Abstract
Intracellular protein interaction domains are essential for eukaryotic signaling. In T cells, the CD2BP2 adaptor binds two membrane-proximal proline-rich motifs in the CD2 cytoplasmic tail via its GYF domain, thereby regulating interleukin-2 production. Here we present the structure of the GYF domain in complex with a CD2 tail peptide. Unlike SH3 domains, which use two surface pockets to accommodate proline residues of ligands, the GYF domain employs phylogenetically conserved hydrophobic residues to create a single interaction surface. NMR analysis shows that the Fyn but not the Lck tyrosine kinase SH3 domain competes with CD2BP2 GYF-domain binding to the same CD2 proline-rich sequence in vitro. To test the in vivo significance of this competition, we used co-immunoprecipitation experiments and found that CD2BP2 is the ligand of the membrane-proximal proline-rich tandem repeat of CD2 in detergent- soluble membrane compartments, but is replaced by Fyn SH3 after CD2 is translocated into lipid rafts upon CD2 ectodomain clustering. This unveils the mechanism of a switch of CD2 function due to an extracellular mitogenic signal.
Keywords: CD2/GYF domain/lipid rafts/NMR/SH3 domain
Introduction
Proline-rich sequences are amongst the most abundant motifs in eukaryotic cells and play a pivotal role in many signaling pathways (Rubin et al., 2000). They mediate the assembly of molecular complexes by interacting with versatile recognition domains contained in various intracellular proteins (Kay et al., 2000). The currently identified signaling domains that recognize proline-rich sequences are the SH3 domain (Mayer et al., 1988; Stahl et al., 1988), the WW domain (Bork and Sudol, 1994), the EVH1 domain (Niebuhr et al., 1997), profilin (Carlsson et al., 1977) and the GYF domain (Nishizawa et al., 1998; Freund et al., 1999). Strikingly, these domains recognize multiple different sequences and can compete for the same target sites in vitro (Kay et al., 2000 and references therein). Whether the observed in vitro binding promiscuity is physiologically relevant in the cellular environment is a matter of much debate. Post-translational modifications of sequences adjacent to the proline-rich core motif, such as, for example, serine phosphorylation (Lu et al., 1999) or arginine methylation (Bedford et al., 2000), might control the interaction of certain recognition domains with the proline-rich target sequences. At the cell membrane, subcellular compartmentalization of intracellular signaling molecules into lipid microdomains (rafts) introduces an additional regulatory mechanism for the formation of molecular assemblies (Simons and Toomre, 2000). However, the roles of membrane partitioning of proline-rich sequences, and the mechanisms of protein–protein and protein–lipid interactions that regulate their localization have remained elusive.
In T cells, the formation and maintenance of signaling-competent assemblies have been shown to depend on the recruitment of signaling molecules into lipid rafts (Xavier et al., 1998). Key regulators for intracellular signaling become lipid modified (Koegl et al., 1994; Zhang et al., 1998) and thereby efficiently enter the center of the junction between T cells and antigen-presenting cells, often referred to as the immunological synapse (Grakoui et al., 1999). The human T-cell adhesion molecule CD2 modulates the threshold for T-cell receptor signaling by binding to its counter-receptor CD58 on antigen-presenting cells (Wang et al., 1999 and references therein) and has been shown to translocate inducibly into lipid microdomains (Yang and Reinherz, 2001). CD2 contains five proline-rich stretches in its cytoplasmic tail that are responsible for the direct physical interaction of CD2 with various intracellular proteins. The protein CD2AP has been shown to play a pivotal role in T-cell polarization and cytoskeletal rearrangements (Dustin et al., 1998), while the two proteins CD2BP1 (Li et al., 1998) and CD2BP2 (Nishizawa et al., 1998) have been implicated in adhesion and signal transduction, respectively. CD2BP1 serves as an adaptor to recruit PTP-PEST to CD2, thereby downregulating adhesion and fostering motility (Li et al., 1998). CD2BP2 was shown to contain the novel recognition domain GYF that interacts with CD2 and thereby enhances the production of interleukin-2 (IL-2) on cross-linking of CD2 but not the T-cell receptor (Nishizawa et al., 1998; Freund et al., 1999). There is also evidence that the src family kinase Fyn interacts with CD2 (Gassmann et al., 1994; Lin et al., 1998; Fukai et al., 2000) and thereby connects CD2 signaling with the mitogen-activated protein (MAP) kinase pathway, and the activation of phospholipase Cγ (PLCγ) and the immune cell-specific adaptor proteins Vav and LAT (Fukai et al., 2000).
The production of IL-2 after CD2 ectodomain cross-linking has been demonstrated to depend on a membrane-proximal and highly conserved tandem PPPPGHR repeat sequence of CD2 (Chang et al., 1990). Since Fyn and CD2BP2 are involved in signaling events leading to IL-2 generation, both have to be linked either directly or indirectly to this segment of the CD2 cytoplasmic tail. In the case of CD2BP2, it has been shown that the GYF domain is responsible for the binding of CD2BP2 to the PPPPGHR motifs of CD2 (Freund et al., 1999). By now solving the structure of the CD2BP2 GYF domain–CD2 tail segment complex, we reveal ligand-binding features distinct from those of other proteins that dock to proline-rich sequences. In addition, we demonstrate that the SH3 domain of Fyn but not of Lck can bind competitively to the same PPPPGHR motifs of the CD2 tail under limiting CD2 concentrations in vitro. In T cells, however, we show that Fyn is localized exclusively to rafts, whereas CD2BP2 is restricted to the non-raft cytosolic cellular fraction. Our study reveals how proline-rich sequence recognition in concert with subcellular compartmentalization co-direct the assembly of protein complexes in a general way, and as related to the CD2 signalosome in T cells specifically. Furthermore, the structural comparison of two domains from unrelated fold families that bind to the same proline-rich segments provides the basis for understanding the mechanisms of and the reasons for the existence of promiscuous binding of these interaction modules.
Results and discussion
Structure of the CD2BP2 GYF domain in complex with the CD2 motif SHRPPPPGHRV
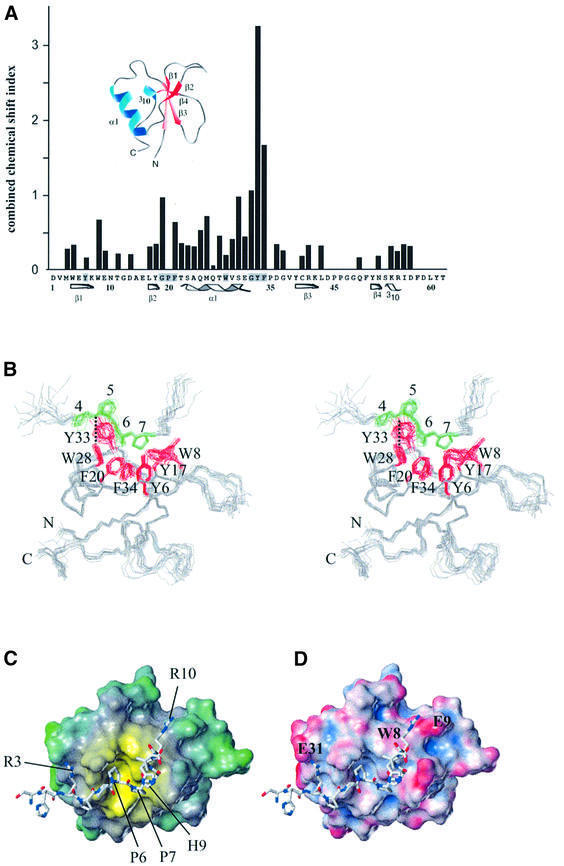
Yeast two-hybrid analysis had suggested that the minimal binding sequence for the GYF domain of CD2BP2 contains the two conserved PPPPGHR motifs of the CD2 cytoplasmic domain (PPPPGHR-X7-PPPPGHR). Subsequently, NMR experiments with a truncated version of the CD2 tail or a 31mer peptide comprising the two proline motifs confirmed a specific interaction (Nishizawa et al., 1998; Freund et al., 1999). To examine whether a single copy of the PPPPGHR motifs can bind CD2BP2 and defines the minimal binding epitope, the peptide SHRPPPPGHRV from the CD2 cytoplasmic tail was added in equimolar amounts to 15N-labeled CD2BP2 GYF domain. 15N–1H correlation spectra were recorded to search for changes of resonance positions that would indicate ligand binding. Indeed, a significant portion of the GYF domain surface residues were affected by the addition of this peptide. The pattern of chemical shift changes (Figure 1A) reveals the binding face of the proline-rich motif on the GYF domain. Most of the strongly shifted resonances belong to those residues that are highly conserved among putative GYF domains, falling into the region of the unique GPF–helix–GYF sequence of the protein domain (Figure 1A). The binding site of the GYF domain for the SHRPPPPGHR peptide is very similar to the binding surface for the 31mer peptide containing the two PPPPGHR motifs (Freund et al., 1999). Therefore, this 11mer peptide sequence from the CD2 cytoplasmic domain was used for detailed structural investigations by NMR spectroscopy.
