Abstract
Fission yeast Klp5 and Klp6 belong to the microtubule-destabilizing Kin I family. In klp5 mutants, spindle checkpoint proteins Mad2 and Bub1 are recruited to mitotic kinetochores for a prolonged duration, indicating that these kinetochores are unattached. Further analysis shows that there are kinetochores to which only Bub1, but not Mad2, localizes. These kinetochores are likely to have been captured, yet lack tension. Thus Klp5 and Klp6 play a role in a spindle– kinetochore interaction at dual steps, capture and generation of tension. The TOG/XMAP215 family, Alp14 and Dis1 are known to stabilize microtubules and be required for the bivalent attachment of the kinetochore to the spindle. Despite apparent opposing activities towards microtubule stability, Klp5/Klp6 and Alp14/Dis1 share an essential function, as either dis1klp or alp14klp mutants are synthetically lethal, like alp14dis1. Defective phenotypes are similar to each other, characteristic of attachment defects and chromosome mis-segregation. Furthermore Alp14 is of significance for kinetochore localization of Klp5. We propose that Klp5/Klp6 and Alp14/Dis1 play a collaborative role in bipolar spindle formation during prometaphase through producing spindle dynamism.
Keywords: Kin I-XKCM/kinetochore/metaphase/spindle checkpoint/TOG-XMAP215
Introduction
Accurate chromosome segregation is vital for cell proliferation and genome stability in all organisms. In eukaryotic cells, spindles, mitosis-specific structures of polarized microtubules, play a fundamental role in capturing, pulling and segregating sister chromatids. Central to the spindle function is the establishment of bipolarity. A surveillance mechanism, called the spindle assembly checkpoint, plays a pivotal role in genome stability (Amon, 1999). This checkpoint monitors spindle bipolarity and detects errors in the attachment of the kinetochores to the spindles. The checkpoint is activated upon spindle damage or a kinetochore defect that causes mitotic kinetochores to appear either unattached or tensionless (Li and Nicklas, 1995; Chen et al., 1996; Zhou et al., 2002b). Genes (MAD1-3, BUB1, 3 and MPS1) involved in this checkpoint are evolutionarily conserved, and in vertebrates many of these genes are implicated in tumorigenesis (Cahill et al., 1998). These checkpoint proteins localize specifically to the mitotic kinetochores (Chen et al., 1996; Li and Benezra, 1996; Taylor and McKeon, 1997; Waters et al., 1998). In addition to kinetochore localization, Mad2 binds directly to the Slp1/Cdc20/Fizzy protein, thereby inhibiting activation of the APC/C ubiquitin ligase (Amon, 1999).
Careful time-lapse analysis has revealed that the mitotic phases in fission yeast are composed of three stages, phase 1 (prophase), phase 2 (from prometaphase to anaphase A) and phase 3 (anaphase B) (Nabeshima et al., 1998; Mallavarapu et al., 1999). Fission yeast is a genetically amenable organism and, as in vertebrates, the kinetochores free from attachment can be visualized by specific Mad2 and Mad3 localization (Garcia et al., 2001; Ikui et al., 2002; Millband and Hardwick, 2002; Vardy et al., 2002). Bub1 is also shown to localize to the mitotic kinetochores in both normally dividing mitotic cells and mutants in which the Mad2 checkpoint is activated (Bernard et al., 1998; Toyoda et al., 2002).
Molecular mechanisms underlying the formation of bipolar spindles are still poorly understood. We have been exploring this question by characterizing two classes of microtubule-binding proteins. These include Alp14 (also called Mtc1) and Dis1 (Garcia et al., 2001; Nakaseko et al., 2001), the conserved Dis1/TOG family MAPs (microtubule-associated proteins; XMAP215 in Xenopus and ch-TOG in mammals) (Ohkura et al., 2001). These two proteins are structurally and functionally redundant and act as a bridge between the plus end of the mitotic spindles and the kinetochore. Consistent with this, Alp14 and Dis1 localize to the kinetochores only during mitosis (Garcia et al., 2001; Nakaseko et al., 2001). In addition to its structural role as a linker, Alp14 functions as a regulatory component of the spindle checkpoint pathway. A part, if not all, of the function of Alp14/Dis1 is attributable to their microtubule-stabilizing activities, in which all the members of the Dis1/TOG family are involved (Kinoshita et al., 2002).
