Abstract
Here we take advantage of the well-characterized and simple nervous system of Caenorhabditis elegans to further our understanding of the functions of RNA editing. We describe the two C.elegans ADAR genes, adr-1 and adr-2, and characterize strains containing homozygous deletions in each, or both, of these genes. We find that adr-1 is expressed in most, if not all, cells of the C.elegans nervous system and also in the developing vulva. Using chemotaxis assays, we show that both ADARs are important for normal behavior. Biochemical, molecular and phenotypic analyses indicate that ADR-1 and ADR-2 have distinct roles in C.elegans, but sometimes act together.
Keywords: chemotaxis/double-stranded RNA/inosine/neuronal/vulva
Introduction
The unexpectedly small number of genes in metazoa emphasizes the importance of post-transcriptional pathways in creating diversity and complexity (Baltimore, 2001). For pathways where the need for complexity is very high, such as those involved in neurotransmission, an astounding number of alternative RNA splicing events are possible (Black, 2000). Similarly, another form of RNA processing, RNA editing by adenosine deamination, appears to be particularly abundant in mammalian brain, where one out of every 17 000 nucleotides in mRNA appear as inosine, the base created by this form of RNA editing (Paul and Bass, 1998).
The enzymes that catalyze adenosine to inosine conversion in RNA are known as adenosine deaminases that act on RNA, or ADARs (reviewed in Bass, 2002). Inosine is read as a guanosine by the translational machinery, and when ADARs target codons, the amino acid specified by the codon often is changed. Thus, ADARs create diversity by allowing multiple protein isoforms to be synthesized from RNA encoded by a single gene. Although the amount of inosine found in mammalian brain indicates that there are many more edited RNAs to be discovered (Paul and Bass, 1998), several interesting examples have already been characterized. At the Q/R site of mammalian glutamate receptor B (gluR-B) mRNAs, ADARs convert a glutamine codon (CAG) to an arginine codon (CIG) to alter the flow of calcium into glutamate-gated ion channels (reviewed in Seeburg et al., 1998). RNA editing at multiple sites within the serotonin 2C receptor mRNA allows the production of at least seven different protein isoforms from a single gene (Burns et al., 1997). Here, editing is thought to modulate the affinity with which the receptor binds to a G protein (Niswender et al., 1999). Although it is not yet clear just how large a role ADARs play in the regulation of gene expression, since there are 18 possible non-synonymous codon changes that can be created by adenosine deamination, the possibilities seem almost endless. ADARs can also create splice sites (Rueter et al., 1999) and deaminate adenosines in 5′- and 3′-untranslated regions (UTRs; Morse and Bass, 1999; Morse et al., 2002). The function of inosines in 5′- and 3′-UTRs is not yet known, but they are proposed to alter the stability, localization or translatability of the mRNA.
Studies of mice and flies that lack or have reduced levels of ADAR activity emphasize the importance of RNA editing for neuronal function. Mice containing a homozygous deletion for one of their two ADARs (ADAR2) are prone to seizures and die shortly after birth (Higuchi et al., 2000). A homozygous deletion of the single ADAR found in Drosophila melanogaster causes behavioral abnormalities and brain lesions in old age (Palladino et al., 2000).
The seizures observed in mice lacking ADAR2 result from an absence of editing at the Q/R site of gluR-B. However, in other cases, phenotypes have not been correlated directly with specific RNA editing events. It is intriguing to consider that even subtle behaviors might correlate with specific RNA editing events. Although making this correlation is difficult in an organism as complex as a mammal, we believe it will be simpler in Caenorhabditis elegans, which has a well-characterized and simple nervous system. As a first step in our C.elegans studies, we describe the two ADAR genes found in worms, adr-1 and adr-2, and characterize strains containing homozygous deletions in each, or both, of these genes. Our analyses indicate that both genes have distinct roles in RNA editing in C.elegans, and are essential for normal function of the nervous system.
Results
Caenorhabditis elegans has two genes with similarity to ADARs
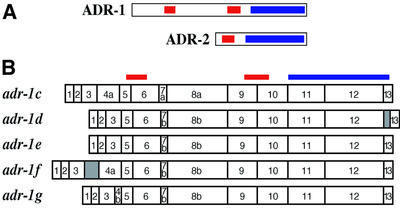
When the first mammalian ADAR was cloned, database searches revealed a C.elegans gene (T20H4.4) with high sequence similarity to the ADARs (Kim et al., 1994). cDNA analyses showed that this gene, called adr-2 (Figure 1A), encodes a protein with a single double-stranded RNA-binding motif (dsRBM) followed by the highly conserved C-terminal catalytic domain found in all ADARs (Hough et al., 1999). adr-2 is the second gene in an operon of six genes. adr-2 mRNAs are heterogeneous at their 5′ termini since multiple spliced leader variants are used during processing from the polycistronic transcript.
Fig. 1. Caenorhabditis elegans has two ADARs. (A) ADR-1 (adr-1c) and ADR-2 ORFs are shown, with boxes indicating dsRBMs (red) and the catalytic domain (blue). (B) Five splice forms of adr-1 are shown, with the relative positions of dsRBMs and catalytic domain indicated at the top. Notable features are as follows, with nucleotide positions given relative to the start of cosmid H15N15 (accession No. Z96100). The adr-1c cDNA (accession No. AY150815) includes exons 7a and 8a which are created by alternative 3′ splice sites (4877 and 4989 nucleotides). Compared with the b forms of these exons, 7a and 8a have nine and six additional nucleotides, respectively, and code for the addition of three amino acids (LLQ) or two amino acids (LQ), respectively. Exon 4 is absent from adr-1d and adr-1e cDNAs (accession Nos AY150816 and AY150817, respectively), resulting in a deletion of 66 amino acids that does not disrupt the ORF. adr-1d has an unspliced intron between exons 12 and 13 that inserts 19 amino acids into the catalytic domain near the C-terminus. adr-1f (accession No. AY150818) has an unspliced intron between exons 3 and 4, resulting in a premature stop in the intron, 301 nucleotides from the AUG codon. In adr-1g, an alternative 5′ splice site for intron 4 (4250 nucleotides) results in a 58 nucleotide exon 4b, truncated by 140 nucleotides compared with exon 4a; this predicts a frameshift and a premature stop codon in exon 5, 346 nucleotides from the 5′ end of adr-1g (accession No. AY150819). In theory, short forms of ADR-1 could be synthesized from adr-1f and adr-1g by using downstream methionines.
When the C.elegans genome was near completion, a second gene with similarity to ADARs, H15N14.1a/b, was revealed. This gene, called adr-1 (Figure 1A), has two dsRBMs. Northern analyses with adr-1 probes showed two bands of 3.1 and 2.9 kb (data not shown), suggesting the presence of alternative splice forms for adr-1. Subsequently, we identified eight independent cDNA clones, representing five splice variants (Figure 1B). A 5′ RACE and cDNA sequencing analysis showed that adr-1 was trans-spliced with SL1 (R.Hough, unpublished data).