Fig. 1. The GYF domain–ligand complex. (A) The combined chemical shift index (see Supplementary data) for the backbone NH groups of all GYF domain residues except proline is shown for the binding of an equimolar amount of the peptide SHRPPPPGHRV. Secondary structure elements are given below the sequence of the CD2BP2 GYF domain and are also present in the three-dimensional fold shown above the chemical shift indices. (B) Stereo view of the superposition of the best 15 NMR structures of the CD2BP2 GYF domain in complex with the peptide SHRPPPPGHRV. The side chains of aromatic residues of the binding site are colored red, and the prolines of the peptide are shown in green. All other side chains are omitted for clarity. (C) Lipophilic potential of the GYF domain. The surface was created by using the hydrophobicity scale of the tripos program package (Heiden et al., 1993). Hydrophobicity scaling is from yellow (most hydrophobic) to green (hydrophilic). The peptide ligand is shown as a bonded structure with labels for residues mentioned in the text. (D) Electrostatic potential of the GYF domain. The amber force field has been used to calculate the charge distribution (Case et al., 1997). Scaling is from acid (red) to basic (blue). Residues of the GYF domain that are important for the interaction with the two arginine side chains of the ligand are marked on the surface.
Figure 1B shows the ensemble of the final 15 lowest energy structures of the GYF domain in complex with the peptide ligand SHRPPPPGHRV of human CD2 (see Table I for the structural statistics). The peptide adopts an extended conformation and forms a polyproline type II helix for residues 4–7. The backbone CO group of the peptide Pro4 is engaged in a hydrogen bond with the side chain NH of W28 of the protein in all the 15 lowest energy structures, and Pro6 of the ligand is oriented almost parallel to the aromatic ring of the GYF residue W28 (Figure 1B). The center of the binding face of the GYF domain accommodates Pro6 and Pro7 of the ligand in a rolling surface depression that is defined by the aromatic residues Y6, W8, Y17, F20, W28, Y33 and F34 of the GYF domain (Figure 1B). The binding pocket’s ‘floor’ is defined by Y6 and F34, while F20, W28 and Y33 define the ‘walls’ on three sides. W8 and Y17 are tilted away from the almost parallel main axes of the conserved aromatic residues and thereby open the fourth side of the binding pocket to let Gly8 of the ligand be accommodated. This glycine allows for a kink in the backbone of the ligand with dihedral angles (φ = 76°, ψ = –80°) that are unfavorable for other residue types. Therefore, this residue terminates the proline helical part of the ligand. This kink-like conformation of the ligand is stabilized by a hydrogen bond between the Pro7 CO group and the NH group of His9 that was found based on characteristic distances in most of the calculated low-energy structures.
Table I. Statistics for the final 15 structures of the GYF domain in complex with the CD2 motif-based peptide SHRPPPPGHRV.
| NOE restraints | |
|---|---|
| Intramolecular distances of the GYF domain | 754 |
| Intraresidue | 134 |
| Medium range (i + 2 to i + 4) | 297 |
| Long range (>i + 4) | 323 |
| Intramolecular distances of peptide | 49 |
| HN to H from single chain construct | 22 |
| All others | 27 |
| Intermolecular NOEs | 33 |
| Hydrogen bond restraints (two per bond) | 34 |
| Deviations from idealized geometry | |
| Bonds (Å) | 0.0015 |
| Angles (°) | 0.36 |
| Impropers (°) | 0.14 |
| Coordinate precision (Å) | |
| R.m.s. deviation of backbone atomsa | 0.40 |
| R.m.s. deviation all heavy atomsa | 0.83 |
aThe residues of flexible regions at a distance from the binding site were omitted in the analysis (residues 1, 9–15, 44–50 and 62 of the GYF domain, and residues 1, 2 and 11 of the peptide ligand).
To analyze the determinants of the GYF domain– peptide interaction, we first calculated the lipophilic surface potential according to the method of Heiden et al. (1993). Figure 1C shows that the most hydrophobic surface region is concentrated in a groove lined by the conserved aromatic residues of the GYF domain. This hydrophobic surface pocket accommodates Pro6 and Pro7 of the ligand. The diameter of this hydrophobic ‘hot spot’ is ∼12 Å and provides a van der Waals contact area complementary to the prolines of the ligand. Figure 1D shows that the hydrophobic residues are embedded in a surface of predominantly negative potential (Case et al., 1997). This allows the charged groups of the Arg3 and Arg10 side chains of the ligand to be positioned close in space to E31 and E9 of the GYF domain, respectively. The side chain of His9 folds back onto the proline helix segment of the peptide, while the aliphatic part of the Arg10 side chain makes hydrophobic contacts with the aromatic ring of W8 from the GYF domain. This explains our previous observation that the HR to DE mutations of the PPPPGHR motifs reduce CD2 affinity and corroborates a role for non-proline residues in determining the specificity of the CD2BP2 GYF domain (Nishizawa et al., 1998). These molecular features are reminiscent of SH3–ligand interactions, which are discussed in detail below.