The other molecules of interest are Kin I-related Klp5 and Klp6 (XKCM1 in Xenopus and MCAK in mammals). In vertebrates, the Kin I family have been shown to be microtubule-destabilizing enzymes rather than conventional motors (Desai et al., 1999; Kline-Smith and Walczak, 2002; Moores et al., 2002; Niederstrasser et al., 2002). We have shown that Klp5 and Klp6 form a heterodimer in the cell, which is required for the timely onset of anaphase A. Like Alp14 and Dis1, Klp5 and Klp6 localize to the mitotic kinetochores. In the absence of Klp5/Klp6 function, cells are viable, but stay much longer (2-fold) in pre-anaphase stage, during which the Mad2 checkpoint is activated (Garcia et al., 2002). Time-lapse analysis of centromere-marking green fluorescent protein (GFP) shows that chromosome alignment (congression) at the spindle equator is never established, instead centromeres move back and forth between the two poles. Given these observations, we have proposed that Klp5 and Klp6 play a crucial role in the establishment of metaphase as microtubule-destabilizing factors at the mitotic kinetochores (Garcia et al., 2002).
Experiments performed in Xenopus egg extracts, based upon analysis of microtubule growth and shrinkage, suggest that XKCM1 and XMAP215 play an antagonistic role in microtubule dynamics. XMAP215 plays a stabilizing role, whilst XKCM1 destabilizes microtubules (Tournebize et al., 2000). Despite these in vitro studies, the functional relationship between these two classes of microtubule-binding proteins in vivo remains to be established.
In the current study, we address two questions. First, we examine the molecular basis of mitotic defects in klp5 and klp6 mutants, which result in Mad2 checkpoint activation. Two types of attachment defects activate the spindle checkpoint: complete loss of the attachment and inefficient attachment that results in the lack of tension at the kinetochores (Li and Nicklas, 1995; Chen et al., 1996). We set up experiments to distinguish these two possibilities. Secondly, we ask if there is an in vivo functional relationship between Alp14/Dis1 and Klp5/Klp6. We will present somewhat unexpected results, i.e. Klp5 and Klp6 share an essential, rather than opposing, function with Alp14 and Dis1 in the establishment of bivalent attachment of the kinetochores to the mitotic spindles.
Results
The Kin I homologue Klp5 is required for efficient capture of the kinetochores by the mitotic spindles
The onset of anaphase A is delayed in klp5 and klp6 mutants, which is attributable to the activation of the Mad2-dependent spindle checkpoint (Garcia et al., 2002). We addressed the reason for this checkpoint activation. In order to examine whether or not kinetochore capture occurred at the appropriate time in these mutants, Mad2 localization was examined. A derivative of GFP, YFP, was fused to the C-terminus of Mad2 under the endogenous promoter in wild-type and klp5 mutants. In order to assign the spindle length in mitotic cells, another GFP derivative, CFP, was also tagged to the C-terminus of Cut12, a component of the centrosome-equivalent spindle pole body (SPB) (Bridge et al., 1998).
In wild-type cells, Mad2 localizes to the kinetochores transiently only during prophase equivalent to ∼1% in the total population, in which the spindle length is <2 µm (Figure 1A and C). On the contrary, in the klp5 mutant, >10% of exponentially growing cells showed bright Mad2 dots. Typical examples of these Mad2 dots are shown in Figure 1B. Statistical measurement of 120 mitotic klp5 mutants with Mad2 at the kinetochores showed that the spindle length varied from 1 to 4 µm and sometimes was longer (Figure 1C). This phase corresponds to the cell cycle stage before onset of anaphase B, when the Mad2 checkpoint is activated in this mutant (Garcia et al., 2002). It should be noted that these mitotic mutant cells often showed chromosome segregation defects (rows 4–6 in Figure 1B). Upon mitotic exit, Mad2 localization altered such that instead of discrete dots, broad nuclear staining characteristic of interphase cells was observed (row 7). Thus, in the absence of Klp5, in parallel with mitotic delay, Mad2 localizes to the mitotic kinetochores.
