Abstract
Insertional RNA editing in Physarum polycephalum is a complex process involving the specific addition of non-templated nucleotides to nascent mitochondrial transcripts. Since all four ribonucleotides are substrates for the editing activity(s), both the site of insertion and the identity of the nucleotide to be added at a particular position must be specified, but the signals for these events have yet to be elucidated. Here we report the occurrence of sporadic errors in RNAs synthesized in vitro. These mistakes, which include omission of encoded nucleotides as well as misinsertions, occur only on templates that support editing. The pattern of these misediting events indicates that editing site recognition and nucleotide addition are separable events, and that the recognition step involves features of the mitochondrial template that are required for editing. The larger deletions lack all templated nucleotides between editing sites, suggesting that the transcription/editing apparatus can ‘jump’ from one insertion site to another, perhaps mediated by interactions with editing determinants, while smaller omissions most likely reflect misalignment of the transcript upon resumption of templated RNA synthesis.
Keywords: co-transcriptional/discontinuous transcription/insertional RNA editing/misediting/site recognition
Introduction
Transcripts from a growing number of genes in a wide variety of organisms are being found to be subject to discrete site-specific alterations leading to the expression of functional RNAs and/or variant gene products (Gott and Emeson, 2000; Keegan et al., 2001). Many of these changes occur post-transcriptionally either by base substitution (Gerber and Keller, 2001; Seeburg, 2002) or by addition and deletion of nucleotides via cleavage and religation of the RNA (Madison-Antenucci et al., 2002; Stuart and Panigrahi, 2002). However, RNA editing can also occur by incorporation of extra nucleotides at the growing end of the nascent transcript. Such co-transcriptional editing is employed by a number of viruses, including paramyxoviruses (Thomas et al., 1988; Vidal et al., 1990b; Pelet et al., 1991) and Ebola virus (Volchkov et al., 1995; Sanchez et al., 1996), as a strategy for expressing multiple gene products from their compact genomes.
A subset of the mitochondrial (mt) RNAs in Physarum is also edited by the co-transcriptional addition of nucleotides that are not templated in a conventional manner (Cheng et al., 2001), resulting in the restoration of conserved open reading frames in mRNAs and structural features within tRNAs and rRNAs (Miller et al., 1993). In contrast to the changes in viral RNAs, which occur by polymerase stuttering at a single position within the genome (Vidal et al., 1990a), Physarum editing events include the insertion of eight different mono- and di-nucleotide combinations at hundreds of distinct sites in diverse sequence contexts (Miller et al., 1993; Gott, 2001; Takano et al., 2001). The mechanism by which these site-specific changes are accomplished is currently unknown. At a minimum, insertional editing in Physarum mitochondria must require: (i) recognition of an editing site; (ii) addition of non-encoded nucleotide(s); and (iii) extension of the non-paired nucleotide(s) in a template-directed fashion. In addition, because non-encoded nucleotides are added to the 3′ end of the nascent RNA chain (Cheng et al., 2001), transcription elongation must be interrupted and then restarted at the appropriate sites. Hence, it is likely that one or more of these steps is accompanied by and/or requires a conformational change in the transcription/editing complex.
Physarum insertional editing has been studied in partially purified mitochondrial transcription elongation complexes (mtTECs) (Cheng and Gott, 2000); run-on transcripts from these preparations are partially edited. In recent work investigating the features of the transcription template required for editing (Byrne and Gott, 2002), we made use of a series of recombinant templates generated by digestion of DNA present in mtTECs followed by either ligation to exogenous DNA or self-ligation of the mtTEC DNA fragments. In both cases, RNAs transcribed in vitro from mtTEC DNA upstream of the ligation junction were partially edited. Interestingly, however, only DNA derived from native mtTECs supported editing downstream of the junction; the segments of run-on transcripts synthesized from either cloned Physarum DNA or deproteinized mtDNA were completely unedited. These studies led us to conclude that some feature of the native template is required for addition of non-encoded nucleotides and that editing signals are local (Byrne and Gott, 2002).
Surprisingly, some of the RNAs transcribed from chimeric templates contained misinsertions and other errors at editing sites, interspersed with unedited and correctly edited sites. The frequency of the mistakes seen in these experiments was higher than expected based on previous work, which had shown that RNA editing in Physarum mitochondria is a highly accurate process (Visomirski-Robic and Gott, 1995). Indeed, essentially all cDNA clones derived from total Physarum mtRNAs are fully edited (Gott et al., 1993; Miller et al., 1993; Rundquist and Gott, 1995; Wang et al., 1999), although extremely rare, partially edited clones can be selected out in biased searches (Wang et al., 1999). We therefore examined the extent of misediting that occurs both in vivo and in vitro from the intact genome. Here we describe the sequence errors that we have observed at C insertion sites, which include addition of the wrong nucleotide, incorporation of a second untemplated nucleotide, omission of encoded nucleotides adjacent to editing sites and, strikingly, occasional loss of all templated nucleotides between adjoining editing sites (‘inter-site deletions’). All such sequence deviations are collectively referred to by the term ‘misediting’ in this work. Importantly, we do not observe deletions or insertions of non-templated nucleotides at locations other than editing sites, nor does misediting occur on templates that fail to support nucleotide addition, strongly suggesting that these events are due to errors that take place after the recognition step of the editing process.