Proline-rich sequence recognition by the GYF domain family
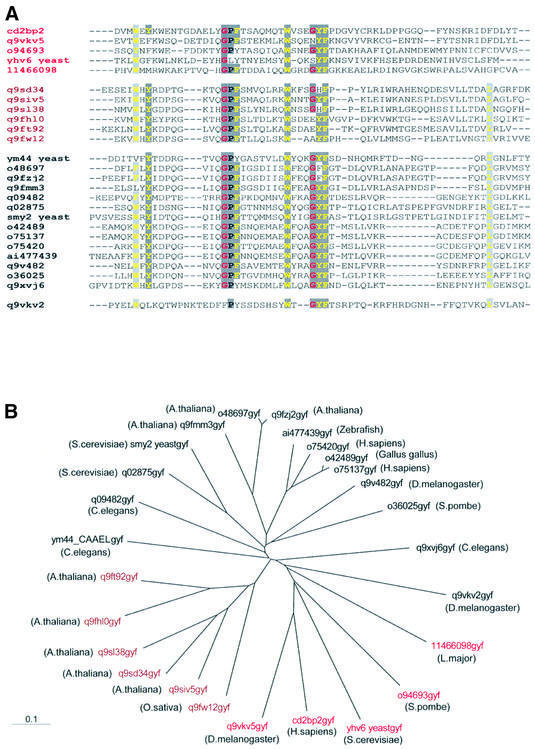
The three-dimensional structure of the complex of the GYF domain of the intracellular CD2BP2 protein and the peptide segment SHRPPPPGHRV of the membrane-proximal CD2 cytoplasmic tail sets the basis for a molecular understanding of this novel interaction module. By searching sequence databases, we have now identified GYF domains in 26 eukaryotic proteins. The alignment of representative domains from diverse organisms is shown in Figure 2A. The high conservation of residues in the bulge–helix–bulge motif GPF-X7-W-X3-GYF (Figure 1A) strongly suggests that these domains recognize proline-rich sequences in various contexts. From the sequence alignment and on the basis of the GYF–ligand structure presented in this study, we propose that the CD2BP2 domain defines a subclass of GYF domains. This subclass contains a tryptophan at position 8 and a relatively long loop connects the first and second β-strand of the domain. Our structure (Figure 1B–D) shows that the bulky side chain of W8 favors a glycine at position 8 of the SHRPPPPGHRV peptide to prevent steric hindrance. Furthermore, favorable hydrophobic interactions are present between the aliphatic groups of Arg10 in the ligand and the GYF W8 side chain. Most GYF domains not belonging to the CD2BP2 subclass contain an aspartate instead of the tryptophan at position 8 (Figure 2A), and the loop connecting β-strands 1 and 2 is shortened. The W8 to D8 substitution certainly changes the lipophilic potential of the binding site and suggests that a different spectrum of ligands can be bound by this type of GYF domain. The notion that the GYF domains containing the tryptophan in position 8 form a functionally relevant group is supported by the finding that the proteins containing the CD2BP2-type GYF domain share large regions of sequence homology N-terminal to the GYF domain. Furthermore, in this class, the GYF domain is always present at the C-terminus of all the respective proteins (Schultz et al., 1998). Figure 2B shows a phylogenetic tree for all putative GYF domains. The CD2BP2 subclass (colored red) is present in evolutionarily distant species ranging from man to fly to yeast. This broad sequence conservation implies that the CD2BP2 subclass has separated early in evolution. For classifications regarding the larger class of non-CD2BP2-type GYF domains, further functional data have to be obtained, but it is worth noting that a number of GYF domains that form a phylogenetic subgroup have only been found so far in plants (brown labels in Figure 2).
Fig. 2. Sequence comparison and subclass definition of GYF domains. (A) Multiple sequence alignment of GYF domains available from the public database. The color coding of the Chroma program was used as follows: single residues with a conservation of >80% are printed as colored characters (glycine residues in red, hydrophobic residues in yellow) on a dark gray background, and the occurrence of an aromatic or a hydrophobic amino acid in >80% of the 26 GYF domains aligned leads to a yellow character on a light gray background. The red and brown color of the protein accession numbers suggest the subclasses as derived from analysis of the multiple sequence alignment. (B) Phylogenetic tree of the GYF domain as calculated with the program ClustalW (version 1.7) using the multiple sequence alignment obtained from the pileup option of the gcg program package. The individual GYF domains are labeled with the DDBJ/EMBL/GenBank accession numbers according to (A) with the species in which they occur in parentheses.
Among all GYF domains shown in Figure 2, there is an absolute conservation of W28, consistent with the critical importance of the hydrogen bond formed between its side-chain NH group and a main-chain carbonyl group at the beginning of the polyproline helix of the ligand (Figure 1B). Y33 is exposed to the solvent and thereby directs the proline helix towards the hydrophobic center of the binding pocket. In a few cases, this Y33 is substituted by a phenylalanine or a histidine side chain. Despite these substitutions within the GYF sequence, the respective domains are very likely to assume the same bulge–helix– bulge motif as the CD2BP2 GYF domain, and we therefore suggest those proteins to be named GYF homology domains. Note how there is limited homology within the C-terminal 20–30 amino acids of various GYF domains, supporting the idea that this part of the protein is structurally but not functionally important.
Sequence repetition enhances the overall affinity for the GYF domain–CD2 interaction
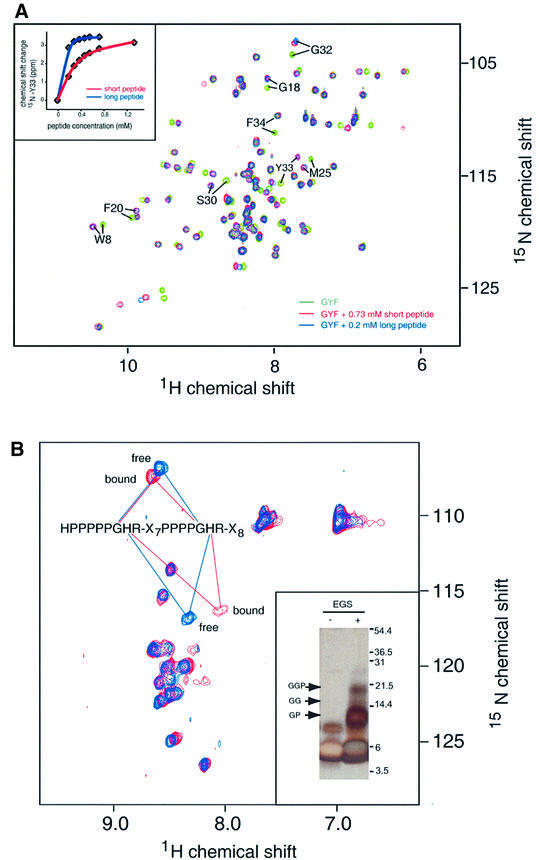
The interaction of the CD2BP2 GYF domain with segments of CD2 containing either one or two of the conserved PPPPGHR motifs was investigated by NMR shift mapping experiments. Figure 3A shows a superposition of the 15N–1H correlation spectra of the unligated GYF domain (green), the GYF domain bound by a short peptide of the amino acid sequence SHRPPPPGHRV (red) or the GYF domain bound by the longer peptide of the sequence HPPPPPGHRSQAP-SHRPPPPGHRVQ HQPQKR (blue). Similar chemical shift changes are observed for GYF domain NH resonances by adding either the short or the long peptide. Since the chemical shift is a very sensitive measure of the chemical environment, the precise overlap for almost all resonances of the red and blue spectra in Figure 3A demonstrates that the binding surface of the GYF domain is almost identical for the two peptides. Additional chemical shift changes for the long peptide, indicative of a second binding epitope, are not observed. However, the peptide concentration for the short peptide has to be significantly higher (0.73 mM) in order to measure chemical shift changes comparable with those observed for the long peptide at 0.2 mM (Figure 3A). The NMR titration curves of the short peptide containing a single PPPPGHR motif and the long peptide comprising two PPPPGHR motifs are shown for the NH resonance of Y33 of the GYF domain in the inset in Figure 3A. Assuming a simple two-state binding model for the shorter peptide SHRPPPPGHRV, an apparent KD value of 190 ± 22 µM was determined by NMR and confirmed by fluorescence titration experiments (KD = 203 ± 19 µM). However, the significantly lower overall apparent KD for the longer peptide (10–20 µM) suggests that a simple two-state binding model is not applicable. To test the possibility that the binding of the GYF domain to the long peptide involves both PPPPGHR motifs of CD2, we followed the shift of NH resonance of the 15N-labeled peptide (long sequence) upon addition of the unlabeled GYF domain (Figure 3B). The blue spectrum shows the backbone amide resonances of the free peptide in aqueous solution. The red spectrum is obtained after the addition of a 3-fold excess of unlabeled GYF domain. The observation that only a small subset of NH cross-peaks changes upon GYF binding confirms that the interaction is specific. In agreement with our structural data, the GH moieties of the peptide are most strongly affected upon binding (Figure 3B). Furthermore, the NH resonances of these GH moieties from both proline-rich repeats are degenerate and shift simultaneously upon addition of the unlabeled GYF domain. This indicates that both proline-rich motifs of this peptide contribute to the binding of the GYF domain, suggesting that two CD2BP2 GYF domains interact with the two PPPPGHR motifs of a single CD2 tail.
