis. (6–9) Rotation of the reconstruction in navette orientation around its transverse axis. (B) Longitudinal slices through the reconstructed volume. (C) Transverse slices through the reconstructed volume. The slice thickness is 0.8 nm.
Discussion
TPP II was purified from dechorionated Drosophila eggs using a three-step chromatographic procedure. The importance of reducing conditions is highlighted by the morphological changes occurring under non-reducing conditions. Because the solubility of molecular oxygen in water is close to the range of commonly used concentrations of DTT or mercaptoethanol of ∼1 mM, the preservation of reducing conditions requires a higher concentration of reducing agents to prevent autoxidation of thiol groups reliably. In fact, the electron microscopic appearance of the mammalian TPP II particles in the presence of 0.5 mM mercaptoethanol shown by Geier et al. (1999) is reminiscent of Figure 1C in the present work, which was obtained in the absence of reducing agent. The concentration of 10 mM DTT used here preserves the native structure of TPP even in small amounts of sample and during prolonged storage above 0°C.
The purified protein was examined in the electron microscope, both negatively stained and embedded in a non-supported layer of vitreous ice. Like human TPP II isolated from red blood cells, the Drosophila homolog also forms a giant oligomeric structure. Here we demonstrate the twisted double strand architecture of the giant complex, in which each of the single strands is subdivided into 11 segments, with a spacing of 6 nm. Since TPP II is a homo-oligomer, the structure of this giant complex must be repetitive. In fact, the striated structure implies that the strands are stacks of disc-shaped subunits. Recent experimental data corroborate this idea: under certain conditions in which Drosophila TPP II becomes marginally stable, such as pH 5.1–5.3, distinct subcomplexes are found that decay further at even lower pH. These circular subcomplexes have a diameter of ∼13 nm, which corresponds to the width of the helical strands in the three-dimensional reconstruction, and they display a noticeable central cavity (data not shown). Single strands also occur under conditions where the double-stranded complex is destabilized. However, in contrast to the precisely defined supercomplexes, the observed length of single strands is variable. The decomposition of the TPP II holocomplex into subcomplexes is accompanied by a drastic drop in specific activity. This correlates with the data of Tomkinson (2000), who reported that the reversible decay of mammalian TPP II into putative dimers results in a 10-fold reduced specific activity. This equilibrium is considered as a possible means of regulation. Whereas putative dimers believed to represent the building blocks of the holocomplex are rectangular (8 × 9 nm) in mammalian TPP II (Harris and Tomkinson, 1990), the subcomplexes of Drosophila TPP II (data not shown) are roughly circular and correspond in diameter to the single strands.
The three-dimensional structure of the holocomplex explains what seemed previously to be moving constrictions that led to the peristaltic movement model proposed by Geier et al. (1999). The projections through the reconstructed volume clearly demonstrate how the views described by Geier et al. (ovoid views with the constrictions at different locations of the complex) can be produced from a twisted double-stranded structure.
Whereas structural homologs of TPP II such as pyrolysin (Voorhorst et al., 1996) and other high molecular weight subtilases do not appear to form oligomeric complexes, the suggested functional homolog in archaea, tricorn (TRI), a hexameric toroid of 720 kDa from Thermoplasma acidophilum, forms a supermolecular structure (Tamura et al., 1996). However, in contrast to TPP II, TRI forms an icosahedral capsid (14.4 MDa) in vivo and its specific activity is not increased by superoligomerization (Walz et al., 1997). Like TPP II, tricorn also cleaves AAF-AMC, albeit at an ∼10-fold lower rate and at 60°C (Tamura et al., 1996).
For TPP II, it has been suggested that in order to protect the cell from uncontrolled proteolysis, only the supercomplex has maximum activity (Geier et al., 1999) and that the formation of a self-compartmentalizing complex (Lupas et al., 1997) may be a prerequisite for acquiring additional proteolytic competence such as endopeptidase activity (Geier et al., 1999; Yao and Cohen, 1999). Since it is conceivable that the formation of the supermolecular assembly leads to the formation of a secluded proteolytic compartment, the question arises of where the TPP II structure is localized in this compartment. Geier et al. (1999) suggested a central channel for human TPP II, but the present three-dimensional reconstruction of Drosophila TPP II represents a twisted double strand, which does not enclose a central channel. However, the segments forming the single strands apparently possess inner cavities, which are likely to form a continuum in the fully assembled complex.