Results
Misediting in RNAs synthesized from rearranged mtTEC fragments
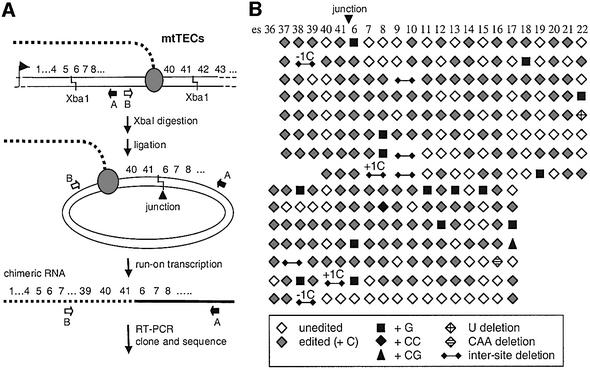
We have recently developed methods to create chimeric transcription/editing templates in order to investigate the requirements for nucleotide insertion (Byrne and Gott, 2002). One of the chimeric templates that supports efficient editing in vitro is shown schematically in Figure 1A. This template was generated by digesting DNA present in mtTECs with XbaI and ligating the resulting sticky ends, leading to the production of genomic rearrangements that include a circularized fragment derived from the atp gene (Byrne and Gott, 2002). In this construct, the template sequences just downstream of atp editing site 41 (es41) are ligated to DNA upstream of atp editing site 6 (es6), with the transcription/editing machinery assembled on mtDNA in vivo serving as the source of the mtRNA polymerase and potential editing factors. Run-on transcription from this template generates atp RNAs of rearranged sequence, which can be amplified by RT–PCR using primers that face away from each other in the intact genome (Figure 1A). Twenty-five independent clones were generated using two distinct sets of primers that together encompass a region of chimeric atp mRNA covering 23 C insertion sites (es36–41 and es6–22). Ten of these clones contained a mixture of edited and unedited sites (Byrne and Gott, 2002; data not shown), as expected based on previous in vitro labeling experiments (Cheng and Gott, 2000). Interestingly, however, the remaining 15 clones contained one or more errors at editing sites.
Fig. 1. Misedited sites are interspersed among correctly edited and unedited sites in transcripts derived from circularized DNA fragments of mtTECs. (A) Generation of circularized mtTEC fragments. (B) Patterns of misediting at 23 C insertion sites within the rearranged atp mRNA. Sites that have been correctly edited are indicated by a gray diamond, unedited sites by an open diamond. One completely unedited clone that could have been derived from either unedited RNA or residual DNA was also isolated in this experiment (data not shown). Symbols for misedited sites indicate the type of nucleotide changes at a given site. Insertions of nucleotides other than C are indicated by black symbols: black square, G insertion; black diamond, CC insertion; black triangle, CG insertion. Nucleotide deletions at editing sites are indicated by diamonds with a plus sign (U deletion) or hatched diamonds (CAA deletion). Deletions encompassing regions between adjacent editing sites are given by a line with small diamonds at either end; +1C indicates that the clone has one more C than the genomic sequence in the region of the deletion, –1C indicates that the clone has one fewer C than the genomic sequence in the region of the deletion.
The types and patterns of misediting observed in these chimeric atp mRNAs are represented schematically in Figure 1B, with each C insertion site represented as a symbol. Because each cDNA is ∼450–560 bp long, sequences between editing sites are not shown; sequence contexts of editing sites are presented in Table I and discussed below. Out of a total of 507 C insertion sites assayed, 309 (61%) were correctly edited, 163 (32%) were unedited, 15 (3%) had a G insertion, 1 had a CC insertion and 1 a CG insertion. In addition, we observed two cases involving omission of encoded nucleotides adjacent to an editing site (–U and –CAA) and eight deletions that lacked all nucleotides between adjacent editing sites (inter-site deletions, described below). In total, 6.9% (35 of 507) of the editing sites in these chimeric RNAs were misedited in some way. It should be noted that this frequency may underestimate the occurrences of these events somewhat, as we cannot determine the precise point at which in vitro RNA synthesis begins in these unlabeled run-on transcripts. These errors were remarkably widespread: 19 of the 23 atp editing sites examined in these clones were involved in one or more examples of misediting. Consistent with our previous conclusion that sites are edited independently (Visomirski-Robic and Gott, 1997; Byrne and Gott, 2002), unedited, correctly edited and misedited sites are interspersed in diverse patterns. However, both in this experiment and those described below, non-templated nucleotide insertions are found only at editing sites and each of the observed deletions includes nucleotides immediately adjacent to editing sites. Thus, these errors are clearly related to editing.
Table I. Misediting occurs at C insertion sites in many contexts.