Caenorhabditis elegans adr-1 is highly expressed in the nervous system and developing vulva
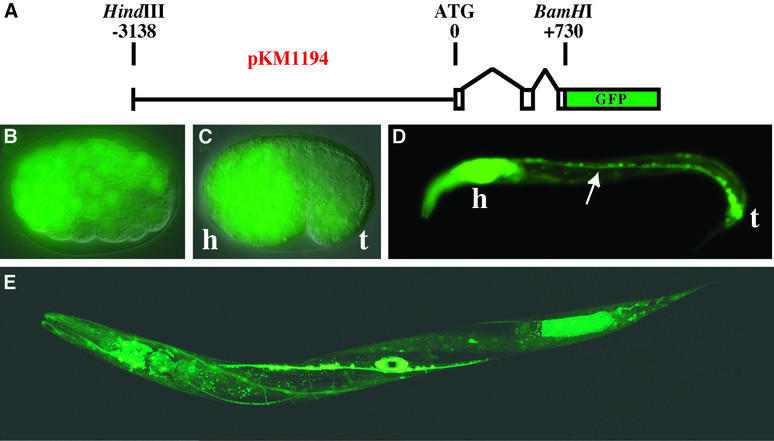
To gain information about where ADARs are expressed in C.elegans, the green fluorescent protein (GFP) open reading frame (ORF) was fused to an ∼4 kb fragment that contained putative upstream regulatory and promoter sequences, as well as the first two exons and a portion of exon three of adr-1 (Figure 2A). The adr-1::GFP fusion construct, designated pKM1194, was co-injected into the gonad of wild-type worms (N2) with pPRF4, a dominant marker (rol-6) that causes worms to roll. F1 rollers were screened for GFP expression, and two independent lines, KM163 and KM164, were established. A third independent line without the roller marker, KM165, was also isolated. The expression patterns of all strains were identical.
Fig. 2. Embryonic and larval expression of adr-1 in C.elegans. (A) The diagram shows the pKM1194 reporter construct used to generate adr-1::GFP expression lines (KM163–5). Nomarski/GFP merged images show that adr-1::GFP is expressed by late gastrulation (B, ≥100 cell stage), and by the comma stage (C) is predominantly neuronal (h, presumptive head; t, tail). Neuronal expression continues into larval stages as shown for an L1 worm (D), where expression was observed in the ventral nerve cord (arrow), along with intense expression in the head ganglia and nerve ring (h) and the tail neurons (t). A confocal image of a late L4 worm (E) shows expression in most cells of the nervous system, as well as in the developing vulva (see also Figure 3). Other tissues with weak expression include the pharynx and body wall muscle. We were unable to determine whether the expression in posterior intestinal cells is true adr-1 expression or an artifact of transgenic expression (Mello and Fire, 1995).
We observed that adr-1::GFP expression began early in embryogenesis, as evidenced by fluorescence of the late gastrula (Figure 2B) and comma stage (Figure 2C) embryos. By the comma stage, expression was highest in developing neuronal tissue in the head (h), tail (t) and ventral side. Neuronal expression continued through embryogenesis, and by the L1 larval stage was the prominent pattern (Figure 2D). The adr-1::GFP reporter continued to be expressed in most, if not all, cells of the nervous system, at all larval stages (e.g. L4, Figure 2E) and in adult animals. Interestingly, an oval of fluorescence in the middle of the L4 worm suggested that adr-1 was also expressed in the developing vulva (Figure 2E). A more detailed analysis of this expression is shown in Figure 3. Although adult worms continued to express adr-1 in the nervous system, vulva expression was observed only during morphogenesis of this organ, and not in adult animals.
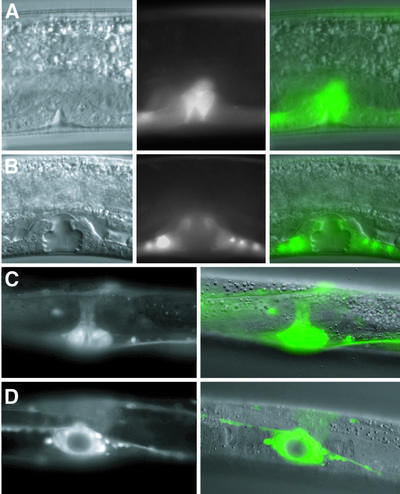
Fig. 3. adr-1 expression during vulva morphogenesis. Nomarski, GFP and merged images (left to right; KM165) show that adr-1::GFP is expressed in proliferating vulval precursor cells that are beginning to invaginate during late L3 (A), and continues to be localized to the developing vulva at the L4 Christmas tree stage (B). GFP and Nomarski/GFP merged images of a late L4 vulva (KM164) are shown with lateral (C) and ventral (D) views. All 22 cells derived from vulval precursor cells continue to express GFP into late L4. After the L4/adult molt, adr-1::GFP expression is no longer seen in the vulva.
We were unable to obtain information about the expression of adr-2 using GFP constructs, probably because of technical difficulties associated with its presence in an operon. Some information is available online from Yuji Kohara’s NEXTDB, a project to produce in situ hybridization expression patterns sytematically for every C.elegans gene. While these studies do not provide a comprehensive analysis of adr-2 expression, they do show that adr-2 mRNA is expressed ubiquitously in early embryos, as well as in the germline of early adults (Y.Kohara, personal communication).
Isolation of animals containing homozygous deletions in adr-1 and adr-2
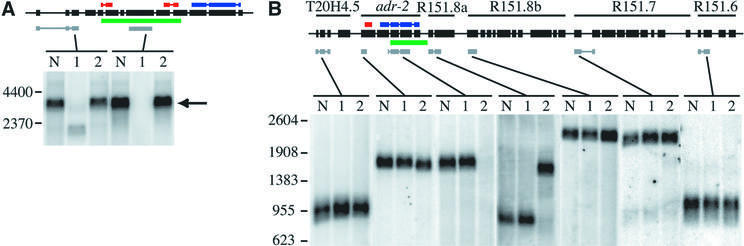
To gain insight into the role of ADARs in C.elegans, we isolated strains containing homozygous deletions in each of the ADAR genes. Mutant strains were isolated from a library of ethyl methanesulfonate (EMS)-mutagenized worms using a PCR screen (Dernburg et al., 1998), and were each back-crossed eight times to an unmutagenized genetic background to eliminate extraneous mutations (Anderson, 1995). As shown in Figure 4A, we isolated a C.elegans strain, adr-1(gv6), with a 1560 bp deletion beginning in exon 5 and ending in exon 10. This deletion is predicted to be a null mutation, since it removed both dsRBMs and created an in-frame stop codon 56 nucleotides (19 amino acids) downstream of the deletion point. As predicted, northern analyses of mRNA derived from the adr-1(gv6) animals showed a shorter, ∼5-fold less abundant mRNA, whose length correlated with the size of the predicted deletion.
Fig. 4. Northern analyses of adr deletion strains. Black boxes with a line through the center represent the genomic structure of the adr-1 gene (A) or the six-gene adr-2 operon (B). Colored bars show the location of dsRBMs (red), catalytic domain (blue), sequences deleted in the mutants (green) and the regions of cDNA sequences used to probe northerns (gray). Gels were loaded with poly(A)+ RNA from three C.elegans strains: wild-type (lane N), adr-1(gv6) (lane 1) and adr-2(gv42) (lane 2). (A) Hybridization with a probe to the 5′ region of adr-1 shows a band corresponding to the full-length transcript (arrow) in wild-type and adr-2(gv42) samples and a shorter transcript in the adr-1(gv6) deletion strain; the strong hybridization signal of this experiment did not allow resolution of alternative splice forms. A probe to sequences within the deleted region shows equivalent expression in wild-type and adr-2(gv42) lanes and no signal in the adr-1(gv6) lane. (B) Hybridization with probes to various regions of the six-gene operon shows that transcripts from T20H4.5, R151.8b, R151.7 and R151.6 are unaffected by the deletion in adr-2(gv42); all genes are named as previously specified (Hough et al., 1999). A probe to the 5′ region of adr-2 hybridizes to a slightly shorter transcript in the adr-2(gv42) deletion strain, while a probe within the deleted region shows normal hybridization to wild-type and adr-1(gv6) RNA but no signal in the adr-2(gv42) RNA. A probe to R151.8a, the gene just downstream of adr-2, confirms that adr-2(gv42) contains a chimeric transcript that fuses adr-2 to R151.8a. The chimeric RNA is predicted to result in a chimeric protein with the first two exons of adr-2 in-frame with the protein encoded by R151.8a. An RT–PCR product spanning the junction of the adr-2::R151.8a fusion was cloned and sequenced, and confirmed results of the northern analyses.