Fig. 3. Binding of proline-rich CD2 peptides to the GYF domain. (A) Overlap of the 15N–1H correlation spectra of the isolated GYF domain (green), the GYF domain upon addition of 0.73 mM of the short CD2 peptide segment (red) and the GYF domain after addition of 0.2 mM of the long CD2 peptide sequence (blue). The eight NH resonances with the largest chemical shift difference between the bound and unbound state are labeled according to residue type and number. The inset shows the change of the 15N chemical shift for Tyr33 of the GYF domain as a function of the ligand concentration of either the short (SHRPPPPGHRV) or the long peptide (HPPPPGHRSQAPSHRPPPPGHRVQHQPQK). The short peptide corresponds to the CD2 sequence 294–304 and the long peptide to the sequence 281–310. (B) 15N–1H correlation spectrum of the isotopically labeled long peptide before (blue) and after (red) addition of a 3-fold excess of unlabeled GYF domain. The resonances of the glycine and histidine residues of the two PPPPGHR motifs are overlapping and highlighted within the spectra. Inset: SDS–PAGE of EGS cross-linking reaction products. The GYF domain (∼26 µM) and long CD2 tail peptide (∼140 µM) were cross-linked in 50 µl of reaction buffer with EGS. The molecular weights of migration markers are shown on the right of the gel. The positions of various cross-linked complexes are indicated with arrows, where G indicates the GYF domain and P indicates the CD2 tail peptide. The identity of cross-linked bands was confirmed by MALDI-TOF mass spectrometry using the molecular weights of the monomers. The GGP band (∼20 kDa) shows the presence of a complex with two GYF domains and one CD2 tail peptide. Lane 1, GYF and CD2 tail peptide mixture without cross-linker; lane 2, GYF and CD2 tail peptide mixture with cross-linker.
To test this hypothesis, we have performed chemical cross-linking experiments using ethylene glycolbis(succinimidylsuccinate) (EGS), which links amino groups of lysine side chains and N-terminal residues. There are four reactive groups in GYF (the N-terminus and three lysines) and two in the long CD2 tail peptide (the N-terminus and one lysine). The inset in Figure 3B shows an SDS–polyacylamide gel of the GYF domain after treatment with EGS, and a control. Migration markers are on the right-hand side of the gel. The GYF monomer is at the bottom. The positions of the GYF–peptide dimer (GP), the GYF–GYF homodimer (GG) and the GYF–GYF–peptide trimer (GGP) are indicated. The appearance of the GGP trimer proves that two copies of GYF bind the tandem repeat of proline-rich sequences of the CD2 cytoplasmic tail. Note that the cross-linking reaction was not run to completion so that only a fraction of the trimers was cross-linked completely. Hence, the gel also shows monomers and dimers.
The SH3 domain of Fyn but not of Lck binds specifically to the GYF domain-binding site of CD2
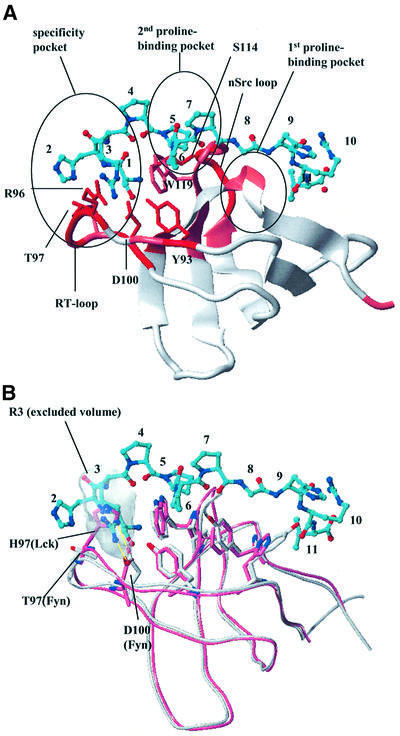
Functional studies have shown that the two PPPPGHR motifs of CD2 are responsible for IL-2 production and Ca2+ flux (Chang et al., 1990). The src-family kinase Fyn has been implicated in these events, since Fyn knock-out mice are severely impaired with regard to CD2-stimulated signaling (Fukai et al., 2000) and it has been suggested that the Fyn SH3 domain interacts directly with the CD2 cytoplasmic domain (Lin et al., 1998). To test the hypothesis that Fyn SH3 can also bind to the PPPPGHR motifs in CD2, we performed chemical shift mapping experiments where the 15N-labeled Fyn SH3 domain was mixed with increasing amounts of the peptide containing either one or two of the CD2 PPPPGHR motifs. Upon addition of the ligand, several NH resonances showed significant chemical shift changes. From the concentration dependence of the chemical shift changes, we estimate an apparent KD of ∼30 µM for the long peptide. The Fyn residues affected include W119 and Y132, which are part of the classical binding site for proline-rich peptides. Modeling the peptide conformation on the basis of the crystal structures of 1Fyn and 1Abo (Musacchio et al., 1994) and using the NMR chemical shift mapping data, we docked the CD2 ligand SHRPPPPGHRV to the Fyn SH3 domain (Figure 4A). Only the second of the two proline-binding pockets of the Fyn SH3 domain is making extensive contacts with the prolines of the CD2 peptide. The most significantly shifted resonances are from residues of the RT loop (R96, T97 and D100), the nSrc loop residue S114 and the conserved Y93. All these residues map to the specificity pocket of Fyn and support a critical role for RT loop residues in the Fyn SH3–CD2 interaction. The model presented here includes a salt bridge between Arg3 of the CD2 SHRPPPPGHRV peptide and D100 of the Fyn SH3 domain (Figure 4A). A similar salt bridge between the arginine of the ligand and Asp100 of Fyn has been observed in experimental structures (Yu et al., 1994).
Fig. 4. Fyn SH3 domain binding to the CD2 sequence SHRPPPPGHRV. (A) Model of the complex of the Fyn SH3 domain with the SHRPPPPGHRV ligand of CD2. The ribbon diagram of the SH3 domain is color coded according to the chemical shift perturbations observed in the NMR shift mapping experiments (red, combined chemical shift index >1.0; pink, combined chemical shift index 0.5–1.0). The side chains of residues making direct contacts to the ligand are displayed in red and marked by residue type and number. The ligand is shown as a bonded structure in cyan. Functionally important elements (RT and nsrc loops, specificity pocket and the two proline-binding pockets) of the SH3 domain fold are highlighted. (B) Structural superposition of the Lck (light red) and Fyn SH3 domain (white). The model of the Fyn SH3 domain in complex with the SHRPPPPGHRV peptide from CD2 was overlapped with the Lck SH3 structure (1Lck; Eck et al., 1994). The side chains of residues in the conserved proline-binding pockets are shown to align extremely well for this part of the interaction site. The side chains of RT loop residues that contribute to the observed binding specificity by interacting with (in the case of Fyn) or sterically hindering (in the case of Lck) the insertion of Arg3 of the peptide into the specificity pocket are also displayed. The shaded area defines the exclusion volume of Arg3. As in (A), the ligand is shown as a bonded structure with its carbons colored cyan.
