Abstract
Cytosolic Sec1/munc18-like proteins (SM proteins) are recruited to membrane fusion sites by interaction with syntaxin-type SNARE proteins, constituting indispensable positive regulators of intracellular membrane fusion. Here we present the crystal structure of the yeast SM protein Sly1p in complex with a short N-terminal peptide derived from the Golgi-resident syntaxin Sed5p. Sly1p folds, similarly to neuronal Sec1, into a three-domain arch-shaped assembly, and Sed5p interacts in a helical conformation predominantly with domain I of Sly1p on the opposite site of the nSec1/syntaxin-1-binding site. Sequence conservation of the major interactions suggests that homologues of Sly1p as well as the paralogous Vps45p group bind their respective syntaxins in the same way. Furthermore, we present indirect evidence that nSec1 might be able to contact syntaxin 1 in a similar fashion. The observed Sly1p–Sed5p interaction mode therefore indicates how SM proteins can stay associated with the assembling fusion machinery in order to participate in late fusion steps.
Keywords: membrane fusion/SM protein/SNARE/syntaxin/X-ray structure
Introduction
Intracellular membrane fusion processes are regulated by a set of conserved proteins (Jahn and Südhof, 1999; Chen and Scheller, 2001) including soluble N-ethylmaleimide-sensitive factor (NSF)-attachment protein receptors (SNAREs) (Söllner et al., 1993a), the Sec1-/munc-18-like (SM) family (Hata et al., 1993; Garcia et al., 1994) and Rab-family proteins (Zerial and McBride, 2001). SNAREs are diverse membrane-anchored proteins which usually contain an N-terminal regulatory domain followed by an ∼60 residue α-helical SNARE motif adjacent to a transmembrane region (Jahn and Südhof, 1999). They are classified into t-SNAREs (also Q-SNAREs) such as syntaxin-like SNAREs and SNAP-25 (containing two SNARE motifs), which is normally encoded by two light chains (Fukuda et al., 2000), and v-SNAREs (also known as R-SNAREs) (Fasshauer et al., 1998). Four SNARE motifs, three t-SNAREs and one v-SNARE, assemble into a four-helical bundle during SNARE pairing (Sutton et al., 1998). t- and v-SNAREs must be positioned on opposing membranes to be functional so as to bring two membranes into close proximity and form membrane fusion-active trans-SNARE complexes (Hanson et al., 1997; Nichols et al., 1997; Sutton et al., 1998; Weber et al., 1998). After completion of membrane fusion, NSF and α-SNAP disassemble the resulting cis complexes (Söllner et al., 1993b). Although SNARE complex assembly was reported to be promiscuous in vitro (Fasshauer et al., 1999), evidence from liposome fusion assays suggests that the interaction of membrane-anchored SNAREs is rather specific, and only physiologically relevant complexes fuse liposomes efficiently (McNew et al., 2000a).
SM proteins are a small family of cytosolic proteins with seven members in higher eukaryotes and only four members in yeast (Chen and Scheller, 2001). This includes Sec1p, which controls exocytosis, Sly1p, which is active at Golgi and endoplasmic reticulum (ER) membranes, Vps45p, which acts in the trans-Golgi network (TGN) and endosomal transport processes, and Vps33p, which restricts vacuolar/lysosomal sorting processes (Jahn and Südhof, 1999; Chen and Scheller, 2001). The functional importance of SM proteins in intracellular traffic is evident from genetic studies, which show the accumulation of vesicles at corresponding membranes and indicate a positive regulatory function for SM proteins (Novick and Schekman, 1979; Ossig et al., 1991; Cowles et al., 1994; Harrison et al., 1994; Verhage et al., 2000).
Originally, neuronal Sec1 (nSec1) was shown to bind to syntaxin (Hata et al., 1993; Garcia et al., 1994) and block SNARE complex formation (Pevsner et al., 1994) while syntaxin 1 adopts a closed conformation (Calakos et al., 1994; Dulubova et al., 1999; Misura et al., 2000). In the fusion-incompetent closed conformation, the SNARE motif folds back on to the three-helical bundle formed by the N-terminal regulatory domain Habc (Misura et al., 2000). Comparison of liganded and unliganded nSec1 structures shows no major conformational changes upon syntaxin 1 interaction (Bracher et al., 2000). The observed binding mode suggested a strictly negative regulatory function for nSec1 (Misura et al., 2000) in contrast to genetic and biochemical data that postulate a positive function (Dresbach et al., 1998; Wu et al., 1999), which led to the general acceptance of a dual role for neuronal SM proteins in exocytosis (Chen and Scheller, 2001).
The SM protein Sly1p was first identified because of a genetic mutation (Sly1-20) that suppresses the requirement for the Rab protein Ypt1p in ER to Golgi transport (Dascher et al., 1991). This was also the first indication that SM proteins, SNAREs and Rab proteins are functionally coupled. In the Golgi, a single syntaxin, Sed5p (vertebrate syntaxin 5), seems to participate in most fusion events (Tsui et al., 2001; Parlati et al., 2002). Sly1p was shown to bind to the syntaxin Sed5p with high affinity (Grabowski and Gallwitz, 1997) and contributes to the specificity of formation of the SNARE complex in vitro as Sly1p–Sed5p interaction prevented non-physiological SNARE complex formation (Peng and Gallwitz, 2002). Although Sed5p contains an N-terminal regulatory domain, which was suggested to form a three-helical bundle, an N-terminal peptide of 20 amino acids is sufficient for Sly1p interaction, and Sly1p also recognizes a similar peptide motif from the ER syntaxin Ufe1p (Kosodo et al., 1998; Yamaguchi et al., 2002). A similar binding mode was also reported for the interaction of Vps45p and the TGN syntaxin Tlg2p (Dulubova et al., 2002). In contrast, yeast Sec1p interacts with the syntaxin Sso1p only when participating in a functional trans-SNARE complex (Carr et al., 1999) whereby Sec1p seems to act at a late step in fusion (Grote et al., 2000). Together, these data indicate that SM proteins interact with syntaxins in at least three different ways, requiring either a closed conformation, an N-terminal peptide region or the engagement in SNARE complex formation.
Here we present the crystal structure of yeast Sly1p in complex with a 45 amino acid peptide derived from the N-terminal region of Sed5p [Sed5p(1–45)]. Sed5p(1–45) binds in a helical conformation to domain I of Sly1p, in a region which is highly conserved among all Sly1p homologues. The structure of Sly1p underlines the evolutionary conservation of the SM protein family and shows the same three-domain arch-shaped assembly as observed for neuronal Sec1 homologues (Bracher et al., 2000; Misura et al., 2000). The binding mode of the SM protein syntaxin shows for the first time how SM proteins can be recruited to fusion sites and participate in downstream regulatory events.
Results
Structure determination
Sly1p forms a stable complex with either the complete cytosolic part of Sed5p (residues 2–324) or the N-terminal region (residues 2–186) containing the Habc domain (Kosodo et al., 1998). Limited proteolysis experiments of Sly1p–Sed5p(2–186) complexes with chymotrypsin suggested that one Sed5p cleavage site, N-terminal to the Sed5p Habc domain, was protected in complex with Sly1p, resulting in a 5 kDa fragment of Sed5p which was still associated with Sly1p after gel filtration chromatography. Based on our results and the recently reported minimal N-terminal Sly1p-binding region of Sed5p (Yamaguchi et al. 2002), we expressed Sed5p residues 1–45 (including the C-terminal chymotrypsin site).
The Sly1p–Sed5p(1–45) complex was crystallized in space group P41212, with one molecule per asymmetric unit, and the structure was solved by single wavelength anomalous diffraction (SAD) using selenomethionine-derivatized Sly1p-containing crystals (see Table I for data collection and phasing statistics). Subsequent maximum likelihood density modification procedures resulted in a good experimental electron density map (Figure 1A), which allowed the unambiguous tracing of the polypeptide chains. The final model contains Sly1p residues 10–662 and Sed5p residues 1–21 [plus an additional N-terminal pentapeptide (GAMAG)] as well as 20 water molecules. Sly1p loop regions comprising residues 245–256, 299–313, 362–381 and 510–527 were disordered. The final model has been refined using diffraction data to 3.0 Å resolution to an R-factor of 0.257 and an Rfree of 0.290 (see Table I, refinement statistics).