| atp editing site contextsa | Misediting event(s) observed | |
|---|---|---|
| es1 | GACCCGUCAAU.GGUCAAAUUAUUUC | +G, Δ1/2 (2) |
| es2 | AUUAUUUCUGU.AAAGAUGGUGUUGC | +G, Δ1/2 (2) |
| es3 | UUGUUACAGGA.UUGAUAAUATTCAA | Δ3/4 |
| es4 | GUAGAAUUUAU.UCUAAGGGUUUAAC | Δ3/4 |
| es5 | AAGGGUUUAAC.GGUAUGGCUCUUAA | |
| es6 | GCUGAACAAGU.GGUUGUAUUAUUUU | +G (3) |
| es7 | GGUGAUGAUAC.UCUGUUCAACAAGA | Δ7/8 |
| es8 | GAUGAUUCUGU.CGUGCUUUAAAUAC | +G (2), +CC, Δ7/8, –C |
| es9 | GCUUUAAAUAC.UUAGUCAAAACCCU | Δ9/10 (3) |
| es10 | UUAGUCAAAAC.CCUGUAGGUUAUGG | Δ9/10 (3) |
| es11 | GGUCGUGUUGU.GAUGGUAUUGGUAA | +G (2) |
| es12 | AUUGGUAAUUU.AUCGAUGGUGGUGA | +G |
| es13 | GGUGAAACUAU.GCUUUUGAGGAAUA | +G |
| es14 | UAUCUCAAUGU.GAACGUAAAGCUCC | |
| es15 | CCUGGUGUUAU.ACUCGUGAAUCUGU | +G |
| es16 | CUGUUACUGAA.CAAUGUUAACUGGU | –CAA |
| es17 | GGUUAUAAAAU.GUUGAUUCUAUGUU | +G, +CG |
| es18 | AUGUUACCUAU.GGACGUGGUCAAAG | +G |
| es19 | UUGGUGAUCGU.AAACAGGUAAAACU | +G |
| es20 | ACUAUUGCUAU.GAUACUAUUCUUAA | |
| es21 | CAACGUUAUAC.AAUGAAGAAGAUAU | |
| es22 | GAUAUUGAUCU.UAUUGUGUGUAUGU | +G, –U (2) |
| es36 | GGUUCUUUAAC.GCUUUACCUAUUGU | |
| es37 | GAAACACAAGC.GGUGAUUUAUCUGG | Δ37/38 |
| es38 | CUGGUUAUAUC.CAACUAAUAUUAUU | +G (2), Δ37/38, Δ38/39 (2) |
| es39 | AUUUCUAUUAC.GAUGGACAAAUUUU | Δ38/39 (2), +C/–G, +C/–GAU |
| es40 | UGGAAAAAGAU.UAUUCUUUAAAGCU | Δ40/41 (2), –U, +C/–U (2) |
| es41 | UUAAAGGUAUC.GUCCAGCUGUUAAU | Δ40/41 (2) |
| es42 | GUUUCUAGAGU.GGUUCUAAAGCUCA | |
| es43 | GCUCAACCAUA.GCUUUACGUUUAGU | +G |
| es44 | AUCAACUUGCU.AAUAUCGUGAAUAU | +G, Δ44/45 |
| es45 | GAAUAUUCUGU.UUUGCACAAUUUGA | +G (2), –U, Δ44/45 |
| es46 | GAUAAUGAUAU.GAUGAUGUUACAAG | +G |
| es47 | UAAUAGAGGUG.UUUAUUAACUGAAA | +G (2), +CC, +U |
| es48 | UCCUAAUAUGC.AAUGCAAUUAUAUA | |
| es49 | GUUUUAAUUAU.UUAGCUGGUGCUUU | +G |
aSummary of misediting events observed at atp sites described in Figures 1–4. Sites of C insertion are indicated with a dot. Note that when a C insertion site is flanked by one or more encoded C residues, the position at which the nucleotide is added cannot be precisely determined. The extent of this ambiguity is denoted by an underline. The alignments chosen for editing sites 8 and 38 are based on sites of G misinsertion. Inter-site deletions are indicated by Δ (endpoints). Numbers in parentheses indicate the number of independent clones with a given misediting event.
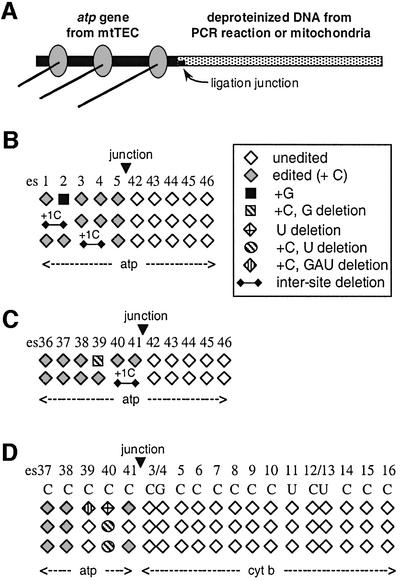
We have also examined transcripts derived from a series of chimeric templates involving the ligation of deproteinized DNA fragments to XbaI-digested mtTEC DNA (Figure 2A). These DNA fragments were either generated by PCR from cloned atp sequences (Figure 2B and C) or by digestion of deproteinized mtDNA (Figure 2D), as described previously (Byrne and Gott, 2002). Of eight chimeric cDNA clones isolated in the experiment shown in Figure 2B, three contained sites of misediting (Figure 2B; data not shown). Similarly, misediting was observed in 2 of 13 clones in which the sequence of the atp gene was reconstructed (Figure 2C; Byrne and Gott, 2002; data not shown) and in 3 of 14 clones from chimeric templates using mtDNA (Figure 2D; Byrne and Gott, 2002). The patterns of misediting observed in these experiments are shown schematically in Figure 2. Strikingly, none of the 301 sites transcribed from deproteinized DNA was either edited or misedited, despite the fact that these same transcripts contained both edited and misedited sites in the upstream portion derived from mtTEC DNA. Thus, it appears that the same signals are required for both editing and misediting.
Fig. 2. Deproteinized DNA fails to support either editing or misediting. (A) Generation of chimeric templates by ligation of XbaI-cleaved mtTECs to exogenous, deproteinized DNA fragments with compatible sticky ends. (B–D) Patterns of misediting within RNAs derived from chimeric templates. Downstream exogenous atp DNA for (B and C) was made by PCR from cloned sequences. Downstream exogenous DNA for (D) is an AvrII–EcoNI restriction fragment of deproteinized mtDNA that contains a portion of the cytochrome b (cytb) gene. Hatched square, C insertion followed by G deletion; hatched circle, C insertion followed by U deletion; vertically hatched diamond, C insertion followed by GAU deletion; other symbols as in Figure 1.