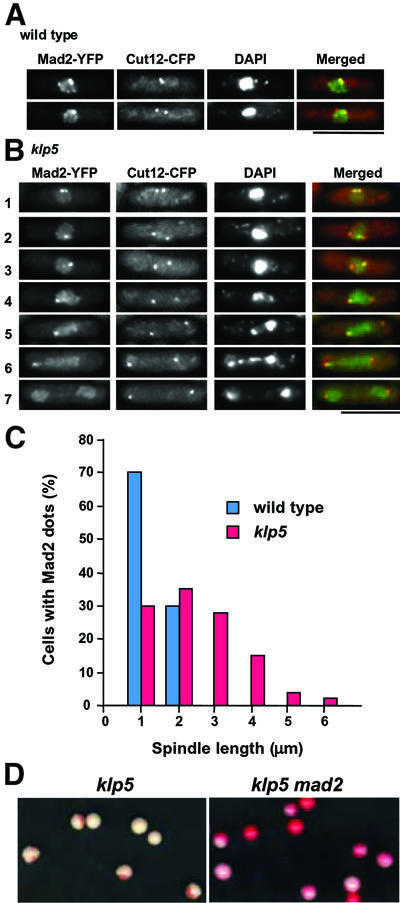
Fig. 1. Mad2 localization at the kinetochores in klp5 mutants and enhanced chromosome loss in klp5mad2 double mutants. (A and B) Localization pattern of Mad2 and Cut12 during the cell cycle. Wild-type (A) or klp5 mutants (B), in which Mad2 and Cut12 (the SPB marker) were tagged with YFP and CFP, respectively, under their endogenous promoters, were grown and the localization patterns of these two proteins were examined. Nuclear DNA was stained with DAPI. In wild-type cells, only early mitotic cells show Mad2 at the kinetochores, whilst in klp5 mutants, Mad2 localizes to the kinetochores for a prolonged period. Representative images in different cell cycle stages are shown (1 and 2; early mitosis; 3–6, prometaphase/metaphase; 7, post-anaphase). Merged images are shown on the right (Cut12–CFP, red; Mad2–YFP, green). The bar indicates 10 µm. (C) Distribution of the spindle length in mitotic cells containing Mad2 dots. The distance between two Cut12 signals (equivalent to the length of the mitotic spindles) was measured in a total 40 or 120 mitotic cells in wild-type (left, blue columns) or klp5 mutants (right, red), respectively, which contain Mad2 at the kinetochores. (D) Minichromosome loss in klp5mad2 mutants. klp5 (left) or klp5mad2 (right) mutant cells carrying minichromosomes, which had been grown in minimal medium without adenine (selective conditions for minichromosomes), were plated on rich media plates and incubated at 30°C for 4 days. Colonies of adenine auxotrophs were red.
Whilst observing Mad2 localization with respect to Cut12 spots, we noticed that at least one of the Mad2 dots almost always co-localized to one of the SPBs (45 out of 50 cells). This tendency was particularly evident in mitotic cells where two SPBs were separated accompanied by chromosome segregation defects (see rows 4–6 in Figure 1B). This suggested that chromosomes with unattached kinetochores are unable to move towards the spindle equator.
Elimination of the Mad2 checkpoint in klp5 mutants exacerbates genome instability
We have shown previously that deletion of the mad2+ gene in the klp5 mutant results in abolishment of mitotic delay (Garcia et al., 2002). Here we examined its consequence in genome stability. A non-essential minichromosome (Niwa et al., 1989) was introduced in the klp5 mutant and the rate of chromosome loss was measured. As shown in Figure 1D, although the klp5 single mutant by itself showed a significant rate of minichromosome loss (left, 9.2 × 10–2 loss per division) (Garcia et al., 2002), in the klp5mad2 double mutant, virtually no Ade+ colonies were retained under non-selective conditions (right, 6.3 × 10–1 loss per division). The Mad2 checkpoint, therefore, plays an auxiliary yet important role in securing genome stability in the absence of Klp5 function. Taking this and previous results together, we concluded that one of the mitotic roles of Klp5 is to facilitate the attachment of the kinetochores to the mitotic spindles during prometaphase.
Bub1 also localizes to the mitotic kinetochores in klp5 mutants
Recently, like Mad2, fission yeast Bub1 was also shown to localize to the kinetochores at early mitosis (Bernard et al., 1998; Toyoda et al., 2002). Given the increased localization of Mad2 to the mitotic kinetochores in the klp5 mutant, we next sought to examine Bub1 localization in this mutant. GFP was fused to the C-terminus of Bub1 under the endogenous promoter in the wild-type and klp5 mutant. In wild-type cells, as reported previously (Bernard et al., 1998; Toyoda et al., 2002), Bub1 signal at the kinetochore was visible only in early mitotic cells (rows 2 and 3 in Figure 2A). In contrast, in klp5 mutants, Bub1 dots were observed in prolonged mitotic cells (Figure 2B). As in the case of Mad2, cells displaying prolonged Bub1, which often consisted of multiple dots, showed chromosome mis-segregation [rows 3–5, see 4′,6-diamidino-2-phenylindole (DAPI) staining].
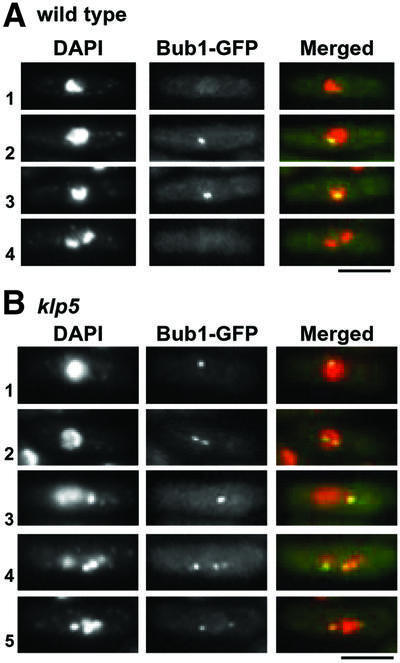
Fig. 2. Bub1 localization at the kinetochores in wild-type and klp5 mutants. (A and B) Wild-type (A) or klp5 mutants (B), in which the endogenous bub1+ gene was tagged with GFP, were grown and Bub1 localization during the cell cycle was examined (in A: 1, interphase; 2 and 3, early mitosis; 4, anaphase; in B: 1 and 2, early mitosis; 3–5, prometaphase/metaphase). Nuclear DNA (DAPI), Bub1–GFP and merged images (DAPI, red; Bub1–GFP, green) are shown. The bar indicates 10 µm.