Table I. X-ray data collection, phasing and refinement statistics.
| X-ray data collection |
|
|
|
|
|
|
|
|
|
| Data set | Soft remote | Peak | |||||||
| Wavelength (Å) | 0.9875 | 0.9789 | |||||||
| Unit cell | |||||||||
| a, b (Å) | 161.06 | 161.39 | |||||||
| c (Å) | 88.21 | 88.45 | |||||||
| Resolution (Å) | 30–2.90 | 25–3.1 | |||||||
| (3.00–2.90) | (3.21–3.10) | ||||||||
| Completeness (%) | 100 (100) | 99.7 (100) | |||||||
| Total reflections | 200 527 | 350 883 | |||||||
| Unique reflections | 26 182 | 40 400 | |||||||
| Rmergea | 0.044 (0.552) | 0.058 (0.491) | |||||||
| <I>/<σI> |
13.3 |
10.9 |
|
|
|
|
|
|
|
| Phasing statistics |
|
|
|
|
|
|
|
|
|
| Resolution shell (Å) | 25–3.2 | 25–10.6 | 10.6–6.88 | 6.88–5.44 | 5.44–4.63 | 4.63–4.10 | 4.10–3.72 | 3.72–3.43 | 3.43–3.2 |
| FOM |
0.32 |
0.39 |
0.46 |
0.45 |
0.43 |
0.38 |
0.29 |
0.21 |
0.14 |
| Refinement statistics |
|
|
|
|
|
|
|
|
|
| Resolution range (Å) | 30–3.0 | ||||||||
| No. of reflections/test set | 23 743/1201 | ||||||||
| R-factorb (%) | 0.2573 | ||||||||
| Rfree (%) | 0.2904 | ||||||||
| Residues | (614 of 723) | ||||||||
| No. of protein/solvent atoms | 4707/20 | ||||||||
| Average B-factor (Å2) | 99.5 (Wilson 92.3) | ||||||||
| R.m.s.d. bond lengths (Å) | 0.009 | ||||||||
| R.m.s.d. bond angles (°) | 1.6 | ||||||||
| Ramachandran plot | |||||||||
| Residues in most favoured and additionally allowed regions (%)c | 97.5 |
Values in parentheses are for last shell limits.
aRmerge = Σhkl Σi |Ii (hkl) – <I(hkl)>|/Σhkl Σi Ii (hkl).
bR-factor = Σhkl ||Fobs| – k|Fcalc||/Σhkl |Fobs|.
cAs defined in PROCHECK (Laskowski et al., 1993).
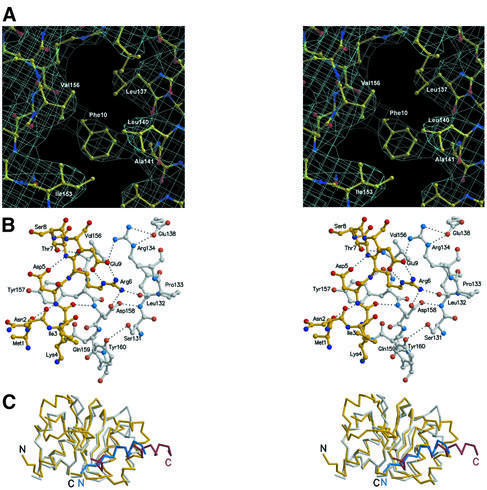
Fig. 1. Close-ups of Sly1p–Sed5p interactions. (A) Stereo diagram of the experimental electron density map of the Sly1p–Sed5p complex. The region shows the conserved Sly1p hydrophobic pocket (residues Leu137, Leu140, Ala141, Ile153 and Val156) that accommodates the Sed5p key residue Phe10. The map is contoured at 0.8σ. (B) Hydrogen bond network at the interface of Sly1p and Sed5p. Residues 1–9 of Sed5p are shown as a ball-and-stick model in yellow; residues 131–134, 138 and 156–160 of Sly1p are shown in grey. Oxygen and nitrogen atoms are shown in red and blue, respectively. Hydrogen bonds are indicated as dashed lines. Note that the region comprising residues 10–21 of Sed5p is involved in hydrophobic interactions only. (C) Superposition of domain I of Sly1p in complex with Sed5p with the corresponding region in s-Sec1 including a helical segment from a neighbouring molecule forming a crystal contact (pdb code 1FVH). The r.m.s.d. for the fragments shown is 1.34 Å within 127 residues (35 identical). The peptide backbones are shown as Cα-traces. The colouring scheme is as follows: Sly1p, yellow; Sed5p, red; s-Sec1 domain I, white; and s-Sec1 residues 321–332, mimicking the Sed5p helical interaction, blue. N- and C-termini are indicated.
Structure of Sly1p
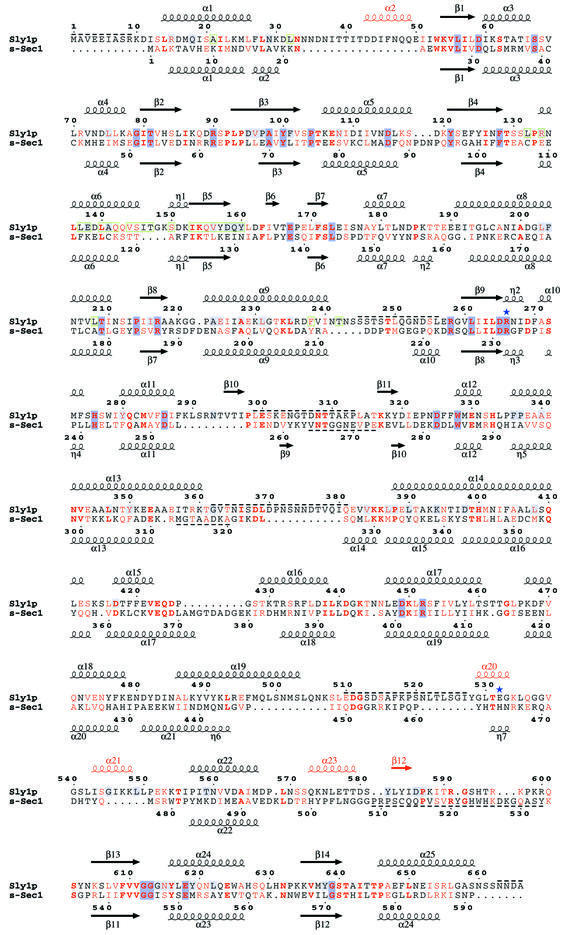
Sly1p is an arch-shaped molecule with overall dimension of 70 × 80 × 60 Å composed of three domains (Figure 2A). Domain I (residues 1–161) is characterized by a typical Rossmann fold containing a parallel five-stranded β-sheet with 2-1-3-4-5 topology lined by α-helices on both sides. Domain II (residues 162–268 and 582–666) also forms an α/β structure encompassing a seven-stranded β-sheet with a central parallel five-stranded β-sheet (1-5-4-3-2 topology) flanked by two antiparallel strands (β6 and β12). β6 and the turn to β7 form part of the interface with domain I, while β12 connects to the central strand β13 via a long, poorly ordered loop. The connection between α9 and β8 (residues 245–256), which is part of an unusual left-hand turn is disordered in the structure (Figures 2A and 3). Domain III is inserted between β9 and β12 and is predominantly α-helical. After three successive short helices (η2, α10 and α11), the peptide backbone forms a β-hairpin containing a disordered loop (residues 299–313) at the tip. This structure is found at the bottom of the crevice of the molecule and is followed by a helical hairpin structure (α13 and α14) protruding outwards, whose connection at the tip is disordered (residues 362–381). The base of domain III is composed of a five-helix arrangement (α15–α19), which is reminiscent of helix repeat proteins. Helices 22 and 23 form part of the interface to domain II, and helix 23 connects back to β12 of domain II. Additionally, α-helices 20 and 21 are part of a surface loop insertion that covers the base and the front of the helical hairpin structure (α13 and α14) (Figures 2A and 3).