The relatively high frequency of misediting in transcripts made in vitro from chimeric templates was seemingly at odds with previous characterization of steady-state RNAs from intact mitochondria (Miller et al., 1993; Rundquist and Gott, 1995). It is unlikely that the misediting events described here were simply the result of limiting substrate concentrations, because these experiments were performed at concentrations of CTP (500 µM) known to support efficient editing in mtTEC preparations (Cheng et al., 2001). However, there are a number of other possible explanations for the observed differences, including: (i) rapid degradation of incorrectly edited RNAs in vivo, leading to under-representation of misedited transcripts in total RNA pools; (ii) nucleotide changes introduced during the assay (i.e. during the reverse transcription and/or PCR steps); and (iii) loss of template-associated editing factors during either mtTEC isolation or genome rearrangement procedures. To distinguish between these possibilities, we cloned and sequenced RT–PCR products using three unrearranged substrates: RNAs present in mtTECs prior to run-on transcription, RNAs from mtTECs after run-on transcription and run-on RNAs synthesized by mock-digested mtTECs (no XbaI present).
Nascent RNAs synthesized in vivo are not detectably misedited
To determine whether a significant level of misediting occurs during RNA synthesis in vivo, we cloned and sequenced individual cDNAs derived from RNA isolated from mtTECs not subjected to run-on transcription. This RNA pool should be highly enriched for transcripts still associated with the mtRNA polymerase (Cheng and Gott, 2000), and therefore representative of the overall accuracy of RNA editing of nascent transcripts in vivo. Sig nificantly, all clones were completely and accurately edited (Figure 3, top). The absence of unedited or misedited sites in the 140 C insertion sites assayed in these cDNA clones indicates that the sequence aberrations described in the previous section are not a function of errors during reverse transcription or the PCR reactions, and argues strongly that misediting within nascent RNAs is quite rare in vivo (<0.7%).
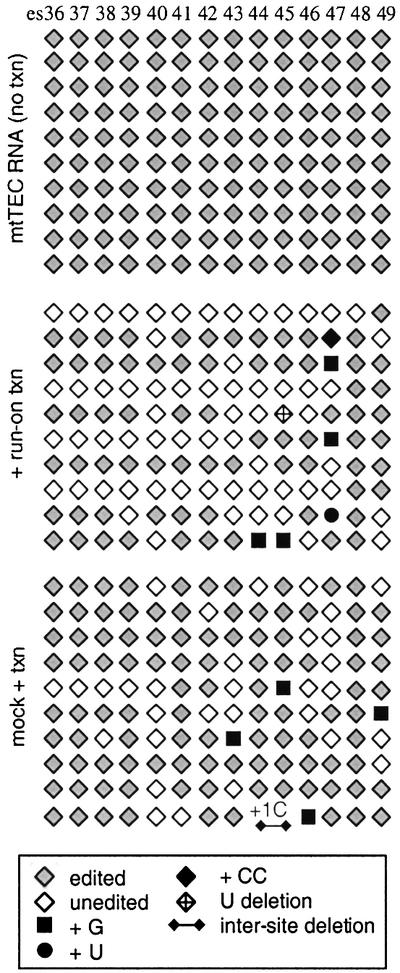
Fig. 3. Misediting occurs during run-on transcription in mtTECs. Editing patterns of mtTEC RNAs at 14 C insertion sites within the atp mRNA are shown using the same symbols as in Figure 1, with black circles indicating U insertions. Top, RNAs isolated from mtTECs not subjected to run-on transcription (i.e. synthesized in vivo); middle, after run-on transcription; bottom, after incubation in conditions used for restriction enzyme digestion, but without the enzyme (mock-treated) followed by run-on transcription. No fully edited sequences were seen after run-on transcription, which is not unexpected, as mtTEC RNA made in vivo may comprise only a small percentage of the RNA present after run-on transcription.
RNAs synthesized by partially purified mtTECs are misedited at a low level
We next asked, using the same assay, whether transcripts made by mtTECs under high nucleotide concentrations in vitro were misedited at a measurable level (Figure 3, middle). Notably, misediting was observed at 5% (7 of 140) of the C insertion sites assessed, including four instances of G insertion, a CC insertion, a U insertion and a U deletion. Similar results were also obtained with mtTECs incubated under the higher salt conditions used for restriction digestion (but without added enzymes) prior to run-on transcription, with four instances of G insertion and an inter-site deletion (Figure 3, bottom). In both sets of clones, there is an apparent bias in the distribution of misedited sites in that no misincorporation is seen before es43. We attribute this to two features of this experiment: (i) as mentioned above, upstream sections of each transcript prior to the first observed unedited or misedited site may have been made in vivo; and (ii) the 5′ portion of the region flanked by these primers includes a number of sites that appear to be less prone to G misinsertions. In comparison, misediting is seen at es37–41 in the 5′ portion of the transcripts represented in Figure 1. This may be due to continued transcription around circular templates, such that a larger fraction of the population of transcripts from this region is made in vitro. Taken together, these data suggest that the decrease in the fidelity of editing is not due to cleavage and ligation of the mtTECs, or to incubation under the conditions used for these manipulations, but is instead a feature of run-on transcription in partially purified mtTECs. This could be due to partial loss or inactivation of factors involved in editing during mtTEC isolation or other alterations of the transcription/editing template under our in vitro conditions.
Misediting contexts
Misediting occurs in a wide variety of contexts, including 25 of the 31 atp sites (81%) encompassed by the cDNA clones depicted in Figures 1–3. Table I displays the sequences surrounding the editing sites, which we have examined in the atp gene in these and other experiments (Figures 1–4; data not shown), and summarizes the misediting events that have been seen at each site in vitro. Several sites seem to be especially prone to misediting, with some favoring nucleotide misincorporation (es6 and 47) and some inter-site deletions (es9 and 10), while others display multiple types of deletion (es39 and 40) or other errors (es8, 38 and 45). Misediting is not specific to the atp mRNA, as we see similar patterns of misediting at C insertion sites within the run-on transcripts from the cytochrome oxidase subunit 1 (coI) and small subunit rRNA (ssu) genes, both in the context of an intact mitochondrial genome and other chimeric templates (data not shown). Moreover, these editing mistakes do not appear to be systematic.