The strain homozygous for a deletion in the adr-2 gene, adr-2(gv42), contained a deletion of 1072 bp beginning in the intron between exons 2 and 3, and terminating in the intergenic space preceding R151.8a, the downstream gene in the operon (Figure 4B). This deletion removed the terminal three exons (245 amino acids), which encode ∼75% of the catalytic domain, leaving the upstream dsRBM intact. Unexpectedly, the adr-2(gv42) deletion resulted in the expression of a chimeric RNA consisting of the first two exons of adr-2 fused to the downstream gene in the operon, R151.8a; this gene encodes a protein with sequence similarity to human GS-1 (Yen et al., 1992), whose function is unknown. Northern analyses confirmed the expression of the chimeric transcript and showed that other transcripts in the operon were unaffected (Figure 4B).
As described in subsequent sections, the adr-1 and adr-2 deletions had significant effects on RNA editing, both in vitro and in vivo. Further, although adr-1(gv6) and adr-2(gv42) mutant animals, as well as the double mutant, appeared normal, closer analysis revealed subtle and interesting phenotypes.
Homozygous deletions in adr-1 or adr-2 alter ADAR activity measured in vitro
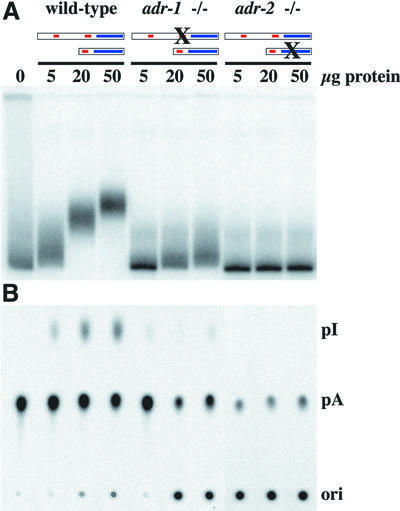
Worms containing homozygous deletions in either adr-1 or adr-2 were grown in culture to make extracts for in vitro biochemical assays. Extracts were prepared from mixed cultures (containing worms at all stages of growth) of wild-type and mutant strains, and two types of assay were performed. The first assay was based on the fact that deamination of an adenosine in an AU base pair creates an IU mismatch; this alters the structure of the RNA and, correspondingly, its mobility on a native gel (Figure 5A). When radiolabeled dsRNA was incubated with extracts made from wild-type animals, the mobility of the RNA progressively decreased as more extract protein was added. In contrast, even at the highest protein concentrations, only a slight change in mobility was observed after incubation with the adr-1(gv6) extract, and a mobility shift was undetectable with the adr-2(gv42) extract.
Fig. 5. In vitro assays of deaminase activity in wild-type and mutant worms. 32P-labeled dsRNA was incubated with various amounts of protein extract, for 2 h at 20°C. In (A), reaction products were electrophoresed on a native gel, and a phosphorimage of the gel is shown. This assay is based on the fact that adenosine deamination changes the structure of a dsRNA and thus its mobility on a native gel. In (B), the conversion of [32P]AMP to [32P]IMP was measured directly by TLC. After incubation, RNA was isolated, digested to mononucleotides and chromatographed on a TLC plate; a phosphor image of the TLC plate is shown. Ori, origin; pI, [32P]IMP; pA, [32P]AMP.
The second assay measured deamination directly, by monitoring the conversion of [32P]AMP to [32P]IMP on a thin-layer chromatography (TLC) plate (Figure 5B). Again, adenosine was readily converted to inosine when dsRNA was incubated with extracts derived from wild-type animals, barely detectable with those derived from adr-1(gv6) animals, and completely undetectable with those derived from adr-2(gv42) animals. Thus, based on results from two in vitro assays, we conclude that a deletion in the adr-1 gene dramatically reduces the amount of ADAR activity measured in vitro, while a deletion in the adr-2 gene eliminates it entirely.
Homozygous deletions in adr-1 or adr-2 alter ADAR activity measured in vivo
The in vitro deaminase assays described above were performed with a synthetic dsRNA that may not mimic endogenous ADAR substrates accurately. To confirm that the changes in ADAR activity observed in vitro reflected changes occurring on endogenous RNA, we isolated mRNA from adr-1(gv6) or adr-2(gv42) mutant worms and monitored the effects of the deletions on editing in vivo. cDNA was prepared from poly(A)+ RNA derived from young adults (day 1) of wild-type and each mutant strain. The cDNA was not cloned, but amplified as a population (unedited and edited) using primers surrounding known RNA editing sites in 3′-UTRs of C.elegans RNAs (Table I; Morse and Bass, 1999; Morse et al., 2002). For each sample, the entire population of PCR products was sequenced and compared with the sequence of PCR products amplified from genomic DNA, which represented the unedited sequence.
Table I. Changes at editing sites in adr-1(gv6) compared with wild type.
| RNA (WormBase ID)a | No. of editing sites |
Total | ||
|---|---|---|---|---|
| Decreased (eliminated)b | Increased (new sites)b | Unchanged | ||
| 36A (C35E7.6) |
90 (12) |
59 (6) |
47 |
196 |
| 9A (ZC239.6) | 16 (6) | 1 | 3 | 20 |
| 16G (Y6D11A.1) | 15 (12) | 0 | 0 | 15 |
| Syntaxin (F56A8.7a) | 6 (2) | 5 | 2 | 13 |
| pop-1 (W03D8.2) | 5 (3) | 1 | 1 | 7 |
| Laminin-γ (C54D1.5) | 4 (1) | 0 | 1 | 5 |
| Total | 136 (36) | 66 (6) | 54 | 256 |
aFor mRNAs of unknown function, substrates are designated as described previously (Morse and Bass, 1999).
bNumbers in parentheses denote a subset of the preceding number.
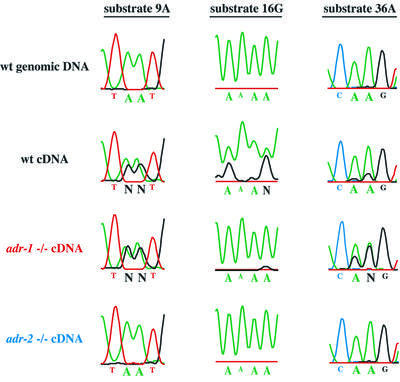
Figure 6 shows sequencing electropherograms corresponding to a portion of the edited regions we sequenced for three of the C.elegans ADAR substrates. Since inosine base-pairs with cytidine, an inosine in an mRNA appears as a guanosine in the corresponding cDNA. As illustrated by data from the wild-type cDNA, editing sites are indicated by a decrease in the peaks of the A trace (green) compared with genomic DNA, and the appearance of a new trace from guanosines in the cDNA (black). In total, we analyzed 256 editing sites, that together included six different C.elegans ADAR substrates (Table I). Analyses were performed multiple times to confirm that the patterns were reproducible. Consistent with the observation that extracts derived from the adr-2(gv42) deletion mutant had no detectable deaminase activity, cDNAs from this mutant showed no sign of editing and instead showed a sequence indistinguishable from that of genomic DNA for all 256 sites assayed. At least in a qualitative sense, the adr-1(gv6) data were also consistent with the amount of deaminase activity observed in extracts of this mutant, which was reduced, but detectable. Deletion of the adr-1 gene led to several different effects that depended on the editing site being considered (see Table I). Some sites were unchanged, showing amounts of editing comparable with wild-type animals (54/256, e.g. Figure 6, substrate 9A), editing at many sites was decreased or eliminated entirely (136/256, e.g. Figure 6, substrate 16G) and editing at other sites actually increased relative to wild-type (66/256, e.g. Figure 6, substrate 36A).