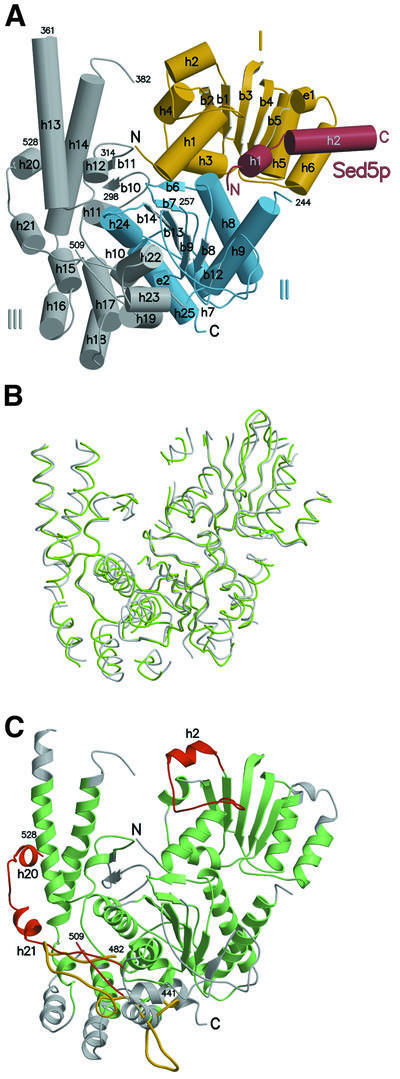
Fig. 2. General architecture of the Sly1p–Sed5p complex and comparison of SM paralogues Sly1p and nSec1. (A) The domain structure of Sly1p is indicated; domain I in yellow, domain II in blue and domain III in grey. Sed5p is shown in red. β-sheets are represented as arrows and α-helices as cylinders. Note that the N-terminus of Sed5p contacts domain II while the remaining part interacts only with domain I. The secondary structure elements are labelled accordingly and disordered regions are indicated. (B) Superposition of Sly1p with squid nSec1 (pdb code 1FVH). The peptide backbones of Sly1p and s-Sec1 are shown as grey and green coils, respectively. Regions superimposing with an r.m.s.d. >3.5 Å have been omitted for clarity. Sly1p is shown in the same orientation as above. (C) Ribbon representation of Sly1p. Regions superimposing to s-Sec1 with an r.m.s.d. <3.5 Å are highlighted in green, and those >3.5 Å in grey. Sequence insertions resulting in independent secondary structure elements are shown in red. The conformation of the loop region between α-helices 21 and 22 in s-Sec1 is shown in yellow for comparison.
Fig. 3. Structure-based sequence alignment of Sly1p with s-Sec1. Secondary structure elements are shown above and below the sequences. Red colouring of secondary structure elements indicates Sly1p-specific insertions. Homologous and identical residues are shown in red and bold red letters. Conservation within the Sly1p group is indicated by a light blue background and within nSec1 and Sly1p groups by a dark blue background. Disordered regions are marked by dashed lines. Green boxes denote residues involved in Sed5p binding. The positions of Sly1p mutations E532K (Sly1-20) and R266K (sly1-ts) are indicated by blue asterisks.
Sed5p interaction with Sly1p
Sed5p folds into an N-terminal α-helical turn (residues 1–4; helix 1) followed by residue 5 with β-type conformation and α-helix 2 (residues 7–21) (Figures 1C and 2A). The extensive interface between Sly1p and Sed5p buries a total surface of 1647 Å2, consistent with a high affinity interaction (Grabowski and Gallwitz, 1997). The N-terminus of Sed5 is in van der Waals contact with residues in the domain linker region and in domain II. Ile3 contacts Tyr160 (conserved), Leu208 and Phe238, while Tyr160 also contacts the side chain of Lys4. In addition, the aromatic ring of Tyr157 stacks on to the π-electron system of the Asp5 carboxyl group (Figure 1B). Residues 5–7, which are conserved among all Sed5p sequences (Figure 4), form an antiparallel extension to the C-terminal flank of the β-sheet in domain I (Figure 1B). A hydrogen bond contact between the hydroxyl group of Thr7 and the backbone amide of Val156 mimics a third β-sheet contact. The carboxyl group of Asp5 forms a 2.83 Å short hydrogen bond to the amide group of Thr7, which defines the geometry of the transition into α-helix 2. Helix 2 of Sed5p is involved in tight hydrophobic packing with helix 6 of Sly1p (Figures 1A and C and 2A). The packing angle of the helices was determined to be 24°, which is close to the theoretical packing angle of 23° for ±4 and ±3 ridges in α-helices (Chothia, 1984). The closest contact is formed by residues Phe10, Ser13 and Val14 in Sed5p, and Sly1p Ala141, which is part of a hydrophobic pocket (Leu137, Leu140, Ala141, Ile153 and Val156) that accommodates Phe10 (Figure 1A). Ala141 and Val156 are conserved, while the others can be substituted by conservative changes (Figure 4). In addition, Sed5p Tyr17 is within van der Waals distance of Gln142 and Ser145 and lines the hydrophobic cluster.
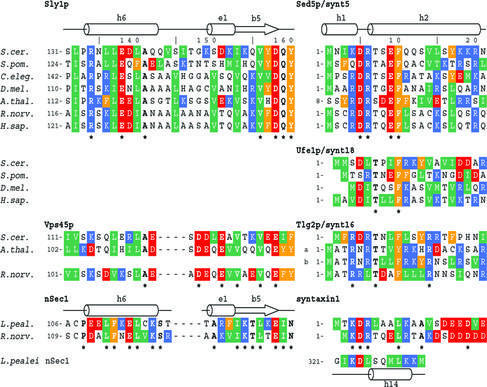
Fig. 4. Sequence conservation of SM protein–syntaxin interaction regions. Sequence alignment of the binding regions of Sly1p, Vps45p and nSec1 homologues (left panel) and their syntaxin family members Sed5p, Ufe1p, Tlg2p and syntaxin 1 (right panel). In addition, the sequence of a loop region of s-Sec1, which is involved in a crystal contact that is largely reminiscent of the Sly1p–Sed5p binding mode, is aligned based on the structural similarity. Residues identical within one group are indicated by an asterisk and by bold letters. Side chain properties are indicated by green (hydrophobic), orange (aromatic), red (negatively charged) and blue (positively charged) background.
An extended hydrogen bond network exclusively composed of absolutely conserved residues supports the hydrophobic packing (Figure 1B). Central to the network is the guanidinium group of Arg6, which interacts with the carboxyl group of Asp158, the carbonyl of Leu132 and the carboxyl group of Glu9. From Glu9, the network is extended to Glu138 via Arg134. The carboxyl group of Asp158 enforces the link to Leu132 by contacting the amide nitrogen. The conserved residue Gln159 stabilizes the binding site geometry by a conserved hydrogen bond to Asn29, which itself contacts Gln155 that links back to the peptide backbone at the carbonyl function of Leu30. A hydrogen bond between the hydroxyl function of Tyr160 and Ser131 may further define the geometry of the binding site.