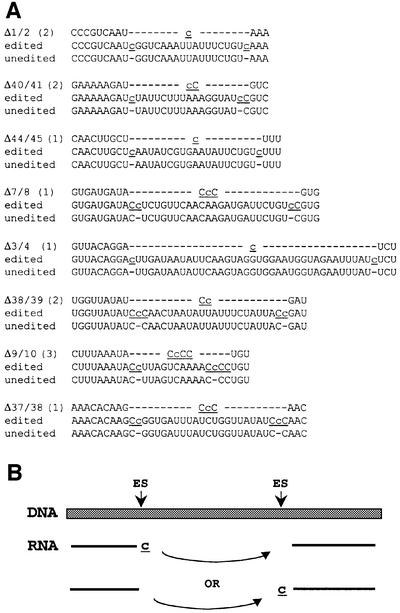
Fig. 4. Deletions between consecutive atp editing sites. (A) Sequences surrounding each of the inter-site deletions are shown aligned with genomic (unedited) and fully edited sequences. The number of independent occurrences of each deletion is given in parentheses. Note that Δ1/2, Δ40/41, Δ44/45, Δ7/8 and Δ3/4 result in the addition of a C residue relative to the sum of the number of Cs encoded immediately adjacent to each pair of editing sites, Δ38/39 results in the net removal of a C, and Δ9/10 and Δ37/38 preserve the number of Cs. The positions of the ambiguous editing sites 8, 38 and 39 are depicted based on other misediting events, as discussed in the text. Deletions include clones described in Materials and methods as well as those shown schematically in Figures 1–3. (B) Model for the generation of inter-site deletions involving jumping of the transcription/editing machinery from one editing site to the next, after (top) or before (bottom) C insertion.
Although there are no obvious contextual features common to all misedited sites in either upstream or downstream regions, certain types of misediting occur more frequently in particular contexts. For instance, all of the U deletions observed thus far occur in the context of a C insertion site flanked by encoded Us (Table I; data not shown). In addition, the small number of U insertions that we have seen, which include two instances at coI es35 and one at coI es43 (Figure 3; data not shown), are all in the context of one or more downstream encoded Us. Another possible instance of context effect involves ‘ambiguous’ insertion sites. Of the 15 different C insertion sites that serve as endpoints of atp inter-site deletions, eight are adjacent to encoded Cs, suggesting that this sequence context may favor the creation of these intriguing deletions by the editing machinery.
Misincorporation of a G at C insertion sites
There is a strong bias to the types of misediting events observed. The preponderance of G insertions, which comprise 49% of the total misediting events seen at atp C insertion sites (Table I), was particularly surprising given that G does not occur among single nucleotide editing events in natural RNAs from either Physarum or closely related organisms. G addition does occur in Physarum mitochondrial transcripts as part of GU or GC dinucleotide insertions (Miller et al., 1993), but is quite rare, occurring at only 4 of 445 known nucleotide insertion sites (<1% of all sites). The single G insertions observed here occur in ‘typical’ C insertion contexts (Table I), following a templated purine–uridine ∼68% (13 of 19) of the time (Miller et al., 1993; Horton and Landweber, 2000).
In all cases where the precise points of both C insertion and G misinsertion can be unambiguously identified (with neither C nor G encoded adjacent to the added nucleotide), the sites of natural editing and misediting coincide exactly, with all other G insertions falling at potential insertion sites. Therefore, despite the fact that an unusual nucleotide is inserted, it is likely that the G misincorporations represent errant processes whereby the insertion site is recognized by the editing apparatus, but there is a mistake in the choice of nucleotide. These observations also suggest that G misinsertions can be informative at sites where the location of C insertion is ambiguous. For example, G misinsertion locates the site of C insertion between the two encoded Cs at atp es38, and before the encoded C at es8. The fact that we have observed G misinsertions at the same position in two independent clones at atp es8 and atp es38 (Figure 1) argues that the site of nucleotide insertion is precisely determined, even at C insertion sites adjacent to encoded Cs.
Omission of encoded nucleotides
To date, we have observed 10 instances of small (1–3 nucleotide) deletions of encoded nucleotides adjacent to editing sites in run-on and chimeric transcripts derived from the atp gene (Table I). Interestingly, two different deletions have been observed after C insertion at es39, involving either the following G or the next three templated nucleotides (GAU). In addition, we have seen two instances of a U deletion downstream of an inserted C at es40, and another variant in which the U deletion at es40 occurs in the absence of C insertion. Although in some cases we cannot distinguish exactly which nucleotide has been omitted due to the sequence context around the editing site, where definitive judgements can be made it is always the nucleotide downstream of the editing site that is absent.
Larger omissions of templated nucleotides have also been observed (Figure 4); each involves all of the encoded nucleotides between adjacent C insertion sites and is denoted by the editing sites that serve as its endpoints (e.g. Δ1/2 lacks the nucleotides between es1 and es2). The eight distinct atp deletions reported here (total of 13 instances) involve 15 different editing sites and range from 10 to 37 nucleotides in length. One cDNA clone displays two deletions separated by a single inter-site interval of 14 nucleotides (Δ7/8 and Δ9/10 from Figure 1). In addition, site 38 acts as a downstream endpoint in one deletion (Δ37/38), but as an upstream endpoint in another (Δ38/39) deletion in a separate transcript. As is true for the other errors at editing sites, there is no evidence that inter-site deletions occur in vivo or during the RT–PCR, cloning or sequencing steps; they are almost certainly a feature of run-on transcription and editing in mtTEC.