In order to follow the in vivo dynamics of Bub1 localization during mitosis, time-lapse live imaging of Bub1–GFP was performed. As shown in Figure 3A, in wild-type cells, Bub1–GFP localizes to the kinetochore for 2–3 min and then disappears. This duration is consistent with that found in a previous report (Toyoda et al., 2002). It is of note that, in agreement with Toyoda et al. (2002), we have never seen split signals of Bub1–GFP in wild-type mitotic cells. In clear contrast, klp5 mutants show two characteristic defects, which were observed in still images described above. First Bub1–GFP localizes to the kinetochore for a much longer period (>5 min, Figure 3B). Secondly, Bub1–GFP signal appears as two or three discrete dots (marked by arrowheads). In addition, live analysis shows dynamic movement of Bub1–GFP in this mutant. As shown in Figure 3B, a single Bub1 dot split into three (1, 1.5 and 2 min), then appeared as a single dot (2.5 min) and again split into two or three dots (3.5, 4 and 4.5 min). This result indicated that in the absence of Klp5, not only Mad2 but also Bub1 localized to the kinetochores and that each sister chromatid displayed rapid oscillation between the two poles before the onset of anaphase A.
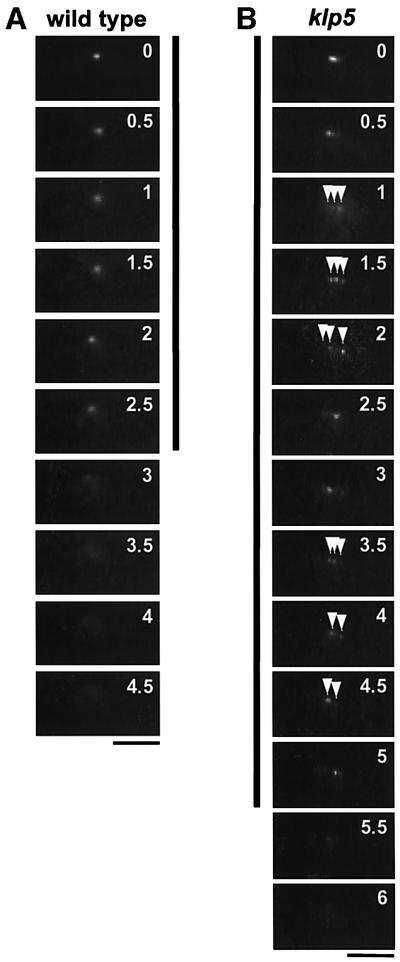
Fig. 3. Time-lapse images of Bub1–GFP during mitosis. (A and B) Live analysis of Bub1–GFP during mitosis. Time-lapse imaging of Bub1– GFP was performed using a conventional fluorescence microscope with 30 s intervals in wild-type (A) or klp5 mutants (B). The period during which Bub1–GFP at the kinetochores was visible is marked with vertical lines, 2.5 min in wild-type and 5 min in klp5 mutants. Multiple Bub1–GFP dots in klp5 mutants are shown with arrowheads. The numbers in the top right corner in each image represent minutes after the start of live imaging. The bar indicates 10 µm.
klp5 mutant may contain two types of the defective mitotic kinetochores, unattached and tension-less
In vertebrates, Bub1, unlike Mad2 which locates to only unattached kinetochores, localizes to those lacking tension or exhibiting weak attachment (Waters et al., 1998; Skoufias et al., 2001; Taylor et al., 2001; Zhou et al., 2002a,b). However, in fission yeast, the relationship between Mad2 and Bub1 regarding kinetochore recruitment had not been addressed. We asked whether the kinetochores to which Bub1 localizes and those to which Mad2 localizes co-exist in a single cell. It is possible that Mad2 and Bub1 localization takes place in order rather than simultaneously, such that Mad2 is recruited to the kinetochores first and then Bub1 follows upon Mad2 dissociation from them. Alternatively, Mad2 and Bub1 may co-localize to the unattached kinetochores. Differential tagging of these two proteins with YFP and CFP was not possible experimentally because of the rapid decay of Bub1–CFP signals. Accordingly, a double-tagged strain, in which Bub1 was fused with a haemagglutinin (HA) epitope, was constructed together with Mad2–YFP, and localization of these two proteins was examined.