Structural conservation of SM proteins
Comparison of the Sly1p structure and the neuronal isoform of the plasma membrane paralogue Sec1, nSec1 (Munc18a), whose structure has been solved in complex with syntaxin 1A (rat nSec1–syntaxin 1A; Misura et al., 2000) and in the free unliganded form (squid s-Sec1; Bracher et al., 2000; Bracher and Weissenhorn, 2001), highlights the overall similarity between SM proteins and exemplifies functional differences. Sly1p and nSec1 share a common fold, and the spatial arrangement of domains I and II, of the α-helical hairpin region (α13 and α14) and of the central region including α-helices 10, 11 and 22 is well conserved with nSec1 (Figure 2B). Within the two structures, the Cα atoms of 358 residues (90 conserved residues; Figure 3) can be aligned with an r.m.s.d. of 1.75 Å using a distance cut-off of 3.5 Å (Figure 2B). Despite several sequence insertions and deletions, the secondary structure elements in domains I and II superimpose with low r.m.s.ds of 1.3 Å (119 residues, 33 conserved) and 1.43 Å (131 residues, 34 conserved), respectively (Figures 2B and 3). Regions of poor sequence conservation such as the β-hairpin (β10 and β11) and the helix repeat motif (α15–α19) in domain III have the same topology as the corresponding elements in nSec1, but their arrangement is distorted and cannot be matched satisfyingly to nSec1 (Figures 2A and 3).
Secondary structure element insertions are found in all domains, including α2 into domain I, which is unique to Sly1p of Saccharomyces cerevisiae and bulges towards domain III (Figures 2C and 3). Furthermore, α20 and α21 are inserted into domain III (Figures 2C and 3). Interestingly, α20 contains the Sly1-20 mutation (Glu532Lys) that has been isolated as an efficient suppressor of the deletion of the essential Rab GTPase Ypt1p (Dascher et al., 1991). α20, which is only poorly defined, masks a conserved hydrophobic region at the bottom of the helical hairpin together with α21 (Figure 5B). While α20 is encoded by variable sequences within Sly1p homologues, α21 is conserved. In contrast, in nSec1, this elaborate connection does not exist and consists of a short coil region attached to the bottom of the helical repeat. Finally, the poorly conserved linker region of domains II and III that was disordered in all structures of nSec1 contains α23 (domain III) and β12 (domain II) (Figures 2C and 3).
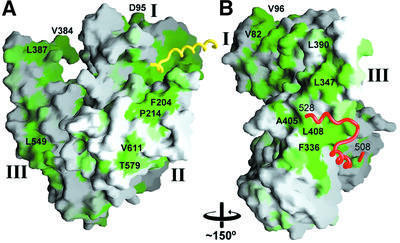
Fig. 5. Surface conservation of Sly1p homologues. The homology score for an alignment of Sly1p homologue sequences was plotted on to the surface of Sly1p using a scale from green (identical) to white (no conservation). Coils denote the backbone of Sed5p (yellow) and of an insertion containing helices 20 and 21 (red). (A) The orientation is similar to Figure 2. (B) Orientation after an ∼150° rotation around the vertical axis. Some conserved residues are indicated for orientation.
The relative orientation of the domains is controlled by hydrophobic packing, as is evident from the interaction between α1 (domain I) and the β6/β7 hairpin (domain II) or the extensive interface between domains II and III (Figure 2). In addition, conserved polar interactions between and within domains provide specific contacts that might be important during folding (residues marked in Figure 3). In domain II, two conserved salt bridges between Arg258 and Asp586 as well as Arg266 and Glu618 interconnect the N- and C-terminal parts. The guanidinium group of Arg266 extends the polar network to the backbone carbonyl groups at residues Asp269 and Gly613. The importance of Arg266 is underlined by the temperature sensitivity caused by the mutation Arg266Lys (sly1-ts) (Cao et al., 1998). A conserved salt bridge between Glu167 and Trp328 links domains II and III. Within domain III, conserved hydrogen bonds were found between His276 and Asp449, Asp325 and His400, Lys394 and Asp398, and Asp286 and Arg452, which is in contact with the backbone at Phe274 and Ser275 (Figure 3).
Even though SM proteins have a common fold, their sequence diversity allows for multiple functional differences. Within the Sly1p family, relatively few strictly conserved surfaces can be identified. This includes the binding pocket for the syntaxin peptide (Figure 5A) and the outer face of the helical hairpin (α13 and α14), which is partially covered by the helical protrusion (α20 and α21) (Figure 5B). Otherwise, the conserved surface regions are rather scattered. Notably, the lowest surface conservation maps to the helix repeat part of domain III (α15–α19) (Figure 5).
Conservation of the SM protein–syntaxin peptide interaction mode
The sequence and functional homology between Sed5p and Ufe1p (Yamaguchi et al., 2002) suggests a very similar interaction of Sly1p with Ufe1p (Figure 4). The conserved residue Phe9 (position 10 in Sed5p) of Ufe1p presumably binds into the hydrophobic pocket lined by residues Leu137, Leu140, Ala141, Ile153 and Val156, and Ufe1p Asp4 and Thr6 could fulfil the same function as in the Sly1p–Sed5p complex. Although the N-termini of Ufe1p/syntaxin 18 and Sed5p/syntaxin 5 are similar, some key residues of Sed5p are not conserved in Ufe1p. Notably, the polar residues Arg6 and Glu9 are replaced with leucine and isoleucine. Only the Ufe1p homologue from Schizosaccharomyces pombe exhibits sequence conservation with Sed5p in these positions but lacks aspartic acid and valine in positions 3 (5 in Sed5p) and 12 (14) (Figure 4). In the complex of Sly1p with Ufe1p, the loss of polar interactions is probably therefore compensated by hydrophobic interactions.
The structural similarity between SM proteins together with biochemical evidence, which suggests that Tlg2p residues 1–33 are sufficient for Vps45p interaction (Dulubova et al., 2002), allow the extrapolation of the Sly1p–Sed5p interaction to the Vps45p–Tlg2p complex. All residues forming the hydrophobic binding pocket for Sed5p residue Phe10 in Sly1p (Leu137, Leu140, Ala141, Ile153 and Val156) are conserved in Vps45p (Figure 4). The charged residues of Sly1p involved in the hydrogen bond network (Sly1p Arg134, Glu138 and Asp158) are also conserved or substituted by conservative residues (Figure 4). However, the sequence alignment indicates that Vps45p contains a four-residue deletion between α-helix 6 and β5 (Figure 4). The N-terminal sequence of its cognate syntaxin Tlg2p is very similar to that of Sed5p, including conservation of Arg3, Thr6 and Phe9 (or Tyr9). Sed5p residue Glu9, however, is substituted by hydrophobic residues as in the case of the Ufe1p sequences (Figure 4). Therefore, most of the principal interactions, the extension of the β-sheet, hydrophobic helix–helix packing and hydrophobic interactions with Phe10 observed in the Sly1p–Sed5p complex seem to be feasible, and Vps45p paralogues are likely to bind to the N-terminus of Tlg2p in a similar way to that observed in the Sly1p–Sed5p complex.
Surprisingly, the N-terminal sequences of neuronal syntaxin 1 show a high degree of conservation and some similarity to Sed5p homologues, including a predicted helix formation propensity (Misura et al., 2002) (Figure 4). Indirect evidence for a potential interaction of the N-terminal region of neuronal syntaxin 1 with nSec1 arises from a crystal contact in two slightly different crystal forms of s-Sec1, the nSec1 homologue from squid (pdb codes 1EPU and 1FVH). In these crystal forms, part of the loop region connecting α-helices 13 and 15 (corresponding to α13 and α14 in Sly1p) interacts with domain I of a neighbouring molecule. This internal s-Sec1 sequence is remarkably similar to the conserved sequence motif at the N-terminus of syntaxin 1. A short coil region starting at residue 321 aligns in an antiparallel fashion to β-strand 5, and the following α-helix 14 packs against α-helix 6 in a way which strikingly resembles the interaction between Sly1p and Sed5p, and both helical segments overlay with a low r.m.s.d. of 0.82 Å (Figure 1C). Leu329 (may correspond to syntaxin Leu9), which occupies the corresponding Sed5p Phe10 position, binds into a conserved hydrophobic pocket (Cys116, Phe112, Ala121, Ile124 and Leu127) (Figure 4). The aliphatic part of the side chain of Lys330 shields the hydrophobic cluster from solvent, and the carboxyl group of Asp324 contacts the backbone amide at 326 similarly to the Sed5p backbone interaction between Sed5p Asp5 and Thr7 (Figure 1B). Together, these data indicate that nSec1 may be able to contact syntaxin 1 in the same way as observed in the crystal contact and the Sly1p–Sed5p structure.