Repeated independent occurrences of inter-site deletions have been observed for four pairs of sites: three instances of Δ9/10 and two each of Δ1/2, Δ38/39 and Δ40/41. Although the numbers are small, it is noteworthy that we have not yet seen any variability in the number of C residues among the instances of a given deletion, i.e. the final sequence across the point of deletion between any two sites is always identical. It is therefore likely that there is a single favored outcome for each pair of sites, and it is easiest to envisage that a set number of Cs is added by the editing activity at any given site pair. We also note that seven nucleotides adjacent to the downstream deletion endpoints in Δ7/8 and Δ44/45 are identical, with additional homology extending to 9 of 11 nucleotides (GAnnATTCTGTc). Ambiguous C insertion sites may also be more prone to both initiating and terminating inter-site deletions (Table I; Figure 4). Thus, template sequence or context may influence initiation and/or resolution of these deletions.
All of the atp deletions observed thus far contain at least one C residue at the point of deletion. The three deletions bordered by unambiguous sites (Δ1/2, Δ3/4 and Δ44/45) have clearly defined ends, with a single C presumably added by the editing activity. Although the origins of the C residues at the other inter-site deletions are ambiguous, inferences from other misediting events reduce the number of likely possibilities for some of the deletions. In the case of Δ38/39, for example, the positions of both insertion sites are inferred from the G misinsertion and small deletion data discussed previously, so the insertion of a single C by editing is again proposed. Importantly, similar scenarios can be envisioned for each of the other deletions depicted in Figure 4A. Thus, despite differences in the number of Cs present at the deletion junctions and the ambiguity of the deletion endpoints, a unifying model can be drawn that is consistent with all of our data (Figure 4B). In this model, the transcription/editing machinery reaches an editing site, where it potentially pauses and/or undergoes a conformational change, adds a non-templated C and then jumps to the next editing site prior to resuming transcription. Note that we cannot distinguish between this possibility and a similar scenario involving jumping from one editing site to the next prior to C insertion; each is equally plausible based on currently available data.
Discussion
In these and other experiments, we see a low level of sporadic misediting, which occurs during run-on transcription in vitro, but not at readily detectable levels in vivo. These errors occur only at editing sites and only on templates that support editing. Thus, rather than representing random mistakes by the RNA polymerase, these sequence alterations are clearly editing associated.
The data presented here indicate that the initial step in the editing process, site recognition, can occur independently of correct nucleotide insertion (or any nucleotide insertion at all), and is thus unlikely to depend upon interactions with the correct editing substrate. We have previously shown that editing does not occur on deproteinized DNA in chimeric templates, indicating that some feature of the native mtTEC is needed for editing to occur (Byrne and Gott, 2002). Intriguingly, misinsertions and deletions are also absent on portions of chimeric templates that fail to support editing, despite the fact that both editing and misediting occur upstream of the ligation junction within the same transcripts (Figure 2; Byrne and Gott, 2002). The fact that there are not even small deletions on these templates argues that the lack of editing on naked DNA is likely to be due to the inability of the transcription/editing apparatus to recognize editing sites. Thus, we hypothesize that recognition of precisely defined sites of nucleotide insertion involves template-associated factors.
The occasional misinsertion of G, U or dinucleotides at C insertion sites (Figures 1–3; Table I), combined with the absence of added nucleotides outside of editing sites, clearly indicates that these errors occur after the sites are correctly recognized by the transcription/editing machinery. The mtTEC preparations used here are isolated by passing a soluble mitochondrial extract through a gel filtration column and collecting the excluded volume (>2 × 107 kDa) (Cheng and Gott, 2000). This partially purified fraction contains the mitochondrial genome with transcribing RNA polymerases, nascent RNAs and other associated molecules, but may have lost a subset of editing factors. However, because misedited sites can be followed by correctly edited sites on individual transcripts, our data suggest that misediting is not solely due to dissociation of factors required for post-recognition functions that might travel with the polymerase.
This work clearly demonstrates that the mitochondrial RNA polymerase is capable of extending nascent RNAs containing either a misinserted nucleotide or a dinucleotide at sites normally edited by single C insertion, indicating that resumption of transcription after an editing event does not require a unique three-dimensional configuration at the 3′ end of the RNA chain. However, omission of encoded nucleotides may reflect problems resuming transcription in a template-directed manner in vitro, either after an inserted nucleotide or while the transcription/editing machinery is ‘stalled’ at an editing site prior to nucleotide addition. Among the small deletions, it is striking that all U omissions occur in the same context, a C insertion site flanked by encoded Us, and it is possible that this context may be mechanistically significant for these deletions. In the absence of C insertion, nucleotides could be skipped upon resumption of transcription if the 3′ end of the RNA anneals to template sequences just downstream of the editing site (e.g. an upstream U for U deletions at es22, es40 and es45 or, in the case of the CAA deletion at es16, an upstream AA). Encoded nucleotides could also be deleted if a C residue added via editing was extended as if it were a misincorporated nucleotide rather than an ‘inserted’ residue, given that RNA polymerases can continue transcription from a 3′ end that contains a mispaired nucleotide (Erie et al., 1993).