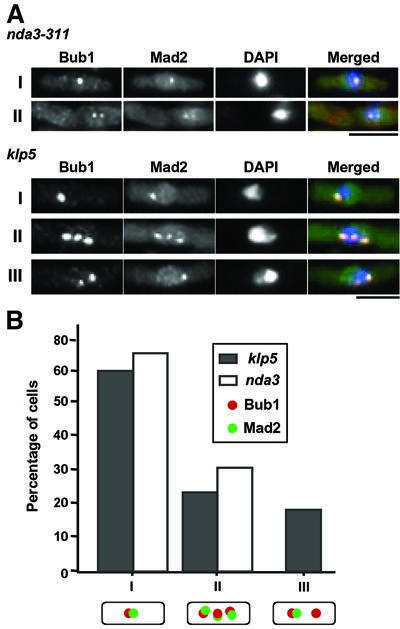
Immunofluorescence microscopy showed characteristic mitotic patterns of Bub1 and Mad2 localization as described below. We examined 100 mitotic cells, which displayed Bub1 and/or Mad2 dots at the kinetochores, and scored co-localization patterns. This analysis enabled us to classify these mitotic cells into three categories. The first type shows single Bub1 and Mad2 dots in an overlapped manner (I in Figure 4B). Nearly 60% of the cells showed this pattern (Figure 4B). In the second type, multiple Bub1 and Mad2 dots (mostly two or three) co-localize (II), which makes up of 22% of the counted population. In the third type (18%), cells display multiple Bub1 dots (two or three), where only one of the Bub1 dots co-localizes with one Mad2 dot (III in Figure 4A). Cells containing only Bub1 dots without Mad2 dots were rare (2%), and we did not detect any cells displaying only Mad2 dots without Bub1 dots. In contrast, mitotically arrested cells in which the kinetochores are unattached (using nda3-311 mutants, defective in β-tubulin), both Bub1 and Mad2 always co-localized to the kinetochores, and no type III cells were observed (Figure 4A and B, type I and II).
Fig. 4. Localization of Bub1 and Mad2 in klp5 mutants. (A) Classification of Bub1 and Mad2 localization patterns in nda3-311 or mitotic klp5 mutants. nda-311 mutants incubated at 18°C for 10 h (upper two rows) or exponentially growing klp5 mutants (lower three rows), in which both strains contained Bub1-HA and Mad2–YFP, were fixed with formaldehyde and processed for immunofluorescence microscopy. Cells which displayed clear nuclear dots for Bub1 and/or Mad2 were examined, and localization patterns were scored (type I, single dot and co-localization; type II, multiple dots and co-localization; type III multiple Bub1 dots and a single Mad2 dot, which co-localized with one of the Bub1 dots). The bar indicates 10 µm. (B) Percentage of three types of mitotic cells. One hundred mitotic cells which displayed Bub1 and/or Mad2 dots were examined in klp5 (closed columns) or nda3-311 mutant cells (open columns) and scored for individual patterns.
From these data, we deduced that types I and II cells contain chromosomes with unattached kinetochores, as Mad2 localized to them. In contrast, type III cells may contain two classes of the mitotic kinetochores in a single cell, one unattached like those in types I and II, and the other attached, indicated by the dissociation of Mad2. Nevertheless, in these chromosomes, tension between the kinetochore and the spindle fails to be produced, resulting in recruitment or retainment of Bub1 at these kinetochores. Thus, Klp5 is required to ensure the bivalent attachment of the kinetochore to the spindle, probably via dual mechanisms: one is efficient capture and the other is stabilization of attachment, which helps to generate tension.
Klp5 functions independently of sister chromatid cohesion
Fission yeast mutants defective in sister chromatid cohesion have been reported to show split and prolonged localization of Bub1 at the mitotic kinetochores (Toyoda et al., 2002). In addition, like klp5 and klp6 mutants, the Mad2 checkpoint is activated in these mutants. We addressed the functional relationship between Klp5/Klp6 and cohesin molecules at the kinetochores and centromeres. First, we examined whether or not sister chromatid cohesion was lost prematurely in the klp5 mutant. For this purpose, we used a new construct for centromere-marking GFP–LacI (cen2-GFP) (Straight et al., 1996; Nabeshima et al., 1998), in which lac operator sequences were integrated in the genome only 5 kb distant from cen2 (A.Yamamoto, unpublished results). Immunofluorescence microscopy showed that sister chromatid cohesion is retained normally in klp5 mutants; we did not see any cells that displayed precocious segregation of chromosome II centromeres during mitosis (Figure 5A). However, we noticed that, like Mad2 (see Figure 1B), Cen2–GFP signal tends to locate in close proximity to one of the SPBs (using anti-Sad1 antibody; Sad1 is an SPB marker) (Hagan and Yanagida, 1995). Next, Klp5 localization to the kinetochores was examined in a mutant defective in the MCD1/SCC1 homologue, a temperature-sensitive rad21-K1 strain (Tatebayashi et al., 1998). As shown in Figure 5B, Klp5, as in wild-type cells (Garcia et al., 2002), localized to the mitotic kinetochores in this mutant at 36°C.