Discussion
The yeast SM protein Sly1p and its mammalian homologue act at ER and Golgi membranes by forming complexes with its resident syntaxins Ufe1p and Sed5p (Sogaard et al., 1994; Grabowski and Gallwitz, 1997; Yamaguchi et al., 2002). The crystal structure of Sly1p presented here is the second structure of an SM family protein, and the conserved overall fold indicates their evolutionary conservation. Despite their structural similarities, substantial functional versatility might arise from sequence insertions with additional secondary structure elements as demonstrated by the Sly1p structure.
Docking of vesicles is largely controlled by Rab proteins, a family of vesicle membrane-associated small GTPases, and includes Rab effector molecules that may interact with SM proteins as well as syntaxins (Peterson et al., 1999; Siniossoglou and Pelham, 2001; Zerial and McBride, 2001). A genetic Sly1p mutation, Sly1-20, has been shown to uncouple ER to Golgi protein transport from Rab Ypt1p function (Dascher et al., 1991). The E532K mutation locates to the poorly defined helix 20 that packs loosely against the outer surface of the helical hairpin (helices 13 and 14). The residues corresponding to α20 in Sly1p exhibit low sequence conservation, and Glu532 seems to be only conserved among yeast and lower plant Sly1p sequences (S.cerevisiae, S.pombe and Arabidopsis thaliana), while all other Sly1p homologues show great sequence diversity in this region. Indeed, the region comprising α20 maps to a highly variable region of SM proteins, which, for example, has no equivalent in neuronal Sec1. Since the external surface of the helical hairpin (α13 and α14) that is partly shielded by α20 and α21 is among the best conserved surface regions within Sly1p homologues, the insertion might act as a lid controlling a function of Sly1p. In the mutant Sly1-20, Glu532Lys, the lid may be open permanently and thereby bypass the regulatory step guarded by the Rab GTPase Ypt1p.
Biochemical studies suggested that the N-terminal half of Sly1p (Grabowski and Gallwitz, 1997) and an N-terminal fragment of Sed5p were sufficient for binding (Grabowski and Gallwitz, 1997; Kosodo et al., 1998). Therefore, the binding was considered to be different from the interaction of neuronal syntaxin 1 with nSec1, which requires almost the entire cytosolic region of syntaxin 1 (Dulubova et al., 1999; Yang et al., 2000; Misura et al., 2000). Recently, the Sly1p-binding region was defined to a short 20 amino acid N-terminal peptide motif present in Sed5p and Ufe1p (Yamaguchi et al., 2002). The same group reported that the SM protein Vps45p recognizes a conserved N-terminal sequence motif in its cognate syntaxin Tlg2p/syntaxin 16, which indicated that, at least for transport between ER and Golgi and through the TGN/early endosome, SM proteins are recruited by a conserved mechanism (Dulubova et al., 2002). Our crystal structure of Sly1p in complex with a 45 residue peptide derived from the N-terminus of Sed5p shows that Sed5p interacts predominantly with domain I of the arch-shaped Sly1p. In the crystal structure, only the previously anticipated binding region is ordered, and the structure suggests that the binding to Sly1p induces helix formation, since the region is unstructured in solution (Yamaguchi et al., 2002). This is in agreement with secondary structure prediction that suggested a propensity for α-helix formation at the N-termini of syntaxin-type SNARE proteins including Sed5p, Ufe1p and Tlg2p, as well as neuronal syntaxin 1 (Misura et al., 2002).
The N-terminal residues of Sed5p (1–10) form a conserved sequence signature among all Sed5p homologues (Yamaguchi et al., 2002). The most important residue in the Sly1p–Sed5p interaction is Phe10, which binds into a highly conserved hydrophobic pocket on Sly1p. Mutation to alanine abrogated Sed5p–Sly1p interactions (Dulubova et al., 2002), indicating a nucleation role for Phe10 in adopting the helical binding mode. Beyond residue 10, only key hydrophobic residues are conserved, which participate in the helical packing. Similarly important, according to the observed sequence conservation, are polar interactions, which form an extensive network. The N-terminal sequence of Ufe1p represents only a variation of the Sed5p sequence signature, where hydrophobic contacts substitute conserved polar side chains. It is therefore most likely that Sly1p binds the N-terminal region of Ufe1p (Yamaguchi et al., 2002) in the same way as Sed5p.
Interestingly, the binding mode can be extended to the interaction of Vps45p with the N-terminal region of Tlg2p, which shows substantial sequence homology to Sed5p including the proposed TXXF motif (Dulubova et al., 2002). In addition, the mapping of Vps45p residues on to the Sly1p structure shows a marked conservation of the peptide-binding region, which strongly indicates a similar interaction site on domain I. Together, the biochemical studies and our structural studies underline a common SM/syntaxin binding mode for these two transport systems (Dulubova et al., 2002; Yamaguchi et al., 2002). However, the affinities of Sed5p for Sly1p and of Tlg2p for Vps45p are mutually exclusive (Dulubova et al., 2002). This binding selectivity seems to be encoded in the polar contacts, since the hydrophobic interactions are conserved between both systems. Accordingly, in contrast to Sly1p–Sed5p, Arg3 and Arg13 are conserved in Tlg2p homologues, and Asp124, Glu127 and Glu134 in Vps45p homologues (Figure 4).
Neuronal Sec1 controls syntaxin and SNARE action in a negative (closed conformation; Dulubova et al., 1999; Misura et al., 2000) as well as in a positive way (Dresbach et al., 1998; Wu et al., 1999; Verhage et al., 2000), which implies that nSec1 may stay associated with the SNARE fusion machinery. The analysis of a crystal contact found in our structure of squid nSec1 (Bracher and Weissenhorn, 2001) indicates that neuronal Sec1 can bind a helical peptide in the corresponding position of the Sly1p–Sed5p interaction site. In addition, the crystal contact helix exhibits key contact residues, which are conserved with the N-terminal region of syntaxin (upstream of Habc) that has been suggested to be able to adopt a helical conformation (Misura et al., 2002). It is therefore tempting to suggest that neuronal Sec1 interacts with the N-terminus of syntaxin 1 as observed in the crystal contact, which is comparable with the observed Sly1p–Sed5p complex formation. The interaction may be weak and part of a multiprotein complex, which would then couple nSec1 to the fusion machinery at SNARE assembly and/or post-assembly steps. Such a complex would be in line with its postulated positive regulatory function (Jahn and Südhof, 1999; Chen and Scheller, 2001). In contrast, however, the N-termini of syntaxin homologues involved in non-neuronal exocytosis processes, such as Sso1p from S.cerevisiae and its homologues from S.pombe and A.thaliana show no homology to Sed5p, suggesting functional differences.
Although SNAREs can form multiple complexes in vitro (Fasshauer et al., 1999; Yang et al., 1999), their physiological functionalities depend on intact membrane anchors (McNew et al., 2000b). Interestingly, Sed5p no longer commits itself to non-physiological SNARE complexes in vitro when it is bound to Sly1p, indicating that Sly1p controls SNARE complex formation specificity (Peng and Gallwitz, 2002). This further implies that Sly1p has to interact with other parts of Sed5p during SNARE assembly. We have shown previously that squid nSec1 can dimerize through pseudo coiled-coil interactions of its helical hairpin structure, which might be involved in the control of SNARE complex formation (Bracher and Weissenhorn, 2001). Other possibilities include Sly1p interactions with the N-terminal three-helical bundle (Yamaguchi et al., 2002) and/or the SNARE motif. Such interactions may be weak and transient and therefore difficult to observe. Likewise, Vps45p stabilizes the syntaxin Tlg2p and positively regulates SNARE complex formation (Bryant and James, 2001). There, an SM protein-independent function was restored when the N-terminal half of Tlg2p was deleted. However, Tlg2p does not adopt a closed conformation like neuronal syntaxin 1 and Sso1p (Misura et al., 2000; Munson et al., 2000), and Vps45p-mediated stabilization may involve a novel type of syntaxin conformation (Dulubova et al., 2002).