Fig. 6. Analysis of editing in RNA isolated from wild-type and mutant worms. The figure shows regions of electropherograms generated by sequencing PCR products amplified from genomic DNA, wild-type cDNA or adr mutant cDNAs, for three different ADAR substrates. In genomic DNA (top line), the sites indicated by large type are exclusively adenosines (green peaks) but, in wild-type cDNAs (second line), they are a mixture of adenosines (green peaks) and guanosines (black peaks), indicating that these sites are edited. The relative area under the peaks is a measure of the efficiency of editing. Editing sites shown for each substrate were chosen to illustrate three different effects of the adr-1 deletion and are not necessarily representative of all editing sites in these substrates. N indicates sites with nearly equal amounts of A and G that could not be called by the sequencing software. Note that the failure of the A trace of substrate 16G to go to baseline is typical of regions of homopolymer sequence; in other experiments, editing at these sites was confirmed by sequencing individual cDNA clones (Morse and Bass, 1999).
Taken together, the results of the assays for deaminase activity in vitro, and editing sites in vivo, confirm that adr-1 and adr-2 are involved in converting adenosines to inosines in vivo. At present, we cannot be certain why the deletion in the adr-1 gene leads to a partial loss of activity while the deletion in the adr-2 gene completely eliminates deaminase activity. Our favorite explanation for these results is that ADR-2 has deaminase activity on its own, while ADR-1 requires ADR-2 for activity, and perhaps the two proteins act together as a heterodimer (see also Discussion).
C.elegans ADARs are required for normal chemotaxis
Since the adr-1::GFP construct was expressed to high levels in the nervous system, we wondered whether worms lacking ADARs exhibited any behavioral defects. As a start, we chose to look for chemosensory defects, and focused on the AWA and AWC olfactory neurons, which allow a worm to move towards volatile chemicals (attractants). Previous studies have correlated these neurons with the detection of specific chemicals, and thus we analyzed AWA (diacetyl and pyrazine), AWC (benzaldehyde, 2-butanone and isoamyl alcohol) or both neurons together (thiazole) using well-established chemotaxis assays (Bargmann et al., 1993).
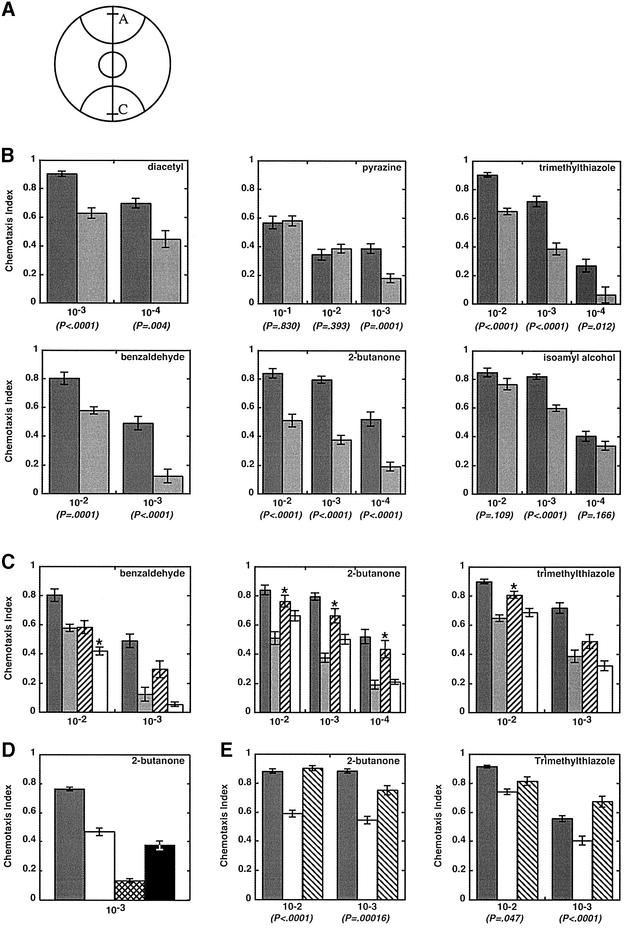
Plates containing a thin layer of agar were marked to create regions A and C as shown (Figure 7A). Immediately prior to the start of the assay, the attractant, diluted to various concentrations in ethanol, was spotted in region A and, as a control, ethanol was spotted in region C. Approximately 200 day 2 adults were placed in the center of the agar plate, allowed to move for 2 h at 20°C, and then counted to determine a chemotaxis index (Bargmann et al., 1993; Figure 7 legend). While the chemotaxis indices from assays of wild-type worms matched previously reported values (Bargmann et al., 1993; see Figure 7 legend with regard to pyrazine), values derived from assays of adr-1;adr-2 double mutants were consistently lower (Figure 7B). Chemotaxis indices were aberrant for chemicals detected by the AWA neuron as well as the AWC neuron.
Fig. 7. Chemotaxis population assays of wild-type and mutant animals. (A) Plates were marked with semi-circles (radius, 28 mm) to delineate attractant (A) and control (C) regions and spotted with 1 µl of attractant 5 mm from one edge (line in region A) and 1 µl of ethanol 5 mm from the opposite edge (line in region C); 1 µl of sodium azide (1 M) was placed at the same position to anesthetize animals reaching those areas. Between 100 and 200 adult animals (day 2) were placed in a circle at the center of the plate. After 2 h, animals were counted to calculate a chemotaxis index: [animals at (A) – animals at (C)]/[total animals on plate]. (B) Bar graphs show data from assays as in (A) for wild-type animals (dark gray) and adr-1;adr-2 double mutants (light gray). Chemotaxis index values are the average of ≥10 independent determinations, and error bars indicate the SEM; P-values show the t-probability from an unpaired (non-directional) two-tailed test. Attractants were diluted in ethanol as specified on the x-axis (v/v), except for pyrazine, which is a solid and was dissolved in ethanol (w/v). Chemotaxis values observed with wild-type animals all matched reported values, except for pyrazine (Bargmann et al., 1993), possibly because pyrazine had to be dissolved, increasing the chance of differences between laboratories. (C) As in (B) except that the wild type (dark gray) and adr-1;adr-2 double mutants (light gray) were compared with the single mutants, adr-1(gv6) (hatched) and adr-2(gv42) (open). Asterisks mark data for the single mutants that were significantly different from the double mutant at P < 0.01. (D) Chemotaxis to 2-butanone (10–3) was compared for wild-type (dark gray), adr-2(gv42) (open), che-2(e1033) (cross hatched) and odr-3(n2150) (black) animals. Chemotaxis values are averages of ≥7 independent determinations; error bars show the SEM. che-2(e1033) (P = 0.0001), but not odr-3(n2150) (P = 0.03), chemotaxis data were significantly different from those of adr-2(gv42). (E) Chemotaxis assays comparing the wild type (dark gray) and adr-2(gv42) (open) with the adr-2(gv42) rescue line B (hatched). P-values show that adr-2(gv42) rescue worms are significantly different from adr-2(gv42), except at the 10–2 dilution of trimethylthiazole. The adr-2(gv42) rescue lines were assayed by counting only the GFP-expressing portion of the population.