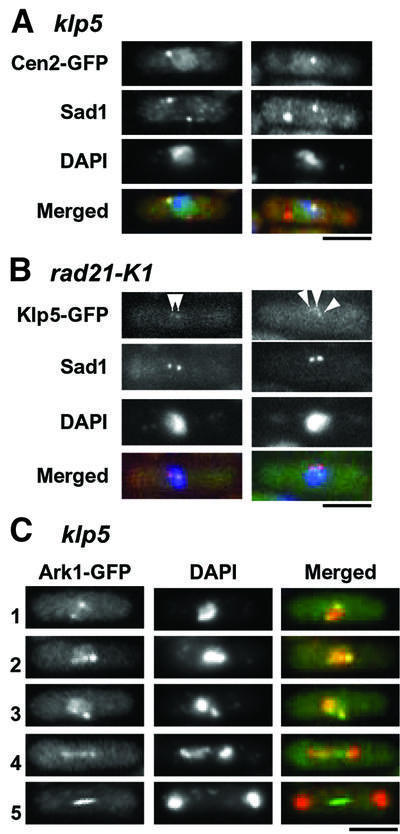
Fig. 5. Independence of Klp5 and Klp6 function from cohesin and the aurora kinase. (A) Sister chromatid cohesion in klp5 mutants. klp5 mutants containing centromere-marking Cen2–GFP were fixed and processed for immunofluorescence microscopy with anti-Sad1 antibody. Nuclear DNA was stained with DAPI. Merged images are shown on the bottom (Cen2–GFP, green; Sad1, red; DNA, blue). Two representative mitotic cells before chromosome segregation are shown. Note that Cen2–GFP overlaps or is proximal to one of the two SPBs. (B) Klp5 localization at the kinetochores in the cohesin mutant. The temperature-sensitive rad21-K1 mutant containing Klp5–GFP was incubated for 6 h at 36°C and localization of Klp5–GFP was examined. Anti-Sad1 antibody was used to visualize the SPBs. Merged images are shown on the right (Klp5–GFP, green; anti-Sad1, red; DAPI, blue). Arrowheads mark Klp5 dots at the kinetochores. (C) Localization of the aurora kinase (Ark1) in klp5 mutants. Ark1 was tagged with GFP in klp5 mutants and its localization during the cell cycle was examined (1 and 2, early mitosis; 3, prometaphase/metaphase; 4, anaphase; 5, late anaphase). Nuclear DNA was stained with DAPI. Merged images are shown on the right (Ark1–GFP, green; DAPI, red). The bar indicates 10 µm.
Aurora kinase (Ark1/Aim1 in fission yeast) localizes, like Klp5 and Klp6, to both the mitotic kinetochores and the spindle midzone, and is required for chromosome segregation (Morishita et al., 2001; Petersen et al., 2001; Leverson et al., 2002; Rajagopalan and Balasubramanian, 2002). In order to address the requirement for Klp5 and Klp6 for aurora kinase localization, the C-terminus of Ark1 was tagged with GFP under the endogenous promoter in the klp5 mutant. As shown in Figure 5C, Ark1 localizes properly to the kinetochores during early and mid mitosis, as well as to the spindle midzone at late anaphase in the klp5 mutant, as in wild-type cells (Morishita et al., 2001; Petersen et al., 2001; Rajagopalan and Balasubramanian, 2002). Thus, loss of Klp5 function does not interfere with Ark1 localization. Taken together, these results suggested that Klp5 acts independently of cohesin and the aurora kinase.