Further alternative or additional SM protein interactions have been described for yeast Sec1p, which does not interact with its cognate syntaxin Sso1p despite its closed conformation structure (Munson et al., 2000). In contrast, it has been shown that Sec1p binds only to fully assembled SNARE complexes regulating post-SNARE complex assembly processes (Carr et al., 1999; Grote et al., 2000). Concerning the fourth yeast SM protein Vps33p, no direct interaction with its cognate syntaxin Vam3p that neither folds into a closed conformation (Dulubova et al., 2001) nor contains an N-terminal helical peptide sequence motif (Misura et al., 2002) was found, although both participate in a larger complex (Rieder and Emr, 1997; Sato et al., 2000; Wurmser et al., 2000).
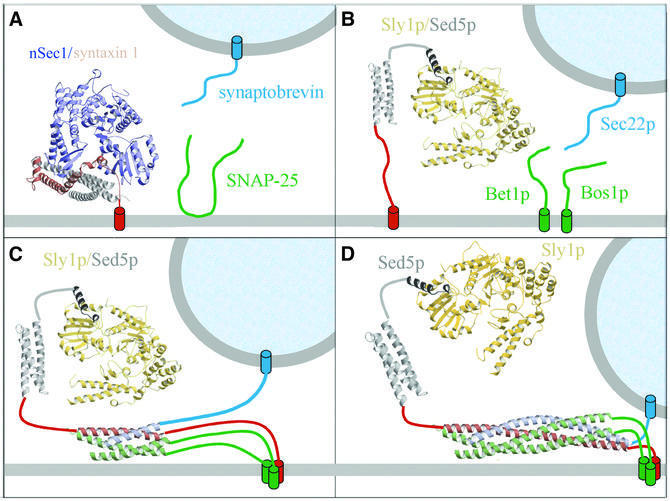
Together, these data indicate a versatile functional role for SM proteins, and we suggest the following model based on our structural data. First, Sly1p and Vps45p are recruited to their respective target membranes by binding to the N-terminus of their respective syntaxin (Figure 6B). This interaction is fundamentally different from the closed nSec1/syntaxin conformation, which prevents SNARE complex formation (Figure 6A). Secondly, the observed interaction allows the SM proteins to stay associated with the assembling SNARE complex, believed to follow a zippering mechanism beginning at the N-termini of the SNARE motifs (Fiebig et al., 1999). This is supported by biochemical evidence suggesting that Sly1p controls the specificity of SNARE pairing (Peng and Gallwitz, 2002) through potential transient interactions (Figure 6C). Thirdly, the binding mode allows the SM proteins to stay associated with syntaxin participating in assembled trans-SNARE complexes during the fusion process (Figure 6D), which is also evident from biochemical studies (Peng and Gallwitz, 2002). In addition, the observed interaction mode does not interfere with the disassembly of cis-SNARE complexes in vitro, thus potentially preparing syntaxin for another round of fusion (Peng and Gallwitz, 2002). Such an interaction might also be plausible for neuronal Sec1 and syntaxin 1, as suggested by the crystal contact. There, nSec1 may stay associated with syntaxin 1 after the transition from the closed (Figure 6A; Misura et al., 2000) to an open con formation (similar to Figure 2B), thereby exerting the pro posed positive regulatory function (Jahn and Südhof, 1999; Chen and Scheller, 2001). In summary, the spatial arrangement of the Sly1p–Sed5p peptide interaction provides multiple opportunities to participate in further protein– protein complexes, which may be involved in regulatory steps from docking to assembly, in membrane fusion and finally in disassembly of cis-SNARE complexes.
Fig. 6. Models of SM protein–syntaxin interactions. (A) Neuronal Sec1 (blue) binds syntaxin (Habc domain grey, SNARE motif red) in a closed conformation (Misura et al., 2000). (B) Sed5p was proposed to contain a Habc domain (grey), which folds into a three-helical bundle (Yamaguchi et al., 2002) (grey) and recruits Sly1p (yellow) to the target membrane through the N-terminal peptide (dark grey) interaction. (C) This allows Sly1p to exert potential regulatory functions such as controlling the specificity of SNARE complex assembly; Sly1p thus may contact the assembling trans-SNARE complex transiently. The secondary interaction mode for neuronal Sec1 and syntaxin 1 proposed herein would allow for a similar function. (D) Finally, Sly1p (as well as nSec1) may stay associated with the assembled trans-SNARE complex during membrane fusion, enabling participation in as yet unknown transient multiprotein interactions. Furthermore, the interaction with Sly1p does not interfere with the disassembly of SNARE complexes in vitro (Peng and Gallwitz, 2002), and thus Sly1p and Sed5p might stay associated for further rounds of docking and fusion.
Materials and methods
Protein expression and purification
Full-length S.cerevisiae Sly1p was expressed as a maltose binding fusion protein (MBP) in Escherichia coli BL21 codon+ cells using a modified pMal expression vector (New England Biolabs) (Bracher et al., 2002). Selenomethionine-labelled Sly1p protein was produced as described (Bracher et al., 2000). Saccharomyces cerevisiae Sed5p residues 1–45 were expressed as a GST fusion protein (vector pETM30; G.Stier, EMBL Heidelberg, Germany) in E.coli BL21 codon+ cells. pETM30 contains a TEV protease cleavage site and an N-terminal (of GST) His6 tag. Cell pellets from E.coli cells expressing Sly1p and Sed5p constructs were mixed and lysed together at 277 K by ultrasonication in a lysis buffer [50 mM Tris–HCl pH 7.5, 200 mM KCl, 20 mM imidazole and 1 mM phenylmethylsulfonyl fluoride (PMSF)]. The cleared lysate was applied to an Ni2+-chelating Sepharose (Amersham Biosciences) column and the MBP-Sly1:His6-GST–Sed5 complex was eluted with 250 mM imidazole in the lysis buffer. 0.1 mM EDTA and 0.5 mM dithiothreitol (DTT) were added, and the fusion proteins were cleaved overnight at 277 K with His6-TEV (1:100; w/w). After reduction of imidazole by ultrafiltration in lysis buffer, His-tagged TEV and GST were removed by Ni2+ affinity chromatography. The final purification step included size exclusion chromatography (Amersham Biosciences) in a buffer containing 20 mM Tris–HCl pH 7.2, 50 mM NaCl.
Crystallization and data collection
Crystals were obtained at 293 K from hanging drops containing a 1:1 mixture of complex at 11 mg/ml and well buffer (3.4–3.6 M Na-formate). For cryogenic data collection, the crystals were equilibrated in 3.6 M Na-formate and 5% glycerol for several days and flash-cooled in a gaseous nitrogen stream at 100 K. Data were collected at beamline BM14CRG at the European Synchrotron Light Source (ESRF, Grenoble, France). The selenomethionine-labelled crystal was collected at wavelengths 0.9875 Å (soft remote), 0.9793 Å (inflection point), 0.9789 Å (peak) and 0.9184 Å (hard remote).