Clearly, the unique structure of the giant TPP II complex raises more intriguing questions. (i) To our knowledge, TPP II is the only protein composed of more than two stacked discs of subunits that has a precisely defined length, so what limits the growth of the helical strands to a length corresponding to one-half helical turn? (ii) What is the structural basis for the drastic increase of activity associated with the formation of the supermolecular complex?
A refinement of the three-dimensional structure will contribute significantly to the understanding of this enigmatic giant protease complex and enable us to detect conformational changes upon substrate binding and to understand how the disc-shaped subcomplexes are organized in the helical strands. The study of the disc-shaped subcomplexes and their structural characterization will be indispensable for elucidating the structural basis of the assembly–disassembly equilibrium and its functional significance for this key enzyme of cellular proteolysis.
Materials and methods
Protein preparation
Drosophila embryos were grown and harvested as described previously (Hölzl et al., 2000). The supernatant of disrupted and dechorionated Drosophila eggs kept at –80°C was thawed out and centrifuged at 18 000 g. The supernatant was filtered through a 0.22 nm filter (Millipore) and applied to a Superose 6 column equilibrated with 20 mM triethanolamine–HCl pH 7.5, 70 mM ammonium sulfate, 1 mM DTT, 5% glycerol, and thermostated at 4°C. Fractions including the maximum of AAF-AMC-hydrolyzing activity at Kav 0.33–0.43 were pooled and loaded on a Sepharose Q-XL column after 50% dilution with 5% glycerol, addition of DTT to 5 mM final concentration and adjustment of the pH to 8.0. Elution was performed using a gradient of ammonium sulfate ranging from 80 to 280 mM, in 20 mM triethanolamine–HCl pH 7.8, 5 mM DTT and 5% glycerol. Active fractions were again pooled, adjusted to 1 M ammonium sulfate using a 4 M stock solution and loaded on phenyl–Sepharose HP (Amersham). Then a gradient of ammonium sulfate descending to 100 mM, in 20 mM triethanolamine pH 7.5, 10 mM DTT and 5% glycerol, was applied. TPP II eluted at ∼200 mM ammonium sulfate. All operations were performed on ice. Samples were kept at 4°C for several weeks or frozen in liquid nitrogen and stored at –80°C. Freezing did not impair the activity and structural integrity of the protein. TPP II activity was assayed by measuring AAF-AMC-hydrolyzing activity at 30°C. Bestatin was added to inhibit aminopeptidase activity.
Electron microscopy and image processing
Carbon-coated copper grids were glow discharged and incubated on 10 µl of the protein sample for 1 min. After washing with water, grids were placed for 15 s on a drop of methylamine tungstate and subsequently blotted. Images of negatively stained TPP II particles were recorded using a Philips CM12 electron microscope at 120 kV at a nominal magnification of 26 900× on AGFA film at a defocus of –1 µm. Images were scanned on a Flextight Precision 2 scanner (Imacon, Denmark) using a pixel size of 15.9 µm. From scanned images, 2659 TPP II particles were selected interactively using the EMAN software (Ludtke et al., 1999).
For ice embedding, 2–3 µl of a TPP II solution were applied on glow-discharged lacey carbon grids and incubated for 1–2 min. Subsequently the grids were washed twice with 20 mM potassium phosphate buffer pH 7.5 in order to dilute the glycerol in the sample. The solution was then blotted and the grids quick-frozen in liquid ethane. Images of TPP II particles were recorded using a JEOL4000 microscope under low-dose conditions at a nominal magnification of 50 000× on KODAK SO-163 films. In order to obtain high-contrast images, we recorded data at a high defocus value with the first zero of the contrast transfer function at 3.0 nm. Negatives of ice-embedded particles were scanned with a Leafscan 45 scanner (Leaf Systems Inc., Westborough, MA) at a pixel size of 20 µm, resulting in a pixel size on the specimen level of 0.4 nm. From scanned images, 1713 particles were selected interactively using the EMAN software package (Ludtke et al., 1999), and subsequently the data set was CTF corrected by phase-flipping the Fourier coefficients in the negative transfer regions.