ADARs are thought to target numerous transcripts in the nervous system (reviewed in Paul and Bass, 1998; Bass, 2002; Morse et al., 2002), so the observed chemotaxis defects probably reflect the combined effects of altering RNA editing on many different mRNAs. Consistent with this, the characteristics of the observed chemotaxis defects were diverse, and varied depending on the chemical assayed and its concentration. For example, the adr-1;adr-2 double mutant showed significant defects in chemotaxis to benzaldehyde, trimethylthiazole and 2-butanone, but a very mild defect in chemotaxis to isoamyl alcohol. While the chemotaxis index for diacetyl was ∼30% lower in the double mutant at all concentrations tested, chemotaxis to pyrazine was equal to wild-type at high concentrations, and only showed a significant defect when pyrazine was diluted to a very low concentration. Similarly, while double mutants tracked quite poorly to benzaldehyde at a dilution of 10–3 (chemotaxis indices ∼20% of wild-type), increasing the concentration to 10–2 allowed mutant animals to chemotax much more like wild-type animals (chemotaxis indices ∼75% of wild-type). This concentration dependence suggests that at least some portion of the chemotaxis defect derives from an inability of the worm to detect the chemical, and can be overcome by increasing concentrations of the chemoattractant.
As a first step in delineating how each of the ADARs contributes to chemotaxis, we tested the response of adr-1(gv6) and adr-2(gv42) single mutants to three of the volatile chemicals, benzaldehyde, 2-butanone and trimethylthiazole (Figure 7C). adr1(gv6) and adr-2(gv42) animals both exhibited chemotaxis defects, emphasizing that both enzymes are important for normal chemotaxis. Consistent with the observation that both adr-2(gv42) and the adr-1;adr-2 double mutant lack any detectable editing activity, in all but one case (asterisks, P < 0.01, see legend) the chemotaxis indices of these animals were not significantly different from each other. For the exception (benzaldehyde 10–2), adr-2(gv6) animals showed a slightly stronger defect than the double mutant, suggesting that, in the absence of adr-2, adr-1 has deleterious effects. Of course, given the subtlety of this difference, further studies will be needed to confirm its significance. The defects associated with the adr-1(gv6) single mutant were relatively mild, and in almost all assays these animals showed a significantly weaker chemotaxis defect than the adr-2(gv42) animals. This is consistent with the observation that editing could be detected in adr-1(gv6) animals, although levels were significantly lower than the wild type.
As mentioned, we carefully optimized our assays to yield chemotaxis indices for wild-type animals that closely matched previously reported values (Bargmann et al., 1993; Roayaie et al., 1998). This allowed us to compare the chemotaxis indices of the adr mutants with those of previously characterized chemotaxis mutants. The chemotaxis defects observed in the adr mutants were similar in magnitude to defects observed with other chemotaxis mutants, and this was confirmed in side by side assays (Figure 7D). For example, when assayed with 2-butanone, the adr-2(gv42) chemotaxis defect is weaker than that observed with che-2(e-1033), a WD40 protein found in the cilia of sensory neurons (Fujiwara et al., 1999), and almost identical to that of odr-3(n2150), a Gα protein important for cilia morphology and signal transduction (Roayaie et al., 1998).
We also isolated two adr-2(gv42) lines expressing a cosmid (T20H4) containing the entire operon that includes the adr-2 gene. In both lines, the cosmid rescued the chemotaxis defects of the adr-2(gv42) animals (Figure 7E; data not shown). As yet, we have not been able to show rescue with transgenes that contain only the operon or the adr-2 gene, possibly because smaller transgenes are more likely to form extrachromosomal arrays that are repetitive (Kelly et al., 1997). Recent evidence suggests that such transgenes are silenced in adr deletion strains (Knight and Bass, 2002).
Are C.elegans ADARs required for normal vulva development?
Consistent with the observation that adr-1 was strongly expressed during vulva development, we found that a fraction of the adr-1;adr-2 double mutants exhibited protruding-vulva (Pvl) phenotypes (Seydoux et al., 1993; Eisenmann and Kim, 2000). To quantify this observation, synchronized populations of young adult worms were monitored by light microscopy. The phenotype typically became apparent between the last larval molt and day 2 of adulthood, and animals with vulva defects were moved to a separate plate and counted. The Pvl phenotype was exhibited with low penetrance, appearing in only 6.7% (n = 450) of the adr-1;adr-2 population. Pvl animals usually had defects in their somatic gonad as well, lacked embryos and subsequently died. Since the defective vulva prohibited egg-laying, animals that did have embryos showed a ‘bag of worms’ phenotype, with larvae developing inside the hermaphrodite (Ferguson and Horvitz, 1985).
The adr single mutants were scored to determine whether one or both genes were responsible for the Pvl phenotype. Surprisingly, we found that the adr-2(gv42) worms, in which editing was undetectable, did not exhibit the phenotype (n = 1866). Rather, the Pvl phenotype derived from the deletion in the adr-1 gene, consistent with the adr-1::GFP expression observed in the vulva. adr-1(gv6) worms showed 5.2% Pvl progeny (n = 2408), similar to the percentage observed in the double mutant. As yet, we have not been able to rescue the Pvl phenotype. This may be due in part to the transgene silencing observed in adr mutants (Knight and Bass, 2002).
Discussion
Here we provide the first analysis of the C.elegans ADARs, ADR-1 and ADR-2. The tools available for C.elegans studies allowed us to add to existing knowledge about ADARs. We were able to analyze expression in whole animals, throughout development, as well as assay effects of ADARs on the function of specific neurons. Editing patterns in mRNA isolated from animals lacking one or both of the ADAR genes, as well as in vitro assays using extracts, suggest that ADR-1 and ADR-2 have distinct but overlapping roles in C.elegans. Phenotypic analyses emphasize this: adr-1, but not adr-2, appears to play a role in vulva development, but both genes are important for normal chemotaxis, albeit to different degrees.
The distinct roles of ADR-1 and ADR-2 in catalysis
ADARs comprise a family of enzymes that all contain a highly conserved C-terminal catalytic domain, and variable numbers of dsRBMs (reviewed in Hough and Bass, 2000). ADARs are unique to metazoa and, although Drosophila has only a single ADAR, most metazoa have multiple enzymes. Studies of mammalian ADAR1 and ADAR2 show that these enzymes have distinct but overlapping functions, and differences can be traced to the substrate specificities intrinsic to each enzyme (Lehmann and Bass, 2000). For example, in vitro, both mammalian ADAR1 and ADAR2 can edit adenosines at the gluR-B R/G site (Melcher et al., 1996), and serotonin A and C sites (Burns et al., 1997). In contrast, the serotonin B site is deaminated only by ADAR1, and the gluR-B Q/R and serotonin D sites are deaminated only by ADAR2 (Melcher et al., 1996; Burns et al., 1997). Analyses of mice lacking or having reduced levels of either ADAR emphasize that these same specificities exist in vivo (Higuchi et al., 2000; Wang et al., 2000).