Structure determination and refinement
The diffraction data were processed using the programs DENZO and SCALEPACK (Otwinowski and Minor, 1997; Table I). The anomalous differences from the highly redundant peak wavelength data to 3.2 Å resolution were used to find 10 of 11 expected Se sites with the program SnB (Weeks and Miller, 1999). Using only the peak wavelength data as pseudo single isomorphous displacement (SIR) data, the sites were refined and phases calculated with SOLVE (Terwilliger and Berendzen, 1999; Table I). Maximal likelihood density modification with RESOLVE (Terwilliger, 2000) yielded an excellent electron density map, which was superior to subsequently calculated maps including the diffraction data collected at other wavelengths. Guided by the peptide backbone of s-Sec1 (pdb code 1FVH) and the selenomethionine positions, the model of Sly1p was built according to the RESOLVE map using the program O (Jones et al., 1991) and electron density was identified corresponding to Sed5p after completion of the Sly1 model. The model was improved by alternating cycles of model building, conjugate gradient minimization and restrained individual B-factor refinement using CNS (Brünger et al., 1998). The coordinates were refined against the soft remote diffraction data between 30 and 3.0 Å resolution using the MLHL maximal likelihood target with the RESOLVE phases as constraint. In the final stage of refinement, a maximal likelihood target and model phases alone were used.
The final model contains Sly1p residues 10–244, 257–298, 314–361, 382–509 and 528–662, the Sed5p residues 1–21 plus a peptide GAMAG at the N-terminus and 20 water molecules (Table I). Due to missing electron density for side chains, 47 solvent-exposed residues were modelled as alanine. The model exhibits good stereochemistry with no outliers in the Ramachandran plot. The coordinates have been deposited in the RCSB Protein Data Bank (accession code 1MQS).
Sequence alignment and structure analysis
Figures were generated using the programs O (Jones et al., 1991), MOLSCRIPT (Kraulis, 1991), Raster 3D (Merritt and Bacon, 1997), ESPript (Gouet et al., 1999) and GRASP (Nicholls et al., 1991). Sequen ces were aligned using Clustal_X (Thompson et al., 1997). Secondary structure elements were assigned using the program DSSP (Kabsch and Sander, 1983). The degree of homology according to the Clustal_X alignment was calculated using the Risler scoring matrix (Risler et al., 1988) and the standard settings of ESPript. The buried surface was calculated with CNS (Brünger et al., 1998). For the superposition of s-Sec1 and Sly1p–Sed5p, the program LSQMAN was used (Kleywegt and Jones, 1994).
Acknowledgments
Acknowledgements
We thank Drs M.Walsh and H.Belrhali for help with data collection at the ESRF CRG beamline BM14, Drs Y.Kosodo and K.Yoda for providing Sly1p and Sed5p cDNA clones, and Dr J.Garin and Ms M.Louwagie for N-terminal sequencing. A.B. was supported by a Marie Curie fellowship from the European Union.
References
- Bracher A. and Weissenhorn,W. (2001) Crystal structures of neuronal squid Sec1 implicate inter-domain hinge movement in the release of t-SNAREs. J. Mol. Biol., 306, 7–13. [DOI] [PubMed] [Google Scholar]
- Bracher A., Perrakis,A., Dresbach,T., Betz,H. and Weissenhorn,W. (2000) The X-ray crystal structure of neuronal Sec1 from squid sheds new light on the role of this protein in exocytosis. Structure Fold. Des., 8, 685–694. [DOI] [PubMed] [Google Scholar]
- Bracher A., Kadlec,J., Betz,H. and Weissenhorn,W. (2002) X-ray structure of a neuronal complexin–SNARE complex from squid. J. Biol. Chem., 277, 26517–26523. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Bryant N.J. and James,D.E. (2001) Vps45p stabilizes the syntaxin homologue Tlg2p and positively regulates SNARE complex formation. EMBO J., 20, 3380–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calakos N., Bennett,M.K., Peterson,K.E. and Scheller,R.H. (1994) Protein–protein interactions contributing to the specificity of intracellular vesicular trafficking. Science, 263, 1146–1149. [DOI] [PubMed] [Google Scholar]
- Cao X., Ballew,N. and Barlowe,C. (1998) Initial docking of ER-derived vesicles requires Uso1p and Ypt1p but is independent of SNARE proteins. EMBO J., 17, 2156–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr C.M., Grote,E., Munson,M., Hughson,F.M. and Novick,P.J. (1999) Sec1p binds to SNARE complexes and concentrates at sites of secretion. J. Cell Biol., 146, 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.A. and Scheller,R.H. (2001) SNARE-mediated membrane fusion. Nat. Rev. Mol. Cell. Biol., 2, 98–106. [DOI] [PubMed] [Google Scholar]
- Chothia C. (1984) Principles that determine the structure of proteins. Annu. Rev. Biochem., 53, 537–572. [DOI] [PubMed] [Google Scholar]
- Cowles C.R., Emr,S.D. and Horazdovsky,B.F. (1994) Mutations in the VPS45 gene, a SEC1 homologue, result in vacuolar protein sorting defects and accumulation of membrane vesicles. J. Cell Sci., 107, 3449–3459. [DOI] [PubMed] [Google Scholar]
- Dascher C., Ossig,R., Gallwitz,D. and Schmitt,H.D. (1991) Identification and structure of four yeast genes (SLY) that are able to suppress the functional loss of YPT1, a member of the RAS superfamily. Mol. Cell. Biol., 11, 872–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresbach T., Burns,M.E., O’Connor,V., DeBello,W.M., Betz,H. and Augustine,G.J. (1998) A neuronal Sec1 homolog regulates neurotransmitter release at the squid giant synapse. J. Neurosci., 18, 2923–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I., Sugita,S., Hill,S., Hosaka,M., Fernandez,I., Südhof,T.C. and Rizo,J. (1999) A conformational switch in syntaxin during exocytosis: role of munc18. EMBO J., 18, 4372–4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I., Yamaguchi,T., Wang,Y., Südhof,T.C. and Rizo,J. (2001) Vam3p structure reveals conserved and divergent properties of syntaxins. Nat. Struct. Biol., 8, 258–264. [DOI] [PubMed] [Google Scholar]
- Dulubova I., Yamaguchi,T., Gao,Y., Min,S.W., Huryeva,I., Südhof,T.C. and Rizo,J. (2002) How Tlg2p/syntaxin 16 ‘snares’ Vps45. EMBO J., 21, 3620–3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasshauer D., Sutton,R.B., Brünger,A.T. and Jahn,R. (1998) Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl Acad. Sci. USA, 95, 15781–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasshauer D., Antonin,W., Margittai,M., Pabst,S. and Jahn,R. (1999) Mixed and non-cognate SNARE complexes. Characterization of assembly and biophysical properties. J. Biol. Chem., 274, 15440–15446. [DOI] [PubMed] [Google Scholar]
- Fiebig K.M., Rice,L.M., Pollack,E. and Brünger,A.T. (1999) Folding intermediates of SNARE complex assembly. Nat. Struct. Biol., 6, 117–123. [DOI] [PubMed] [Google Scholar]
- Fukuda R., McNew,J.A., Weber,T., Parlati,F., Engel,T., Nickel,W., Rothman,J.E. and Söllner,T.H. (2000) Functional architecture of an intracellular membrane t-SNARE. Nature, 407, 198–202. [DOI] [PubMed] [Google Scholar]
- Garcia E.P., Gatti,E., Butler,M., Burton,J. and De Camilli,P. (1994) A rat brain Sec1 homologue related to Rop and UNC18 interacts with syntaxin. Proc. Natl Acad. Sci. USA, 91, 2003–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P., Courcelle,E., Stuart,D.I. and Metoz,F. (1999) ESPript: multiple sequence alignments in PostScript. Bioinformatics, 15, 305–308. [DOI] [PubMed] [Google Scholar]
- Grabowski R. and Gallwitz,D. (1997) High-affinity binding of the yeast cis-Golgi t-SNARE, Sed5p, to wild-type and mutant Sly1p, a modulator of transport vesicle docking. FEBS Lett., 411, 169–172. [DOI] [PubMed] [Google Scholar]