The src kinase Lck is the predominant kinase that phosphorylates the T-cell receptor complex (Iwashima et al., 1994) and it has been hypothesized that it also plays a role in CD2 signaling by the direct interaction of its SH3 domain with the GHRPPPPSHR motif in CD2 in rat (Bell et al., 1996). To investigate the binding of the human Lck SH3 domain to the CD2 signaling motifs, we performed NMR titration experiments with the Lck SH3 domain under conditions identical to those employed for the Fyn SH3 domain. Addition of the Lck SH3 domain caused very small chemical shift changes in a number of residues, but the affinity for the Lck–peptide interaction was very weak and the KD too high to be determined quantitatively (>500 µM) for the short as well as for the long CD2 peptide segment used in this study (data not shown). Figure 4B offers an explanation for the observed difference in binding: the residues with the largest chemical shift changes in Fyn SH3 (R96 and T97) are substituted in the Lck SH3 domain (S96 and H97). In Lck, the side-chain nitrogen of H97 forms an intramolecular hydrogen bond with the carboxylate group of D100. In contrast, the D100 side chain of Fyn forms an intermolecular salt bridge with Arg3 of the peptide in our model structure (Figure 4B), thereby allowing Arg3 to insert deeply into the binding pocket. For the Lck SH3 domain to mediate such a favorable intermolecular interaction, a major rearrangement of the RT loop would be required. Albeit very similar in their overall structure, the specificity of the two domains for the CD2 sequence SHRPPPPGHRV is different. We conclude that Fyn is the primary src kinase directly interacting with the CD2 signaling motifs in Homo sapiens. This is supported by previous findings that established a CD2 pathway independent of the Lck tyrosine kinase but critically dependent on Fyn (Sunder-Plassmann and Reinherz, 1998; Fukai et al., 2000).
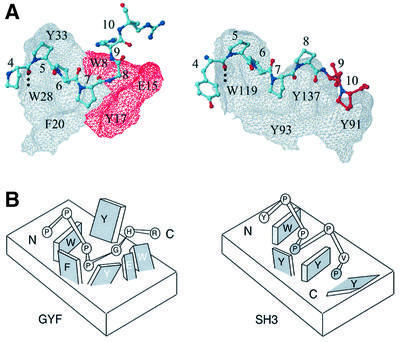
Structural requirements for proline-rich sequence recognition by the CD2BP2 GYF domain in comparison with SH3 domain–ligand interactions
Figure 5 shows a comparison of the CDBP2 GYF domain in complex with the PPPPGHR motif of CD2 and the Fyn SH3 domain in complex with the sequence YPPPPVP from the 3BP-2 sequence of the 1Fyn crystal structure (Musacchio et al., 1994). In both cases, the conserved aromatic residues of the fold family constitute a major interaction site for the prolines of the ligand, and the peptide residues 4–7 overlap almost perfectly. The side-chain NH group of the conserved W28 of the GYF and W119 of the SH3 domain form a hydrogen bond to the carbonyl group of residue 4, and Pro6 is aligned parallel to the aromatic ring of the conserved tryptophan in the two protein–peptide complexes. The high similarity of the ligand conformations ends at position 8 of the respective peptide. Gly8 in the GYF domain ligand terminates the polyproline helix, thereby preventing geometric hindrance between the second half of the peptide and the GYF domain. In contrast, the SH3 domain ligand maintains a near helical conformation throughout the entire seven-residue stretch. In such a conformation, the peptide residues 9 and 10 would clash with the GYF domain surface (shown as a forbidden surface in red in Figure 5A), but allow the SH3 domain ligand to contact the first proline-binding pocket of the SH3 domain. The comparison demonstrates that a ‘classical’ SH3 domain-binding mode cannot be assumed by GYF domain ligands. While SH3 domains provide two hydrophobic binding pockets, each accommodating one proline of the PxxP motif, the CD2BP2 GYF domain residues W8, E15 and Y17 do not allow extension of the proline helix. In sum, the SH3 domain presents a more extended hydrophobic interface with two shallow surface pockets, while the GYF domain contains a single central binding pocket that is deeper than the SH3 domain pockets. To test the importance of the peptide Gly8 for interaction with the GYF domain, a G8A mutant was analyzed; the GYF domain bound this SHRPPPPAHR ligand with a 3-fold lower affinity than the wild type (data not shown). The more stringent geometric requirements at this ligand position for the GYF domain interaction compared with the SH3 domain interaction can be readily explained by the structural comparison in Figure 5.
Fig. 5. Comparison of GYF and SH3 domain–ligand interactions. (A) The CD2BP2 GYF domain in complex with the PPPPGHR sequence from CD2 and the Fyn SH3 domain in complex with the peptide YPPPPVP are shown side by side. The surface of the GYF and the SH3 domain is shown as a grid with the residues Trp8, Glu15 and Tyr17 of the GYF domain displayed in red. Conserved residues important for binding are marked by residue type and sequence number. The peptides are shown as a bonded structure with residues 9 and 10 of the Fyn ligand colored red to highlight the extension of the proline helix as compared with the GYF domain ligand. (B) Schematic model showing the two proline-binding sites for the SH3 (right) and the single pocket in the GYF domain (left). Residues of the protein domains are displayed as rectangular structures, with labels for protein residues as either black (conserved domain residues) or white (non-conserved residues). Peptide residues are shown as circles labeled inside by residue type. The two prolines of the PxxP SH3 recognition motif are shaded gray.
Competition between CD2BP2 GYF and Fyn SH3 in vitro
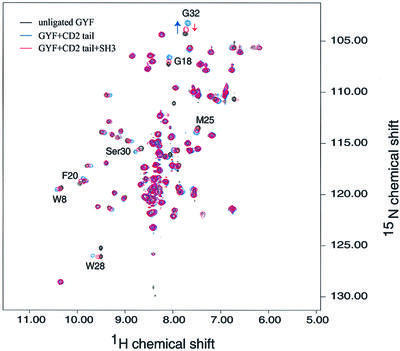
To test whether the CD2BP2 GYF and Fyn SH3 domain can compete for the same binding sequence in the context of the entire CD2 cytoplasmic domain, we performed the following experiment: 15N-labeled GYF domain (0.2 mM) was mixed with a substoichiometric amount of the entire unlabeled CD2 cytoplasmic domain (0.08 mM), and a 15N–1H correlation spectrum was taken (Figure 6). Resonances were shifted relative to free GYF domain in a similar way to our previous experiments, with the shorter segments containing one or two of the proline-rich repeats (see Figure 3). This confirms that the SHRPPPPGHRV sequence is the only element of the CD2 cytoplasmic domain competent to bind the GYF domain of CD2BP2. In a second step, unlabeled Fyn SH3 domain (0.4 mM) was added in 2-fold excess. The corresponding spectrum is colored red in Figure 6 and clearly shows that the NH resonances of binding site residues shift in the direction of unbound GYF domain. The fraction of CD2-bound GYF domain is significantly decreased in the presence of the Fyn SH3 domain (estimated to be 20% instead of 60%). This result shows that, under limiting CD2 concentrations, there is competition between the two domains for the same binding site within CD2.