At present, it is not clear which mammalian enzymes are most similar to which C.elegans enzymes; the names used to differentiate the worm ADARs are not meant to correlate with a specific mammalian enzyme. However, our studies show that, like mammalian ADARs, the two C.elegans ADARs have overlapping, but distinct, functions. When the adr-1 gene is mutated, some editing sites are unchanged, suggesting that they are targeted by adr-2, while others are eliminated, suggesting that adr-1 is necessary for their deamination (see Table I). A subtle difference from observations made in mammals is that the adr-1(gv-6) deletion actually increases editing at some sites, and creates entirely new sites, as if the wild-type adr-1 serves to reduce editing at certain sites.
By far the most significant difference from the mammalian studies is that a deletion in adr-2 eliminates editing altogether. One explanation for this result is that ADR-2 is catalytically active on its own, while ADR-1 requires ADR-2 for its activity. Possibly, the two proteins function as a heterodimer, with ADR-2 acting as the catalytic subunit. In support of this idea, the ADR-1 sequence is significantly different from other ADARs in highly conserved regions of the catalytic domain (reviewed in Hough and Bass, 2000). The consensus HAE(x)41–58PCG(x)44–154SCSDK is followed closely by ADR-2, and almost all ADARs characterized to date. As written above, the underlined H and C residues are proposed to coordinate a catalytic zinc, while the E is thought to serve a proton transfer function; mutations at each of these four residues eliminate deaminase activity (Lai et al., 1995; Maas et al., 1996). The ADR-1 sequence is easily aligned with the ADAR family but, in contrast to other ADARs, its sequence differs substantially from the above consensus [DAI(x)48PPC(x)42CTADK]. Although these amino acid differences do not prove that ADR-1 is inactive, they are consistent with the idea. Importantly, at present, we cannot eliminate the possibility that the adr-2 deletion creates a dominant-negative allele, and that this is the reason why these animals lack ADAR activity. However, we find that heterozygous worms have levels of editing comparable with that of wild-type worms, which argues against this idea (L.Tonkin and B.Bass, unpublished data).
Why are ADARs essential in mammals but not in worms or flies?
Based on our studies in C.elegans, we believe that the primary role of ADARs is a non-essential one, but one that optimizes the function of many biological pathways, and increases an organism’s chance of survival. Biological pathways depend on a multitude of interactions between proteins. The amount of a particular protein–protein interaction, or complex, at a given time often dictates the strength of a downstream signal, or whether a signal will occur at all. We believe that ADARs function in many biological pathways to alter the amount of various protein complexes. ADARs could do this by creating amino acid changes that alter the affinity of the interacting proteins, as occurs in the G protein-coupled 5-HT2C serotonin receptor (Niswender et al., 1999). Since ADARs also act in non-coding regions of mRNAs (Morse and Bass, 1999; Morse et al., 2002), they may sometimes regulate the actual levels of an RNA, or its translatability; in this way, ADARs could alter the amount of a complex by changing the concentration of one of the protein partners. As suggested by a recent analysis (Knight and Bass, 2002), ADARs may also serve to modulate dsRNA-mediated gene silencing pathways, such as RNA interference (RNAi). While the non-essential functions of ADARs may occur in all organisms that express the enzymes, clearly mammals have co-opted ADARs to play essential roles. These functions may have evolved when an otherwise lethal genomic mutation was corrected at the RNA level by an ADAR; here ADARs would be playing a DNA repair role (Gray, 2000).
Do ADARs function in vulva development?
The observation that animals with a deletion in adr-1 have vulva defects, as well as the strong expression of the adr-1::GFP construct in the developing vulva, implicates the ADR-1 protein in vulva morphogenesis. However, because the Pvl defects are subtle, future studies will be needed to confirm this. There are myriad protein–protein and cell–cell signaling events that are crucial for vulva development (reviewed in Greenwald, 1997; see also Sharma-Kishore et al., 1999) and, according to the scenario presented above, ADARs could act at any of these steps. Since animals with a deletion in adr-2 have no detectable editing, it is perplexing that these animals do not have vulva defects. Possibly, adr-1 has functions beyond RNA editing or, alternatively, some adr-1-specific editing may exist in adr-2(gv42) animals but is beyond our limits of detection. The latter is consistent with the observation that the RNAi defects of the adr mutants are less severe in adr-2(gv42) animals compared with the double mutants (Knight and Bass, 2002). Of course, although all mutant animals were back-crossed to wild-type animals eight times, since we have not been able to rescue the Pvl defect, in theory it could derive from a mutation in a very closely linked gene. However, the strong vulva expression of the adr-1::GFP construct in multiple transgenic lines argues against this possibility. Finally, while there are no other genes with obvious sequence similarity to ADARs in the C.elegans genome, ADR-1 activity in the vulva could be mediated by interaction with an as yet unknown factor.
How do adr-1 and adr-2 modulate behavior in worms?
Once an odorant is detected by a sensory neuron, a particular behavioral response is elicited through specific connections to interneurons, other sensory neurons and motor neurons (Bargmann and Kaplan, 1998). The data we have collected so far are not sufficient to indicate where in the chemosensation pathway ADARs are acting. The adr-1::GFP construct is expressed in the sensory neurons and cilia, but also in the ventral nerve cord, motor neurons and interneurons; at present, it is possible that ADARs are acting in any or all of these cells. RNA editing could affect chemosensation by targeting RNAs that encode receptors or signaling molecules within the AWA or AWC neurons, or affect molecules in the downstream cells that mediate the response of these neurons. Two of the C.elegans ADAR substrates analyzed in this study (Table I), unc-64 syntaxin and laminin-γ mRNAs, are important for proper function of the nervous system (Saifee et al., 1998; Kim and Wadsworth, 2000). Editing sites in the 3′-UTRs of both of these substrates are altered in the adr deletion mutants and, in theory, either of these substrates could be involved in the chemotaxis defects we observed. However, since there are probably hundreds of ADAR substrates in the worm nervous system to choose from, future studies will be required to determine this.
Materials and methods
RNA isolation and cDNA synthesis
Synchronized young adults were harvested from 100 ml of liquid cultures (Lewis and Fleming, 1995). Pellets (1 ml of settled worms) were frozen in liquid nitrogen, ground to a fine powder and added to 20 ml of proteinase K reaction mixture [200 mM Tris–HCl pH 7.5, 300 mM NaCl, 25 mM EDTA, 2% SDS, 0.5 mg/ml proteinase K (Roche)] and incubated at 65°C for 30 min. After two organic extractions, nucleic acids were ethanol precipitated twice. DNA was removed by treating with RNase-free RQ1 DNase (Promega; 1 h, 37° C) followed by extraction and precipitation. Poly(A)+ RNA was purified from 1 mg of total RNA using Oligotex mRNA midi (Qiagen) or FastTrack 2.0 mRNA (Invitrogen) kits.
First-strand cDNA was synthesized as described previously (Morse and Bass, 1999). A 100 µl aliquot of reactions contained 10 µg of poly(A)+ RNA and was primed with 5 µg of random hexamer (Life Technologies) or 3 µg of oligo d(T)16 (Perkin-Elmer).