- Grote E., Carr,C.M. and Novick,P.J. (2000) Ordering the final events in yeast exocytosis. J. Cell Biol., 151, 439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson P.I., Roth,R., Morisaki,H., Jahn,R. and Heuser,J.E. (1997) Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell, 90, 523–535. [DOI] [PubMed] [Google Scholar]
- Harrison S.D., Broadie,K., van de Goor,J. and Rubin,G.M. (1994) Mutations in the Drosophila Rop gene suggest a function in general secretion and synaptic transmission. Neuron, 13, 555–566. [DOI] [PubMed] [Google Scholar]
- Hata Y., Slaughter,C.A. and Südhof,T.C. (1993) Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature, 366, 347–351. [DOI] [PubMed] [Google Scholar]
- Jahn R. and Südhof,T.C. (1999) Membrane fusion and exocytosis. Annu. Rev. Biochem., 68, 863–911. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kabsch W. and Sander,C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J. and Jones,T.A. (1994) A super position. CCP4/ESF–EACBM Newslett. Protein Crystallogr., 31, 9–14. [Google Scholar]
- Kosodo Y., Noda,Y. and Yoda,K. (1998) Protein–protein interactions of the yeast Golgi t-SNARE Sed5 protein distinct from its neural plasma membrane cognate syntaxin 1. Biochem. Biophys. Res. Commun., 250, 212–216. [DOI] [PubMed] [Google Scholar]
- Kraulis P. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 924–950. [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–290. [Google Scholar]
- McNew J.A., Parlati,F., Fukuda,R., Johnston,R.J., Paz,K., Paumet,F., Söllner,T.H. and Rothman,J.E. (2000a) Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature, 407, 153–159. [DOI] [PubMed] [Google Scholar]
- McNew J.A., Weber,T., Parlati,F., Johnston,R.J., Melia,T.J., Söllner,T.H. and Rothman,J.E. (2000b) Close is not enough: SNARE-dependent membrane fusion requires an active mechanism that transduces force to membrane anchors. J. Cell Biol., 150, 105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt E.A. and Bacon,D.J. (1997) Raster3D photorealistic graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Misura K.M.S., Scheller,R.H. and Weis,W.I. (2000) Three-dimensional structure of the neuronal-Sec1–syntaxin 1a complex. Nature, 404, 355–362. [DOI] [PubMed] [Google Scholar]
- Misura K.M., Bock,J.B., Gonzalez,L.C.,Jr, Scheller,R.H. and Weis,W.I. (2002) Three-dimensional structure of the amino-terminal domain of syntaxin 6, a SNAP-25 C homolog. Proc. Natl Acad. Sci. USA, 99, 9184–9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson M., Chen,X., Cocina,A.E., Schultz,S.M. and Hughson,F.M. (2000) Interactions within the yeast t-SNARE Sso1p that control SNARE complex assembly. Nat. Struct. Biol., 7, 894–902. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Nichols B.J., Ungermann,C., Pelham,H.R., Wickner,W.T. and Haas,A. (1997) Homotypic vacuolar fusion mediated by t- and v-SNAREs. Nature, 387, 199–202. [DOI] [PubMed] [Google Scholar]
- Novick P. and Schekman,R. (1979) Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 76, 1858–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossig R., Dascher,C., Trepte,H.H., Schmitt,H.D. and Gallwitz,D. (1991) The yeast SLY gene products, suppressors of defects in the essential GTP-binding Ypt1 protein, may act in endoplasmic reticulum-to-Golgi transport. Mol. Cell. Biol., 11, 2980–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Parlati F., Varlamov,O., Paz,K., McNew,J.A., Hurtado,D., Söllner,T.H. and Rothman,J.E. (2002) Distinct SNARE complexes mediating membrane fusion in Golgi transport based on combinatorial specificity. Proc. Natl Acad. Sci. USA, 99, 5424–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng R. and Gallwitz,D. (2002) Sly1 protein bound to Golgi syntaxin Sed5p allows assembly and contributes to specificity of SNARE fusion complexes. J. Cell Biol., 157, 645–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson M.R., Burd,C.G. and Emr,S.D. (1999) Vac1p coordinates Rab and phosphatidylinositol 3-kinase signaling in Vps45-dependent vesicle docking/fusion at the endosome. Curr. Biol., 9, 159–162. [DOI] [PubMed] [Google Scholar]
- Pevsner J., Hsu,S.C., Braun,J.E., Calakos,N., Ting,A.E., Bennett,M.K. and Scheller,R.H. (1994) Specificity and regulation of a synapic vesicle docking complex. Neuron, 13, 353–361. [DOI] [PubMed] [Google Scholar]
- Rieder S.E. and Emr,S.D. (1997) A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol. Biol. Cell, 8, 2307–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risler J.L., Delorme,M.O., Delacroix,H. and Henaut,A. (1988) Amino acid substitutions in structurally related proteins. A pattern recognition approach. Determination of a new and efficient scoring matrix. J. Mol. Biol., 204, 1019–1029. [DOI] [PubMed] [Google Scholar]
- Sato T.K., Rehling,P., Peterson,M.R. and Emr,S.D. (2000) Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol. Cell, 6, 661–671. [DOI] [PubMed] [Google Scholar]
- Siniossoglou S. and Pelham,H.R. (2001) An effector of Ypt6p binds the SNARE Tlg1p and mediates selective fusion of vesicles with late Golgi membranes. EMBO J., 20, 5991–5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogaard M., Tani,K., Ye,R.R., Geromanos,S., Tempst,P., Kirchhausen,T., Rothman,J.E. and Söllner,T. (1994) A rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell, 78, 937–948. [DOI] [PubMed] [Google Scholar]
- Söllner T., Whiteheart,S.W., Brunner,M., Erdjument-Bromage,H., Geromanos,S., Tempst,P. and Rothman,J.E. (1993a) SNAP receptors implicated in vesicle targeting and fusion. Nature, 362, 318–324. [DOI] [PubMed] [Google Scholar]
- Söllner T., Bennett,M.K., Whiteheart,S.W., Scheller,R.H. and Rothman,J.E. (1993b) A protein assembly–disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation and fusion. Cell, 75, 409–418. [DOI] [PubMed] [Google Scholar]
- Sutton R.B., Fasshauer,D., Jahn,R. and Brünger,A.T. (1998) Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature, 395, 347–353. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. (2000) Maximum likelihood density modification. Acta Crystallogr. D, 56, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated structure solution for MIR and MAD. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui M.M., Tai,W.C. and Banfield,D.K. (2001) Selective formation of Sed5p-containing SNARE complexes is mediated by combinatorial binding interactions. Mol. Biol. Cell., 12, 521–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage M. et al. (2000) Synaptic assembly of the brain in the absence of neurotransmitter secretion. Science, 287, 864–869. [DOI] [PubMed] [Google Scholar]
- Weber T., Zemelman,B.V., McNew,J.A., Westermann,B., Gmachl,M., Parlati,F., Söllner,T.H. and Rothman,J.E. (1998) SNAREpins: minimal machinery for membrane fusion. Cell, 92, 759–772. [DOI] [PubMed] [Google Scholar]
- Weeks C.M. and Miller,R. (1999) The design and implementation of SnB v2.0. J. Appl. Crystallogr., 32, 120–124. [Google Scholar]
- Wu M.N., Fergestad,T., Lloyd,T.E., He,Y., Broadie,K. and Bellen,H.J. (1999) Syntaxin 1A interacts with multiple exocytic proteins to regulate neurotransmitter release in vivo. Neuron, 23, 593–605. [DOI] [PubMed] [Google Scholar]
- Wurmser A.E., Sato,T.K. and Emr,S.D. (2000) New component of the vacuolar class C–Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J. Cell Biol., 151, 551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Dulubova,I., Min,S.W., Chen,X., Rizo,J. and Südhof,T.C. (2002) Sly1 binds to Golgi and ER syntaxins via a conserved N-terminal peptide motif. Dev. Cell, 2, 295–305. [DOI] [PubMed] [Google Scholar]
- Yang B., Steegmaier,M., Gonzalez,L.C. and Scheller,R.H. (2000) nSec1 binds a closed conformation of syntaxin1A. J. Cell Biol., 148, 247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M. and McBride,H. (2001) Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol., 2, 107–117. [DOI] [PubMed] [Google Scholar]