Large deletions involving templated nucleotides have also been observed in other transcription and replication systems (Pilipenko et al., 1995; Zhou et al., 1995; Nagy and Simon, 1997; Miller and Koev, 2000; Negroni and Buc, 2001). These have been attributed to polymerase ‘jumping’ (via intra- or inter-molecular template switching or DNA looping), with concomitant transfer of the nascent transcript (Pilipenko et al., 1995; Miller and Koev, 2000; Pasternak et al., 2001). Discontinuous polymerization can be initiated at natural (Negroni and Buc, 2001) or artificial (Zhou and Doetsch, 1994; Rong et al., 1998) template ends, at random breaks (Nagy and Simon, 1997; Negroni and Buc, 2001; Pfeiffer and Telesnitsky, 2001; Chang and Taylor, 2002), or at regions of secondary structure (Nagy and Simon, 1997; Figlerowicz, 2000). Physarum mtDNA is not appreciably nicked at editing sites (A.Rhee, E.M.Byrne and J.M.Gott, unpublished data) and the short deleted regions between editing sites are not predicted to have significant secondary structures. It is therefore unlikely that template discontinuities or secondary structure are involved in initiating or defining the boundaries of the inter-site deletions. By analogy with the more frequent types of polymerase-driven recombination processes, the inter-site deletions would be easiest to understand mechanistically if the 3′ end of the RNA could potentially anneal to template sequences immediately upstream of the deletion endpoint, but there are no significant stretches of conventional homology between the regions upstream of the pairs of editing sites involved (Figure 4). Nevertheless, the putative editing signals might constitute a different sort of homology: if a C residue is inserted at the upstream end of an inter-site deletion and is subsequently transferred from that editing site to the next, at least one nucleotide at the 3′ end of the nascent RNA would be ‘anchored’ for extension by the transcription/editing machinery. In this scenario, deletions could be initiated by occasional stalling of the transcription/editing apparatus after adding the non-templated C. Alternatively, polymerase jumping could be triggered by stalling at editing sites if insertion of non-templated nucleotides was slowed in vitro.
Taken together, these findings have a number of important implications regarding the editing process. The sequence of cDNA clones derived from individual nascent RNAs made in vivo indicates that the process of editing is highly accurate in intact cells, and that the accuracy of editing observed in steady-state RNAs is not simply due to turnover of misedited molecules in vivo. The characteristics and patterns of the editing errors made in vitro indicate that insertional RNA editing in Physarum mitochondria takes place in a series of separable steps, and argue that template associated trans-acting factors are required for editing site recognition. Finally, because non-encoded nucleotides are added to the 3′ end of nascent transcripts (Cheng et al., 2001), the Physarum mitochondrial polymerase must be intimately involved in the process of editing, at least at the initiation and resolution steps. Our data suggest that the transcription/editing machinery can recognize consecutive editing sites without transcribing the intervening template, potentially by interacting with these same editing factors.
Materials and methods
Oligodeoxynucleotides
7atp, 5′-TCAACGTTATCTTTTGAATTCAG-3′; Avr12atp, 5′-ATCCTAGGAGTAATAAAATTAAAAGC-3′; 13atp, 5′-ACTAATTTCGGTGGAGGTTC-3′; and 14atp, 5′-TAGGTAACATAGAATCAAC-3′.
mtTEC isolation, RNA synthesis and cDNA cloning
mtTECs were isolated essentially as described previously in Cheng and Gott (2000) with minor variations in dialysis conditions. Run-on transcription reactions (45–50 µl) (Cheng and Gott, 2000) and RNA isolation, RT–PCR, cloning and sequencing (Byrne and Gott, 2002) were carried out as described previously except where noted. cDNA clones from chimeric RNAs were isolated as described previously by Byrne and Gott (2002), except that RT–PCR primers 13atp and 14atp were used for the lower seven clones of Figure 1, and primer 7atp was used as the upstream primer for the clones in Figure 2B. Run-on transcriptions in Figure 3 were carried out at 500 µM NTPs; RT–PCR primers were Avr12atp and 13atp. For the lower set, mock-restriction cleavage was first performed by incubating mtTEC (5 µg of protein) in the presence of 1× buffer H (Roche) in a final volume of 35 µl at 30°C for 20 min; run-on transcription consequently took place at higher levels of Tris (44 mM pH 7.8) and NaCl (78 mM) than for standard run-ons. Individual cDNAs were sequenced until 10 clones with edited sites had been acquired for each condition. The run-on and mock-treated + run-on sets also included three and five fully unedited clones, respectively.
Acknowledgments
Acknowledgements
We thank Dr Tim Nilsen, Dr JoAnn Wise, Dr Eric Christian, Amy Rhee and Adam Majewski for useful discussions and critical reading of the manuscript, and the anonymous reviewers and Drs Donal Luse, David Setzer and Pieter deHaseth for helpful comments, suggestions and encouragement. This work was supported by NIH grant GM54663 (to J.M.G.).
References
- Byrne E.M. and Gott,J.M. (2002) Cotranscriptional editing of Physarum mitochondrial RNA requires local features of the native template. RNA, 8, 1174–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J. and Taylor,J. (2002) In vivo RNA-directed transcription, with template switching, by a mammalian RNA polymerase. EMBO J., 21, 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.-W. and Gott,J.M. (2000) Transcription and RNA editing in a soluble in vitro system from Physarum mitochondria. Nucleic Acids Res., 28, 3695–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.-W., Visomirski-Robic,L.M. and Gott,J.M. (2001) Non-templated addition of nucleotides to the 3′ end of nascent RNA during RNA editing in Physarum. EMBO J., 20, 1405–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erie D.A., Hajiseyedjavadi,O., Young,M.C. and von Hippel,P.H. (1993) Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science, 262, 867–873. [DOI] [PubMed] [Google Scholar]
- Figlerowicz M. (2000) Role of RNA structure in non-homologous recombination between genomic molecules of brome mosaic virus. Nucleic Acids Res., 28, 1714–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber A.P. and Keller,W. (2001) RNA editing by base deamination: more enzymes, more targets, new mysteries. Trends Biochem. Sci., 26, 376–384. [DOI] [PubMed] [Google Scholar]
- Gott J.M. (2001) RNA editing in Physarum polycephalum. In Bass,B. (ed.), RNA Editing: Frontiers in Molecular Biology. Oxford University Press, Oxford, UK, pp. 20–37.