Fig. 6. In vitro competition of CD2BP2 GYF and Fyn SH3 domains for CD2-binding sites. The spectrum of the isolated 15N-labeled GYF domain (0.2 mM) is shown in black. Substoichiometric amounts of the CD2 tail (residues 245–351 of human CD2) were added and the corresponding spectrum is shown in blue. Then 0.4 mM unlabeled Fyn SH3 domain was added, a third spectrum obtained and displayed in red. Residues that are shifted upon binding of the CD2 tail are marked by residue type and number. The arrow for the NH peak of Gly32 clarifies the movement of this resonance.
Subcellular compartmentalization of CD2BP2 and Fyn in vivo
Our previous results demonstrated that a fraction of human CD2 molecules was recruited to detergent-insoluble membrane lipid rafts upon anti-CD2 cross-linking or CD58 ligand binding (Yang and Reinherz, 2001). This process is physiologically relevant for CD2 signaling function for several reasons. First, many molecules important for T-cell activation, including LAT and src-family kinases, are enriched in lipid microdomains (Harder et al., 1998; Zhang et al., 1998). Secondly, non-mitogenic CD2 antibodies, in contrast to mitogenic CD2 antibodies, fail to recruit CD2 to lipid rafts. Thirdly, disruption of the raft structure impairs the CD2-mediated signaling process as assessed by early T-cell signaling events, such as phosphorylation of cellular substrates or elevation of intracellular free calcium (Yang and Reinherz, 2001). Given the fact that the Fyn SH3 domain can compete in vitro with the binding of the CD2BP2 GYF domain of CD2BP2 to the same proline-rich sequence in the CD2 tail, we examined the subcellular localization of CD2, CD2BP2 and Fyn in T cells. Flag-tagged CD2BP2 stable transfectants of Jurkat T cells were used for lipid raft separation by sucrose gradient centrifugation. The lipid raft compartment is usually localized to fractions 3 and 4 while the detergent-soluble, non-nuclear compartment resides in fractions 9–12 of the sucrose gradient. Figure 7A shows that Fyn is detected exclusively in the lipid raft fraction 4 while CD2BP2 distributes into the non-raft fractions 9–12 in unstimulated cells. Figure 7B shows the result after CD2 cross-linking by a pair of stimulatory anti-T112 and anti-T113 monoclonal antibodies (mAbs). The predominance of LAT in the raft fraction and actin in the detergent-soluble fraction is clearly not affected by cross-linking of CD2 molecules (Figure 7B; Yang and Reinherz, 2001). In contrast, a fraction of CD2 is recruited into lipid rafts after cross-linking, whereas little is found there constituitively (Figure 7B), consistent with previous data (Yang and Reinherz, 2001). Importantly, CD2BP2 is located exclusively in the detergent-soluble cellular fraction, whereas Fyn resides exclusively in the raft fraction, as revealed by western blotting (Figure 7). In addition, neither CD2BP2 nor Fyn protein localization is affected by CD2 cross-linking. Therefore, CD2BP2 and Fyn are localized in distinct cellular compartments in vivo, whereas CD2 shuttles inducibly from non-raft to the raft membrane compartment. Furthermore, anti-Fyn monoclonal antibodies can specifically precipitate CD2 in the raft fractions of CD2-stimulated cells, whereas there is little, if any, CD2 co-precipitated from the raft fraction of non-stimulated T cells (Figure 7C). On the other hand, CD2BP2 remains in the detergent-soluble fraction where it was shown to associate with CD2 in T cells (Nishizawa et al., 1998).
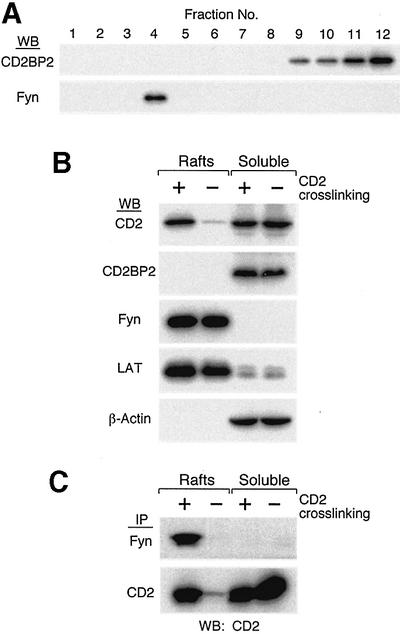
Fig. 7. Distinct cellular compartmentation of CD2BP2 and Fyn proteins in vivo. Flag-tagged CD2BP2 Jurkat T-cell transfectants (Nishizawa et al., 1998) were used for lipid raft preparation according to a previously published protocol (Yang and Reinherz, 2001). (A) The distribution of CD2BP2 as well as Fyn proteins in sucrose density gradients. Each of the 12 fractions are shown from least to most dense (1–12, respectively). CD2BP2 western blotting was performed using the anti-Flag M2 antibody. (B) The distribution of molecules in the lipid raft and detergent-soluble fraction in the presence (+) or absence (–) of prior CD2 cross-linking. Sucrose gradient fraction 10 was used as a representative detergent-soluble fraction, and fraction 4 as the raft fraction. Identical results were obtained with each of two independent Jurkat CD2BP2 stable transfectants. The distribution of CD2, LAT and actin in the sucrose gradient system is consistent with results described previously (Yang and Reinherz, 2001). (C) An equal volume of raft or detergent-soluble fractions from CD2-stimulated (+) or non-stimulated (–) samples was diluted in octyl-glucoside-containing binding buffer, immunoprecipitated (IP) with either anti-Fyn (top row) or anti-T111 as designated (bottom row) and immunoblotted (WB) with anti-CD2 hetero-antisera (both rows).
Functional relevance of CD2-proline-rich sequence recognition and ligand exchange
The experiments with intact cells demonstrate that competitive CD2 binding of CD2BP2 and Fyn is unlikely to happen in a physiological context. In T cells, CD2BP2 localizes to the cytosolic compartment independently of CD2 stimulation, while Fyn is present permanently in the lipid raft fraction through myristylation and palmitylation at its N-terminus (Alland et al., 1994; Shenoy-Scaria et al., 1994; van’t Hof and Resh, 1997; Wolven et al., 1997). In the resting state, CD2BP2 is probably the primary ligand interacting with the proline-rich repeat sequence of CD2. Consistent with this notion, CD2BP2 could immunoprecipitate CD2 in unstimulated Jurkat cells (Nishizawa et al., 1998). As such, CD2BP2 might help to prevent the translocation of CD2 into lipid rafts before CD58 ligation of the CD2 ectodomains. Augmentation of IL-2 production (150–200%) in Jurkat T cells transfected with a CD2BP2 GYF domain-only construct (Nishizawa et al., 1998) is in agreement with this hypothesis: the cellular expression of the isolated GYF domain is likely to release CD2 from CD2BP2 and CD2BP2-associated proteins, thereby fostering raft translocation.
After CD2 ectodomain ligation with either a mitogenic pair of anti-CD2 antibodies or the cognate ligand CD58 (Yang and Reinherz, 2001), a prominent fraction of CD2 is recruited inducibly to the lipid rafts, where the encounter of the Fyn tyrosine kinase takes place. Comparison of human CD2 transgenic mice on Fyn+/+ and Fyn–/– genetic backgrounds shows that the CD2 pathway components regulated by Fyn include the key signaling molecules PLCγ1, Vav, protein kinase C θ (PKCθ), Dok, focal adhesion kinase and Pyk-2. Deficiency in Fyn-dependent PLCγ1 catalytic activity probably contributes to reduced PKCα-dependent ERK activation and reduced Ca2+ mobilization in Fyn–/– mice. Decreased inducible PKCθ catalytic activity and Vav phosphorylation are probably responsible for diminished p38 and JNK activation in Fyn knock-out mice and therefore provide a probable link between Fyn activity and IL-2 production. The fact that Fak and Pyk-2 are target substrates suggests that Fyn may also be an important regulator of T-cell adhesion. Taken together, the herein defined interaction of the Fyn SH3 domain with CD2 in the lipid rafts of T cells is the initial step of downstream signaling events induced by CD2-mediated T-cell activation.