Cloning of H15N14.1a/b
cDNAs were amplified by PCR. Oligos were designed to hybridize with the initiating methionine (LAT072 CGAAATGGATCAAAATCCTAA CTAC), the 3′-UTR (LAT074 CGGCAATGGCTTGAAGATCATA CAC) and the poly(A) tail [QT CCAGTGAGCAGAGTGACG AGGACTCGAGCTCAAGC(T)17]. LAT072/LAT074 PCR products were amplified with AmpliTaq DNA polymerase (Perkin Elmer); LAT072/QT products were amplified with tTh DNA polymerase (Roche). PCR products were cloned into pCRII-TOPO vector (Invitrogen) and characterized using restriction enzymes and automated sequencing (ABI 377) to confirm alternatively spliced forms. A single representative cDNA for each splice form was sequenced to confirm the splicing arrangement.
GFP reporter genes
A reporter construct was generated for adr-1 by genomic PCR. A 3868 bp fragment was amplified with primers MWK382 (GCGAAGCTTGG TGGAGCTACTGGAATGCGGTCTG) and MWK383 (CGCGGATCC TGCTGCTGCTGTTGTTGGCTGAC) and cloned as a HindIII–BamHI fragment into the GFP expression vector pPD95.67 (A.Fire, G.Seydoux, J.Ahnn and S.Q.Xu, personal communication) to generate pKM1194. The reporter gene includes 3138 bp upstream and 730 bp downstream of the predicted translational start site; GFP coding sequences are fused in-frame within the third exon of adr-1. Sequence analysis of the junction and coding regions revealed a single, silent base change (C→T) in exon I at position 60.
Deletion alleles
A single deletion mutant allele was isolated for each ADAR gene from a library created as described previously (Dernburg et al., 1998). The adr-1(gv6) deletion removes 1560 bp of the H15N14.1a/b locus beginning at position 1269 in exon 5 (position 1, A of start codon) and ending at position 2829 in exon 10. The adr-2(gv42) deletion removes 1072 bp of the T20H4.4 locus beginning at position 848 in intron 2 and ending at position 1920 in the 3′-UTR. Both deletion alleles were confirmed by Southern blot analysis of genomic DNA isolated from homozygous mutant populations.
Genomic rescue strains
A 6.7 kb XbaI restriction fragment containing the adr-1 ORF was isolated from the cosmid H15N14 and injected into the adr-1(gv-6) deletion strain with pKM1194 (adr-1::GFP) as a marker. The cosmid T20H4, containing the entire six-gene adr-2 operon, was co-injected with pTG96 (sur5::GFP; Gu et al., 1998) as a marker into the adr-2(gv42) strain.
Northern analyses
Northern blots of poly(A)+ RNA (5 µg/lane) were prepared using standard methods for formaldehyde gels (Sambrook et al., 1989) and hybridized with probe using ULTRAhyb (Ambion). Radiolabeled probes were synthesized from PCR products (∼200–350 bp) corresponding to 5′ ends of messages (except those specific to deleted regions) using Klenow (NEB) in the presence of [32P]dCTP (3000 Ci/mmol; NEN; Sambrook et al., 1989). Blots were imaged with a Molecular Dynamics PhosphorImager.
Extract preparation
Liquid cultures of mixed staged worms were grown and harvested as described above. Two volumes of TGKED [50 mM Tris pH 8.0, 25% glycerol, 50 mM KCl, 0.1 mM EDTA, 0.5 mM dithiothreitol (DTT)] containing 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF) and Complete protease inhibitor cocktail (Roche) were added to 1 vol. of worm pellet and sonicated four times for 10 s; output 5, 100% duty cycle. Lysates were spun at 4°C at 16 000 g for 45 min to pellet cellular debris. Extracts were quantified, aliquoted and flash frozen before storing at –80°C.
ADAR activity assays
An ∼800 bp dsRNA was prepared as described (Bass and Weintraub, 1987). Extracts were diluted with TGKED to give various amounts of protein and mixed with an equal volume of assay buffer for a final concentration of 2 fmol of dsRNA in 40 mM Tris pH 7.9, 5 mM EDTA, 25 mM KCl, 10 mM NaCl, 1.1 mM MgCl2, 5% glycerol, 1 mM DTT, 40 U/µl RNAsin (Promega). Reactions (100 µl) were incubated at 20°C for 2 h and stopped by adding proteinase K, followed by phenol extraction and ethanol precipitation. Nucleic acids were loaded on a 6% native polyacrylamide gel (29:1, Bio-Rad) or processed further for TLC (Lehmann and Bass, 1999).
Amplification and sequencing of cDNA and genomic DNA
All ADAR substrates were identified as described (Morse and Bass, 1999; Morse et al., 2002). Editing was analyzed as described (Morse and Bass, 1999) or within the following regions: C35E7 (top strand, 15503–16390; bottom strand, 16354–17228); F56A8 (36103–36463); and C54D1 (top strand, 15857–16248; bottom strand, 15516–15846).
cDNA and genomic DNA corresponding to ADAR substrates were amplified by two rounds of PCR using nested primer pairs. The 20 µl first round PCRs contained 2 µl of cDNA or 5 µg of genomic DNA. Second round PCRs were 50 µl reactions containing 5 µl of first round PCR products. After PCR-cleanup (Qiagen), PCR products were sequenced in both directions using second round PCR primers.
Population chemotaxis assays
Synchronized populations of day 2 adults were prepared using the alkaline hypochlorite method (Lewis and Fleming, 1995). Animals were cultured in S-basal liquid media at 20°C with HB101 bacteria, and synchronous adult cultures maintained by filtering away embryo and larval stages through miracloth (Calbiochem) on day 1. Population chemotaxis assays were performed as described previously (Bargmann et al., 1993). Agar assay plates (10 cm) contained 25 ml of 1.6% agar, 20 mM potassium phosphate pH 6, 1 mM CaCl2, 1 mM MgSO4. Well-fed day 2 adults were filtered, washed three times in S-basal and once in water, then placed in the center of the plate and assayed (Figure 7 legend).
Acknowledgments
Acknowledgements
We thank E.Jorgenson, V.Maricq and S.Mango and their laboratories for advice and assistance, E.Herrington and R.Littlejohn for technical assistance, and R.Hough for sharing unpublished data. This work was supported by funds to B.L.B. from the National Institute of General Medical Sciences (GM44073). Oligonucleotides were synthesized by the Howard Hughes Medical Institute oligonucleotide synthesis facility at the University of Utah supported by the National Cancer Institute (grant no. 42014) and HHMI. B.L.B. is an HHMI Investigator. Some nematode strains used in this work were provided by the Caenorhabiditis Genetics Center, which is funded by the NIH National Center for Research Resources.
References
- Anderson P. (1995) Mutagenesis. In Epstein,H. and Shakes,D. (eds), Caenorhabditis elegans: Modern Biological Analysis of an Organism. Vol. 48. Academic Press, San Diego, CA, pp. 31–58.