Microtubules in klp5 and klp6 mutants are more resistant to depolymerizing conditions
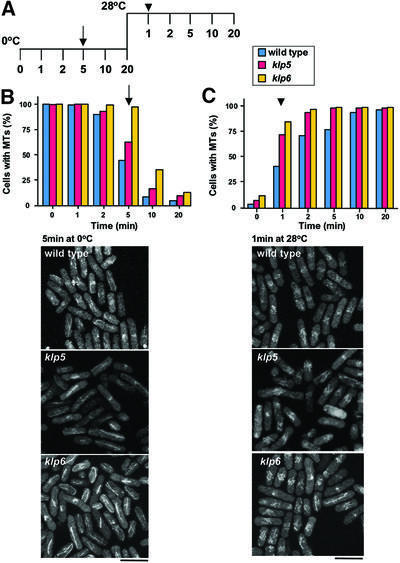
Our previous results and those from others have suggested that Klp5 and Klp6 play a role as microtubule-depolymerizing enzymes, as in the case of vertebrate homologues (West et al., 2001; Garcia et al., 2002). In order to address directly the involvement of Klp5 and Klp6 in microtubule stability, in vivo kinetics of microtubule depolymerization and repolymerization were compared between wild-type and klp5/klp6 mutants. Strains containing integrated GFP-atb2+ (encoding α2-tubulin) (Garcia et al., 2001) were incubated on ice to allow depolymerization of microtubules. The extent of microtubule depolymerization was followed by fluorescence microscopy until 20 min upon cold shock (Figure 6A). As shown in Figure 6B, in wild-type cells, after 5 min on ice microtubules were retained in <50% of cells, and a further 5 min incubation led to the disappearance of microtubules in >90% of cells. In contrast, under the same conditions, microtubules were resistant to cold shock in klp5 or klp6 mutants; 65 or nearly 100% of klp5 or klp6 mutant cells, respectively, retained intact microtubules after 5 min of cold shock, and 20 (klp5) or 40% (klp6) of cells still contained microtubules even after 10 min incubation on ice (Figure 6B).
Fig. 6. Enhanced stability of microtubules in the absence of Klp5 or Klp6. (A) Experimental designs. Wild-type, klp5 and klp6 mutant cells containing GFP-atb2+ (encoding α2-tubulin), were grown at 28°C, incubated on ice for 20 min then shifted up to 28°C and incubated for another 20 min. Samples were taken at the indicated times, and microtubule structures were observed by fluorescence microscopy. Time points at which representative images are shown in (B) and (C) are marked with an arrow and arrowhead, respectively. (B) Depolymerization of microtubules by cold shock. The percentage of cells that contain long microtubules (>1 µm in length) is plotted at each time point. Microtubule structures in wild-type (blue), klp5 (red) and klp6 (yellow) mutants after 5 min on ice are shown below. (C) Recovery of microtubule structures upon temperature shift-up. Images of microtubules after 1 min shift-up at 28°C after cold shock treatment are shown below as in (B). The bar indicates 10 µm.
Recovery of microtubules upon temperature shift-up was then performed after 20 min on ice. In wild-type cells, 1 and 2 min incubation at 28°C resulted in 38 and 73% microtubule recovery, respectively (Figure 6C). In contrast, in the klp5 or klp6 mutant, >70 or 80% of cells, respectively, showed almost intact microtubules after 1 min incubation at 28°C, suggesting that microtubule polymerization is faster in the absence of Klp5 or Klp6. These results indicated that, in the absence of Klp5 and Klp6, microtubules are more resistant to microtubule-depolymerizing conditions and polymerize more rapidly upon recovery from cold shock. The results strongly support the notion that Klp5 and Klp6 are microtubule-depolymerizing factors in fission yeast.
Klp5 and Klp6 share an essential role with Alp14 and Dis1
In Xenopus, XKCM1 is proposed to act antagonistically with XMAP215 in regulating microtubule dynamics (Tournebize et al., 2000), and opposing roles of these two families have also been suggested in budding yeast (Severin et al., 2001). In fission yeast, we and others have shown previously that Alp14 and Dis1 share an essential role and stabilize microtubules, thereby ensuring the interaction between the mitotic spindle and the kinetochore (Garcia et al., 2001; Nakaseko et al., 2001). Our results presented above show that Klp5 and Klp6 are also required for efficient capture of the kinetochores by the spindles. This suggested that, although Alp14/Dis1 and Klp5/Klp6 possess opposing activities towards microtubule stability, these two families might play a collaborative role.
As the first step to address the functional relationship between Alp14/Dis1 and Klp5/Klp6, genetic analysis was performed. As shown previously, Alp14 and Dis1 are functional homologues, in which only double mutants become lethal, whilst single dis1 or alp14 mutants display cold sensitivity or temperature sensitivity, respectively (Garcia et al., 2001; Nakaseko et al., 2001). In contrast, Klp5 and Klp6 are non-essential for cell viability, and either single or double mutants show identical mitotic phenotypes, as they need to form heterodimers for their function in the cell (Garcia et al., 2002). Bearing this in mind, various pairs of double mutants were constructed. As shown in Table I, any combination of mutations in Alp14/Dis1 and Klp5/Klp6 was lethal, i e. alp14klp5, alp14klp6, dis1klp5 or dis1klp6 double mutants were all synthetically lethal at the temperature permissive for dis1 or alp14 single mutants.