Materials and methods
Sample preparation
Recombinant proteins were expressed as described previously (Freund et al., 1999). Details are given in the Supplementary data, available at The EMBO Journal Online.
Binding experiments
Synthetic peptides of the sequence SHRPPPPGHRV or HPPPPPGHRSQAPSHRPPPPGHRVQHQPQK were used in the titration experiments and evaluation of the data was performed as described previously (Freund et al., 1999), and are described further in the Supplementary data.
Subcellular fractionation and western blotting
The anti-human CD2 hetero-antisera M32B specific for the human CD2 ectodomain was prepared in our laboratory (Yang and Reinherz, 2001). Anti-Flag mAb (M2) and anti-β-actin (AC-15) were obtained from Sigma. Anti-Fyn mAb was purchase from Santa Cruz Biotechnology. Polyclonal anti-LAT was from Upstate Biotechnology Inc. All western blotting was performed according to a method detailed previously by Yang and Reinherz (2001). CD2 samples were treated with PNGase F (New England Biolabs) followed the manufacturer’s protocol prior to western blotting. A total of 1.2–1.5 × 108 Jurkat cells transfected with Flag-tagged CD2BP2 (Nishizawa et al., 1998) were either unstimulated (–) or stimulated by anti-CD2 mAb cross-linking (+) (1:100 dilution of anti T112 + T113 ascites) for 10 min. Lipid rafts were prepared from cell lysates by sucrose gradient according to a previous protocol (Yang and Reinherz, 2001). To reduce the sucrose concentration prior to SDS–PAGE analysis, fractions 7–12 were diluted 1:1 (v/v) with distilled water. For immunoprecipitation, raft and soluble fractions were diluted 10 times in binding buffer containing 60 mM N-octyl-glucoside and 1× Tris-buffered saline (TBS) supplemented with protease inhibitors, and incubated with anti-Fyn or anti-T111 mAbs conjugated to Sepharose beads.
SDS–PAGE of cross-linking reaction products
CD2 tail peptide (∼140 µM) and GYF (∼26 µM) were cross-linked in 50 µl of reaction buffer (5 mM sodium phosphate, 150 mM NaCl, pH 7.4). A 1 µl aliquot of EGS dissolved in dimethylformamide was added to the mixture at a final concentration of 2 mM, the reaction was run for 2 h at room temperature and the sample was analyzed by denaturating SDS–PAGE with silver staining. The identity of cross-linked bands was confirmed by MALDI-TOF mass spectrometry using the molecular weights of the monomers.
NMR spectroscopy
All experiments were conducted at 298 K using either UnityInova 500 and UnityInova 750 MHz machines, or Bruker AMX500 and DRX750 instruments. NMR data processing and analysis were carried out as described previously (Nishizawa et al., 1998). Assignment of protein proton resonances was based on the assignments of the unligated GYF domain (Freund et al., 1999) and could be obtained by the use of two-dimensional NOESY/TOCSY spectra in D2O and 15N-edited NOESY- and TOCSY-HSQC spectra in 90% H2O/10% D2O. Peptide proton assignments were extracted from D2O NOESY/TOCSY (150 ms mixing time) spectra of a sample where equal amounts of deuterated 15N-labeled protein and unlabeled SHRPPPPGHRV peptide were mixed at 0.8 mM concentration. Intermolecular nuclear Overhauser effects (NOEs) were obtained either from a 15N-NOESY-HSQC spectrum of the latter sample in 90% H2O/10% D2O or from a D2O sample of a stoichiometric complex of protein–peptide with only the aromatic residues F, W and Y of the protein being protonated. At the concentrations used, 0.8 mM equimolar concentrations and a Kd of ∼0.19 mM, ∼80% of a component is bound. In addition, we obtained intramolecular peptide NOEs from a single chain construct of the topology SHRPPPPGHRV–linker–GYF. Resonance positions in the structured part of the linked construct are essentially identical to those of the bi-molecular complex. Assignments were confirmed by comparing NOESY experiments of the complex and the single chain construct. The GYF domain studied here comprises the sequence 256–341 of CD2BP2 and we have shown previously that the first 25 amino acids are unstructured and flexible in solution. Intramolecular peptide and intermolecular NOE constraints were derived from NOESY spectra with 150 ms mixing times.
Structure calculation
The three-dimensional structures were calculated by simulated annealing from random coordinates at 1000 K with the program XPLOR 3.1 (Brünger, 1993). Force constants for NOE and dihedral angle restraints were 50 kcal/mol/Å2 and 200 kcal/mol/rad2, respectively. No attractive Lennard–Jones or electrostatic terms were used. One thousand steps of gradient minimization were performed with gradually increased weight factors for the individual energy terms. The 15 final lowest energy structures displayed no distance violations greater than 0.3 Å or dihedral restraint violations greater than 5°. The coordinates of the final 15 structures have been deposited at the Protein Data Bank (code 1L27).
Modeling of the Fyn SH3 domain–SHRPPPPGHRV complex
Modeling of the Fyn SH3–ligand complex was based on the crystal structure of 1Fyn and the NMR chemical shift mapping experiments of 15N-labeled Fyn SH3 domain in the presence of the peptide SHRPPPPGHRV. Further details are given in the Supplementary data.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Katharina Thiemke for excellent technical assistance. This work was supported by grants from the NIH (AI37581 to G.W. and AI19807 to E.L.R.). Purchase and operation of the NMR spectrometers used for this research were supported by the NIH (grants GM47467 and RR00995). C.F. was supported by a grant from the Bundesministerium für Bildung und Forschung (0311879).
References
- Alland L., Peseckis,S.M., Atherton,R.E., Berthiaume,L. and Resh,M.D. (1994) Dual myristylation and palmitylation of Src family member p59fyn affects subcellular localization. J. Biol. Chem., 269, 16701–16705. [PubMed] [Google Scholar]
- Bedford M.T., Frankel,A., Yaffe,M.B., Clarke,S., Leder,P. and Richard,S. (2000) Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J. Biol. Chem., 275, 16030–16036. [DOI] [PubMed] [Google Scholar]
- Bell G.M., Fargnoli,J., Bolen,J.B., Kish,L. and Imboden,J.B. (1996) The SH3 domain of p56lck binds to proline-rich sequences in the cytoplasmic domain of CD2. J. Exp. Med., 183, 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P. and Sudol,M. (1994) The WW domain: a signalling site in dystrophin? Trends Biochem. Sci., 19, 531–533. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. (1993) X-PLOR, Version 3.1: A System for X-ray Crystallography and NMR. Yale University Press, New Haven, CT.
- Carlsson L., Nystrom,L.E., Sundkvist,I., Markey,F. and Lindberg,U. (1977) Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J. Mol. Biol., 115, 465–483. [DOI] [PubMed] [Google Scholar]
- Case D.A. et al. (1997) AMBER5. University of California, San Francisco, CA.