The following image processing steps were performed using the EM software package (Hegerl and Altbauer, 1982; Hegerl, 1996). In order to correct for the in-plane rotation of the elongated particles, both data sets first were aligned rotationally using a navette view, the predominant view of both data sets, as a reference. Thereafter, rotational and translational alignment was performed, followed by a sequence of multivariate statistical analysis (van Heel and Frank, 1981) and multireference alignment, which eventually produced characteristic class images for the two data sets.
The basic architecture of the TPP II complex is a twisted double strand. Therefore, we used a curved cylinder model (created with the EM software package; Hegerl and Altbauer, 1982; Hegerl, 1996) with the dimensions of the two-dimensional averages as a reference for multireference alignment. Multireference alignment and three-dimensional reconstruction were performed with the EMAN software package (Ludtke et al., 1999) assuming C2 symmetry. After eight cycles of refinement, the reconstruction was stable and the resolution estimated via Fourier shell correlation (Saxton and Baumeister, 1982) was 3.3 nm according to the 0.5 criterion.
Three-dimensional surface views were created using the AVS Express software (Advanced Visual Systems).
Acknowledgments
Acknowledgements
We thank Dr Harald Hölzl, Marius Boicu and Oana Mihalache (MPI for Biochemistry, Martinsried, Germany) for breeding and collecting Drosophila eggs from fruit flies, and Marietta Peters for assistance with the TPP II preparations. This work was supported in part by a Feodor-Lynen Fellowship from the Alexander von Humboldt Foundation to B.R. and by NIH grant GM51487.
References
- Baumeister W., Walz,J., Zühl,F. and Seemüller,E. (1998) The proteasome: paradigm of a self-compartmentalizing protease. Cell, 92, 367–380. [DOI] [PubMed] [Google Scholar]
- Coux O., Tanaka,K. and Goldberg,A.L. (1996) Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem., 65, 801–847. [DOI] [PubMed] [Google Scholar]
- Geier E., Pfeifer,G., Wilm,M., Lucchiari-Hartz,M., Baumeister,W., Eichmann,K. and Niedermann,G. (1999) A giant protease with potential to substitute for some functions of the proteasome. Science, 283, 978–981. [DOI] [PubMed] [Google Scholar]
- Glas R., Bogyo,M., McMaster,J.S., Gaczynska,M. and Ploegh,H.L. (1998) A proteolytic system that compensates for loss of proteasome function. Nature, 392, 618–622. [DOI] [PubMed] [Google Scholar]
- Glickman M.H. and Ciechanover,A. (2002) The ubiquitin–proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev., 82, 373–428. [DOI] [PubMed] [Google Scholar]
- Harris J.R. and Tomkinson,B. (1990) Electron-microscopic and biochemical-studies on the oligomeric states of human erythrocyte tripeptidyl peptidase 2. Micron Microsc. Acta, 21, 77–89. [Google Scholar]
- Hegerl R. (1996) The EM program package: a platform for image processing in biological electron microscopy. J. Struct. Biol., 116, 30–34. [DOI] [PubMed] [Google Scholar]
- Hegerl R. and Altbauer,A. (1982) The ‘EM’ program system. Ultramicroscopy, 9, 109–116. [DOI] [PubMed] [Google Scholar]
- Hölzl H. et al. (2000) The regulatory complex of Drosophila melanogaster 26S proteasomes: subunit composition and localization of a deubiquitylating enzyme. J. Cell Biol., 150, 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke S.J., Baldwin,P.R. and Chiu,W. (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol., 128, 82–97. [DOI] [PubMed] [Google Scholar]