- Gott J.M. and Emeson,R.B. (2000) Functions and mechanisms of RNA editing. Annu. Rev. Genet., 34, 499–531. [DOI] [PubMed] [Google Scholar]
- Gott J.M., Visomirski,L.M. and Hunter,J.L. (1993) Substitutional and insertional RNA editing of the cytochrome c oxidase subunit 1 mRNA of Physarum polycephalum. J. Biol. Chem., 268, 25483–25486. [PubMed] [Google Scholar]
- Horton T.L. and Landweber,L.F. (2000) Evolution of four types of RNA editing in myxomycetes. RNA, 6, 1339–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan L.P., Gallo,A. and O’Connell,M.A. (2001) The many roles of an RNA editor. Nat. Rev. Genet., 2, 869–878. [DOI] [PubMed] [Google Scholar]
- Madison-Antenucci S., Grams,J. and Hajduk,S.L. (2002) Editing machines: the complexities of trypanosome RNA editing. Cell, 108, 435–438. [DOI] [PubMed] [Google Scholar]
- Miller D., Mahendran,R., Spottswood,M., Costandy,H., Wang,S., Ling,M.-l. and Yang,N. (1993) Insertional editing in mitochondria of Physarum. Semin. Cell Biol., 4, 261–266. [DOI] [PubMed] [Google Scholar]
- Miller W.A. and Koev,G. (2000) Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology, 273, 1–8. [DOI] [PubMed] [Google Scholar]
- Nagy P.D. and Simon,A.E. (1997) New insights into the mechanisms of RNA recombination. Virology, 235, 1–9. [DOI] [PubMed] [Google Scholar]
- Negroni M. and Buc,H. (2001) Mechanisms of retroviral recombination. Annu. Rev. Genet., 35, 275–302. [DOI] [PubMed] [Google Scholar]
- Pasternak A.O., van den Born,E., Spaan,W.J. and Snijder,E.J. (2001) Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J., 20, 7220–7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelet T., Curran,J. and Kolakofsky,D. (1991) The P gene of bovine parainfluenza virus 3 expresses all three reading frames from a single mRNA editing site. EMBO J., 10, 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer J.K. and Telesnitsky,A. (2001) Effects of limiting homology at the site of intermolecular recombinogenic template switching during Moloney murine leukemia virus replication. J. Virol., 75, 11263–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko E.V., Gmyl,A.P. and Agol,V.I. (1995) A model for rearrangements in RNA genomes. Nucleic Acids Res., 23, 1870–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong M., Durbin,R.K. and McAllister,W.T. (1998) Template strand switching by T7 RNA polymerase. J. Biol. Chem., 273, 10253–10260. [DOI] [PubMed] [Google Scholar]
- Rundquist B.A. and Gott,J.M. (1995) RNA editing of the coI mRNA throughout the life cycle of Physarum polycephalum. Mol. Gen. Genet., 247, 306–311. [DOI] [PubMed] [Google Scholar]
- Sanchez A., Trappier,S.G., Mahy,B.W.J., Peters,C.J. and Nichol,S.T. (1996) The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc. Natl Acad. Sci. USA, 93, 3602–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeburg P.H. (2002) A-to-I editing: new and old sites, functions and speculations. Neuron, 35, 17–20. [DOI] [PubMed] [Google Scholar]
- Stuart K. and Panigrahi,A.K. (2002) RNA editing: complexity and complications. Mol. Microbiol., 45, 591–596. [DOI] [PubMed] [Google Scholar]
- Takano H., Abe,T., Sakurai,R., Moriyama,Y., Miyazawa,Y., Nozaki,H., Kawano,S., Sasaki,N. and Kuroiwa,T. (2001) The complete DNA sequence of the mitochondrial genome of Physarum polycephalum. Mol. Gen. Genet., 264, 539–545. [DOI] [PubMed] [Google Scholar]
- Thomas S.M., Lamb,R.A. and Paterson,R.G. (1988) Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell, 54, 891–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal S., Curran,J. and Kolakosky,D. (1990a) A stuttering model for paramyxovirus P mRNA editing. EMBO J., 9, 2017–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal S., Curran,J. and Kolakofsky,D. (1990b) Editing of the Sendai virus P/C mRNA by G insertion occurs during mRNA synthesis via a virus-encoded activity. J. Virol., 64, 239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visomirski-Robic L.M. and Gott,J.M. (1995) Accurate and efficient insertional RNA editing in isolated Physarum mitochondria. RNA, 1, 681–691. [PMC free article] [PubMed] [Google Scholar]
- Visomirski-Robic L.M. and Gott,J.M. (1997) Insertional editing in isolated Physarum mitochondria is linked to RNA synthesis. RNA, 3, 821–837. [PMC free article] [PubMed] [Google Scholar]
- Volchkov V.E., Becker,S., Volchkova,V.A., Ternovoj,V.A., Kotov,A.N., Netesov,S.V. and Klenk,H.D. (1995) GP mRNA of Ebola virus is edited by the Ebola virus polymerase and by T7 and vaccinia virus polymerases. Virology, 214, 421–430. [DOI] [PubMed] [Google Scholar]
- Wang S.S., Mahendran,R. and Miller,D.L. (1999) Editing of cytochrome b mRNA in Physarum mitochondria. J. Biol. Chem., 274, 2725–2731. [DOI] [PubMed] [Google Scholar]
- Zhou W. and Doetsch,P.W. (1994) Transcription bypass or blockage at single-strand breaks on the DNA template strand: effect of different 3′ and 5′ flanking groups on the T7 RNA polymerase elongation complex. Biochemistry, 33, 14926–14934. [DOI] [PubMed] [Google Scholar]
- Zhou W., Reines,D. and Doetsch,P.W. (1995) T7 RNA polymerase bypass of large gaps on the template strand reveals a critical role of the nontemplate strand in elongation. Cell, 82, 577–585. [DOI] [PMC free article] [PubMed] [Google Scholar]