- Chang H.C., Moingeon,P., Pedersen,R., Lucich,J., Stebbins,C. and Reinherz,E.L. (1990) Involvement of the PPPGHR motif in T cell activation via CD2. J. Exp. Med., 172, 351–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustin M.L. et al. (1998) A novel adaptor protein orchestrates receptor patterning and cytoskeletal polarity in T-cell contacts. Cell, 94, 667–677. [DOI] [PubMed] [Google Scholar]
- Eck M.J., Atwell,S.K., Shoelson,S.E. and Harrison,S.C. (1994) Structure of the regulatory domains of the Src-family tyrosine kinase Lck. Nature, 368, 764–769. [DOI] [PubMed] [Google Scholar]
- Freund C., Dotsch,V., Nishizawa,K., Reinherz,E.L. and Wagner,G. (1999) The GYF domain is a novel structural fold that is involved in lymphoid signaling through proline-rich sequences. Nat. Struct. Biol., 6, 656–660. [DOI] [PubMed] [Google Scholar]
- Fukai I., Hussey,R.E., Sunder-Plassmann,R. and Reinherz,E.L. (2000) A critical role for p59(fyn) in CD2-based signal transduction. Eur. J. Immunol., 30, 3507–3515. [DOI] [PubMed] [Google Scholar]
- Gassmann M., Amrein,K.E., Flint,N.A., Schraven,B. and Burn,P. (1994) Identification of a signaling complex involving CD2, ζ chain and p59fyn in T lymphocytes. Eur. J. Immunol., 24, 139–144. [DOI] [PubMed] [Google Scholar]
- Grakoui A., Bromley,S.K., Sumen,C., Davis,M.M., Shaw,A.S., Allen,P.M. and Dustin,M.L. (1999) The immunological synapse: a molecular machine controlling T cell activation. Science, 285, 221–227. [DOI] [PubMed] [Google Scholar]
- Harder T., Scheiffele,P., Verkade,P. and Simons,K. (1998) Lipid domain structure of the plasma membrane revealed by patching of membrane components. J. Cell Biol., 141, 929–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiden W., Moeckel,G. and Brickmann,J. (1993) A new approach to the display of lipophilicity/hydrophilicity mapped on molecular surfaces. J. Comput. Aided Mol. Design, 7, 503–514. [DOI] [PubMed] [Google Scholar]
- Iwashima M., Irving,B.A., van Oers,N.S., Chan,A.C. and Weiss,A. (1994) Sequential interactions of the TCR with two distinct cytoplasmic tyrosine kinases. Science, 263, 1136–1139. [DOI] [PubMed] [Google Scholar]
- Kay B.K., Williamson,M.P. and Sudol,M. (2000) The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J., 14, 231–241. [PubMed] [Google Scholar]
- Koegl M., Zlatkine,P., Ley,S.C., Courtneidge,S.A. and Magee,A.I. (1994) Palmitoylation of multiple Src-family kinases at a homologous N-terminal motif. Biochem. J., 303, 749–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Nishizawa,K., An,W., Hussey,R.E., Lialios,F.E., Salgia,R., Sunder-Plassmann,R. and Reinherz,E.L. (1998) A cdc15-like adaptor protein (CD2BP1) interacts with the CD2 cytoplasmic domain and regulates CD2-triggered adhesion. EMBO J., 17, 7320–7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Hutchcroft,J.E., Andoniou,C.E., Kamoun,M., Band,H. and Bierer,B.E. (1998) Association of p59(fyn) with the T lymphocyte costimulatory receptor CD2. Binding of the Fyn Src homology (SH) 3 domain is regulated by the Fyn SH2 domain. J. Biol. Chem., 273, 19914–19921. [DOI] [PubMed] [Google Scholar]
- Lu P.J., Zhou,X.Z., Shen,M. and Lu,K.P. (1999) Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science, 283, 1325–1328. [DOI] [PubMed] [Google Scholar]
- Mayer B.J., Hamaguchi,M. and Hanafusa,H. (1988) A novel viral oncogene with structural similarity to phospholipase C. Nature, 332, 272–275. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Saraste,M. and Wilmanns,M. (1994) High-resolution crystal structures of tyrosine kinase SH3 domains complexed with proline-rich peptides. Nat. Struct. Biol., 1, 546–551. [DOI] [PubMed] [Google Scholar]
- Niebuhr K. et al. (1997) A novel proline-rich motif present in ActA of Listeria monocytogenes and cytoskeletal proteins is the ligand for the EVH1 domain, a protein module present in the Ena/VASP family. EMBO J., 16, 5433–5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa K., Freund,C., Li,J., Wagner,G. and Reinherz,E.L. (1998) Identification of a proline-binding motif regulating CD2-triggered T lymphocyte activation. Proc. Natl Acad. Sci. USA, 95, 14897–14902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G.M. et al. (2000) Comparative genomics of the eukaryotes. Science, 287, 2204–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J., Milpetz,F., Bork,P. and Ponting,C.P. (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl Acad. Sci. USA, 95, 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy-Scaria A.M., Dietzen,D.J., Kwong,J., Link,D.C. and Lublin,D.M. (1994) Cysteine3 of Src family protein tyrosine kinase determines palmitoylation and localization in caveolae. J. Cell Biol., 126, 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K. and Toomre,D. (2000) Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol., 1, 31–39. [DOI] [PubMed] [Google Scholar]
- Stahl M.L., Ferenz,C.R., Kelleher,K.L., Kriz,R.W. and Knopf,J.L. (1988) Sequence similarity of phospholipase C with the non-catalytic region of src. Nature, 332, 269–272. [DOI] [PubMed] [Google Scholar]
- Sunder-Plassmann R. and Reinherz,E.L. (1998) A p56lck-independent pathway of CD2 signaling involves Jun kinase. J. Biol. Chem., 273, 24249–24257. [DOI] [PubMed] [Google Scholar]
- van’t Hof W. and Resh,M.D. (1997) Rapid plasma membrane anchoring of newly synthesized p59fyn: selective requirement for NH2-terminal myristoylation and palmitoylation at cysteine-3. J. Cell Biol., 136, 1023–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.H., Smolyar,A., Tan,K., Liu,J.H., Kim,M., Sun,Z.Y., Wagner,G. and Reinherz,E.L. (1999) Structure of a heterophilic adhesion complex between the human CD2 and CD58 (LFA-3) counterreceptors. Cell, 97, 791–803. [DOI] [PubMed] [Google Scholar]
- Wolven A., Okamura,H., Rosenblatt,Y. and Resh,M.D. (1997) Palmitoylation of p59fyn is reversible and sufficient for plasma membrane association. Mol. Biol. Cell, 8, 1159–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier R., Brennan,T., Li,Q., McCormack,C. and Seed,B. (1998) Membrane compartmentation is required for efficient T cell activation. Immunity, 8, 723–732. [DOI] [PubMed] [Google Scholar]
- Yang H. and Reinherz,E.L. (2001) Dynamic recruitment of human CD2 into lipid rafts. Linkage to T cell signal transduction. J. Biol. Chem., 276, 18775–18785. [DOI] [PubMed] [Google Scholar]
- Yu H., Chen,J.K., Feng,S., Dalgarno,D.C., Brauer,A.W. and Schreiber,S.L. (1994) Structural basis for the binding of proline-rich peptides to SH3 domains. Cell, 76, 933–945. [DOI] [PubMed] [Google Scholar]
- Zhang W., Trible,R.P. and Samelson,L.E. (1998) LAT palmitoylation: its essential role in membrane microdomain targeting and tyrosine phosphorylation during T cell activation. Immunity, 9, 239–246. [DOI] [PubMed] [Google Scholar]