- Lupas A, Flanagan,J.M., Tamura,T. and Baumeister,W. (1997) Self-compartmentalizing proteases. Trends Biochem. Sci., 22, 399–404. [DOI] [PubMed] [Google Scholar]
- Macpherson E., Tomkinson,B., Bålöw,R.M., Hoglund,S. and Zetterqvist,O. (1987) Supramolecular structure of tripeptidyl peptidase II from human erythrocytes as studied by electron microscopy, and its correlation to enzyme activity. Biochem. J., 248, 259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn S.C., Tomkinson,B. and Taghert,P.H. (1998) Characterization and cloning of tripeptidyl peptidase II from the fruit fly, Drosophila melanogaster. J. Biol. Chem., 273, 19173–19182. [DOI] [PubMed] [Google Scholar]
- Rose C. et al. (1996) Characterization and inhibition of a cholecystokinin-inactivating serine peptidase. Nature, 380, 403–409. [DOI] [PubMed] [Google Scholar]
- Saxton W.O. and Baumeister,W. (1982) The correlation averaging of a regularly arranged bacterial cell envelope protein. J. Microsc., 127, 127–138. [DOI] [PubMed] [Google Scholar]
- Tamura T., Tamura,N., Cejka,Z., Hegerl,R., Lottspeich,F. and Baumeister,W. (1996) Tricorn protease—the core of a modular proteolytic system. Science, 274, 1385–1389. [DOI] [PubMed] [Google Scholar]
- Tamura N., Lottspeich,F., Baumeister,W. and Tamura,T. (1998) The role of tricorn protease and its aminopeptidase-interacting factors in cellular protein degradation. Cell, 95, 637–648. [DOI] [PubMed] [Google Scholar]
- Tomkinson B. (1998) Tripeptidyl-peptidase II. In Barrett,A.J., Rawlings,N.D. and Woessner,J.F. (eds), Handbook of Proteolytic Enzymes. Academic Press, London, UK, pp. 338–340.
- Tomkinson B. (1999) Tripeptidyl peptidases: enzymes that count. Trends Biochem. Sci., 24, 355–359. [DOI] [PubMed] [Google Scholar]
- Tomkinson B. (2000) Association and dissociation of the tripeptidyl-peptidase II complex as a way of regulating the enzyme activity. Arch. Biochem. Biophys., 376, 275–280. [DOI] [PubMed] [Google Scholar]
- van Heel M. and Frank,J. (1981) Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy, 6, 187–194. [DOI] [PubMed] [Google Scholar]
- Voges D., Zwickl,P. and Baumeister,W. (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem., 68, 1015–1068. [DOI] [PubMed] [Google Scholar]
- Voorhorst W.G.B., Eggen,R.I.L., Geerling,A.C.M., Platteeuw,C., Siezen,R.J. and Devos,W.M. (1996) Isolation and characterization of the hyperthermostable serine protease, pyrolysin, and its gene from the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem., 271, 20426–20431. [DOI] [PubMed] [Google Scholar]
- Walz J., Tamura,T., Tamura,N., Grimm,R., Baumeister,W. and Koster,A.J. (1997) Tricorn protease exists as an icosahedral supermolecule in vivo. Mol. Cell, 1, 59–65. [DOI] [PubMed] [Google Scholar]
- Wang E.W., Kessler,B.M., Borodovsky,A., Cravatt,B.F., Bogyo,M., Ploegh,H.L. and Glas,R. (2000) Integration of the ubiquitin– proteasome pathway with a cytosolic oligopeptidase activity. Proc. Natl Acad. Sci. USA, 97, 9990–9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T.T. and Cohen,R.E. (1999) Giant proteases: beyond the proteasome. Curr. Biol., 9, R551–R553. [DOI] [PubMed] [Google Scholar]
- Zwickl P., Seemüller,E., Kapelari,B. and Baumeister,W. (2001) The proteasome: a supramolecular assembly designed for controlled proteolysis. Adv. Protein Chem., 59, 187–222. [DOI] [PubMed] [Google Scholar]