- Baltimore D. (2001) Our genome unveiled. Nature, 409, 814–816. [DOI] [PubMed] [Google Scholar]
- Bargmann C.I. and Kaplan,J.M. (1998) Signal transduction in the Caenorhabditis elegans nervous system. Annu. Rev. Neurosci., 21, 279–308. [DOI] [PubMed] [Google Scholar]
- Bargmann C.I., Hartwieg,E. and Horvitz,H.R. (1993) Odorant-selective genes and neurons mediate olfaction in C.elegans. Cell, 74, 515–527. [DOI] [PubMed] [Google Scholar]
- Bass B.L. (2002) RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem., 71, 817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass B.L. and Weintraub,H. (1987) A developmentally regulated activity that unwinds RNA duplexes. Cell, 48, 607–613. [DOI] [PubMed] [Google Scholar]
- Black D.L. (2000) Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell, 103, 367–370. [DOI] [PubMed] [Google Scholar]
- Burns C.M., Chu,H., Rueter,S.M., Hutchinson,L.K., Canton,H., Sanders-Bush,E. and Emeson,R.B. (1997) Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature, 387, 303–308. [DOI] [PubMed] [Google Scholar]
- Dernburg A.F., McDonald,K., Moulder,G., Barstead,R., Dresser,M. and Villeneuve,A.M. (1998) Meiotic recombination in C.elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell, 94, 387–398. [DOI] [PubMed] [Google Scholar]
- Eisenmann D.M. and Kim,S.K. (2000) Protruding vulva mutants identify novel loci and Wnt signaling factors that function during Caenorhabditis elegans vulva development. Genetics, 156, 1097–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson E.L. and Horvitz,H.R. (1985) Identification and characteriza tion of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics, 110, 17–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M., Ishihara,T. and Katsura,I. (1999) A novel WD40 protein, CHE-2, acts cell-autonomously in the formation of C.elegans sensory cilia. Development, 126, 4839–4848. [DOI] [PubMed] [Google Scholar]
- Gray M. (2000) Speculations on the origin and evolution of editing. In Bass,B.L. (ed.), RNA Editing. Oxford University Press, Oxford, UK, pp. 160–177.
- Greenwald I. (1997) Development of the vulva. In Riddle,D.L., Blumenthal,T., Meyer,B.J. and Priess,J.R. (eds), C.elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 519–542. [PubMed]
- Gu T., Orita,S. and Han,M. (1998) Caenorhabditis elegans SUR-5, a novel but conserved protein, negatively regulates LET-60 Ras activity during vulval induction. Mol. Cell. Biol., 18, 4556–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M., Maas,S., Single,F.N., Hartner,J., Rozov,A., Burnashev,N., Feldmeyer,D., Sprengel,R. and Seeburg,P.H. (2000) Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature, 406, 78–81. [DOI] [PubMed] [Google Scholar]
- Hough R.F. and Bass,B.L. (2000) Adenosine deaminases that act on RNA. In Bass,B.L. (ed.), RNA Editing. Oxford University Press, Oxford, UK, pp. 77–108.
- Hough R.F., Lingam,A.T. and Bass,B.L. (1999) Caenorhabditis elegans mRNAs that encode a protein similar to ADARs derive from an operon containing six genes. Nucleic Acids Res., 27, 3424–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly W.G., Xu,S., Montgomery,M.K. and Fire,A. (1997) Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics, 146, 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. and Wadsworth,W.G. (2000) Positioning of longitudinal nerves in C.elegans by nidogen. Science, 288, 150–154. [DOI] [PubMed] [Google Scholar]
- Kim U., Wang,Y., Sanford,T., Zeng,Y. and Nishikura,K. (1994) Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc. Natl Acad. Sci. USA, 91, 11457–11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S.W. and Bass,B.L. (2002) The role of RNA editing by ADARs in RNAi. Mol. Cell, 10, 809–817. [DOI] [PubMed] [Google Scholar]
- Lai F., Drakas,R. and Nishikura,K. (1995) Mutagenic analysis of double-stranded RNA adenosine deaminase, a candidate enzyme for RNA editing of glutamate-gated ion channel transcripts. J. Biol. Chem., 270, 17098–17105. [DOI] [PubMed] [Google Scholar]
- Lehmann K.A. and Bass,B.L. (1999) The importance of internal loops within RNA substrates of ADAR1. J. Mol. Biol., 291, 1–13. [DOI] [PubMed] [Google Scholar]
- Lehmann K.A. and Bass,B.L. (2000) Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry, 39, 12875–12884. [DOI] [PubMed] [Google Scholar]
- Lewis J.A. and Fleming,J.T. (1995) Basic culture methods. Methods Cell Biol., 48, 3–29. [PubMed] [Google Scholar]
- Maas S., Melcher,T., Herb,A., Seeburg,P.H., Keller,W., Krause,S., Higuchi,M. and O’Connell,M.A. (1996) Structural requirements for RNA editing in glutamate receptor pre-mRNAs by recombinant double-stranded RNA adenosine deaminase. J. Biol. Chem., 271, 12221–12226. [DOI] [PubMed] [Google Scholar]
- Melcher T., Maas,S., Herb,A., Sprengel,R., Seeburg,P.H. and Higuchi,M. (1996) A mammalian RNA editing enzyme. Nature, 379, 460–464. [DOI] [PubMed] [Google Scholar]
- Mello C. and Fire,A. (1995) DNA transformation. Methods Cell Biol., 48, 451–482. [PubMed] [Google Scholar]
- Morse D.P. and Bass,B.L. (1999) Long RNA hairpins that contain inosine are present in Caenorhabditis elegans poly(A)+ RNA. Proc. Natl Acad. Sci. USA, 96, 6048–6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse D.P., Aruscavage,P.J. and Bass,B.L. (2002) RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc. Natl Acad. Sci. USA, 99, 7906–7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niswender C.M., Copeland,S.C., Herrick-Davis,K., Emeson,R.B. and Sanders-Bush,E. (1999) RNA editing of the human serotonin 5-hydroxytryptamine 2C receptor silences constitutive activity. J. Biol. Chem., 274, 9472–9478. [DOI] [PubMed] [Google Scholar]
- Palladino M.J., Keegan,L.P., O’Connell,M.A. and Reenan,R.A. (2000) A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell, 102, 437–449. [DOI] [PubMed] [Google Scholar]
- Paul M.S. and Bass,B.L. (1998) Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J., 17, 1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roayaie K., Crump,J.G., Sagasti,A. and Bargmann,C.I. (1998) The Gα protein ODR-3 mediates olfactory and nociceptive function and controls cilium morphogenesis in C.elegans olfactory neurons. Neuron, 20, 55–67. [DOI] [PubMed] [Google Scholar]
- Rueter S.M., Dawson,T.R. and Emeson,R.B. (1999) Regulation of alternative splicing by RNA editing. Nature, 399, 75–80. [DOI] [PubMed] [Google Scholar]
- Saifee O., Wei,L. and Nonet,M.L. (1998) The Caenorhabditis elegans unc-64 locus encodes a syntaxin that interacts genetically with synaptobrevin. Mol. Biol. Cell, 9, 1235–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Maniatis,T. and Fritsch,E.F. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Seeburg P.H., Higuchi,M. and Sprengel,R. (1998) RNA editing of brain glutamate receptor channels: mechanism and physiology. Brain. Res. Brain Res. Rev., 26, 217–229. [DOI] [PubMed] [Google Scholar]
- Seydoux G., Savage,C. and Greenwald,I. (1993) Isolation and characterization of mutations causing abnormal eversion of the vulva in Caenorhabditis elegans. Dev. Biol., 157, 423–436. [DOI] [PubMed] [Google Scholar]
- Sharma-Kishore R., White,J.G., Southgate,E. and Podbilewicz,B. (1999) Formation of the vulva in Caenorhabditis elegans: a paradigm for organogenesis. Development, 126, 691–699. [DOI] [PubMed] [Google Scholar]
- Wang Q., Khillan,J., Gadue,P. and Nishikura,K. (2000) Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science, 290, 1765–1768. [DOI] [PubMed] [Google Scholar]
- Yen P.H., Ellison,J., Salido,E.C., Mohandas,T. and Shapiro,L. (1992) Isolation of a new gene from the distal short arm of the human X chromosome that escapes X-inactivation. Hum. Mol. Genet., 1, 47–52. [DOI] [PubMed] [Google Scholar]