Table I. Genetic interaction between klp5, klp6, alp14 and dis1.
| Genes deleted |
Multicopy plasmids |
Viable/lethal (L) | ||||
|---|---|---|---|---|---|---|
| klp5 | klp6 | alp14 | dis1 | p(alp14+) | p(dis1+) | ts/cs |
| Δ | + | + | + | – | – | Viable |
| + | Δ | + | + | – | – | Viable |
| + | + | Δ | + | – | – | Viable, ts |
| + | + | + | Δ | – | – | Viable, cs |
| + | + | Δ | Δ | – | – | L |
| Δ | + | Δ | + | – | – | L |
| Δ | + | + | Δ | – | – | L |
| + | Δ | Δ | + | – | – | L |
| + | Δ | + | Δ | – | – | L |
| Δ | Δ | Δ | + | – | – | L |
| + | + | Δ | Δ | – | + | Viable |
| Δ | + | + | Δ | – | + | Viable |
| Δ | + | Δ | + | + | – | Viable |
| Δ | Δ | Δ | + | + | – | Viable |
Deletion mutants are denoted by Δ, whilst wild-type alleles are shown by +. cs and ts denote cold- and temperature-sensitive, respectively.
Synthetic lethality between Dis1/Alp14 and Klp5/Klp6 raised the following two possibilities. The first possibility is, as in Xenopus, these two families play an antagonistic role in mitosis and the reason for lethality of double mutants could be the hyperactivation of Klp5/Klp6 in the absence of Alp14/Dis1. In fact, we have shown previously that overproduction of Klp5 and/or Klp6 results in lethality (Garcia et al., 2002). The second possibility is that Alp14/Dis1 and Klp5/Klp6 are not solely antagonistic to each other, and in addition share an essential function in vivo. These two possibilities could be distinguished by determining whether triple mutants alp14klp5klp6 and dis1klp5klp6 are lethal or not. If the hyperactivation of Klp5/6 were the reason, triple mutants would be viable, whilst if these genes shared an essential function, triple mutants would still be lethal. Further genetic crosses between individual mutants showed that alp14klp5klp6 triple mutants are lethal (Table I). These results indicated that klp5 and klp6 mutants are viable only because either Dis1 or Alp14 is functional. Conversely, alp14 (or dis1) mutants are viable as long as Klp5 and Klp6 are operational. In fission yeast, therefore, the Kin I homologues and the Dis1/TOG family play a collaborating role, rather than an opposing one, in vivo.
Both Klp5/Klp6 and Alp14/Dis1 are required for the establishment of bivalent attachment of the kinetochores to the mitotic spindles
If Klp5/Klp6 and Alp14/Dis1 play a role in common in mitotic progression, double mutants of alp14dis1, alp14klp5 or dis1klp5 might show similar phenotypes. As shown below, this was indeed the case. In order to examine defective phenotypes in these deletion strains, the following strains were constructed: alp14dis1 or dis1klp5 mutants containing episomal plasmids carrying dis1+, or alp14klp5 mutants containing episomal plasmids carrying alp14+. These plasmids were mitotically stable. None theless, at some frequency, plasmids were lost mitotically and defective phenotypes of cells lacking Alp14 and Dis1, Alp14 and Klp5, or Dis1 and Klp5 could be observed (plasmid loss experiments) (Ohkura and Yanagida, 1991). The frequency of plasmid loss was 2–5% under exponentially growing conditions but, upon nitrogen starvation and re-feeding, the rate of plasmid loss was increased significantly (20–30%).
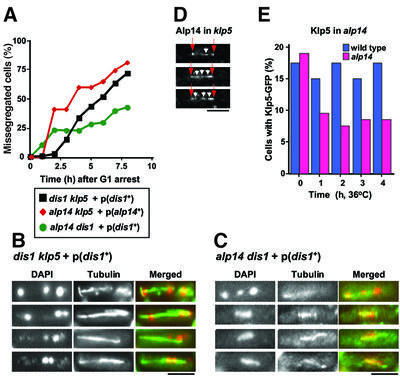
In order to examine defective phenotypes in alp14dis1, alp14klp5 or dis1klp5 mutants, these three strains carrying dis1+ or alp14+ plasmids were starved of nitrogen for 12 h, suspended in rich medium and defective phenotypes were then observed by nuclear staining and immunofluorescence microscopy with anti-tubulin antibody. As shown in Figure 7A, after release into rich medium, mitotic cells displaying chromosome segregation defects were increased dramatically. Representative examples of these defects in dis1klp5 or alp14dis1 cells are shown in Figure 7B and C, respectively. As evident with DAPI and anti-tubulin staining, two strains displayed very similar, if not identical, mitotic defects. Chromosomes were segregated unequally and often three chromosomes with a variable length of spindles were observed. These phenotypes resembled mitotic defects previously described in cold-sensitive dis1 mutants or temperature-sensitive alp14 mutants, which are reminiscent of the failure of the mitotic spindles to capture the kinetochores (Ohkura et al., 1988; Nabeshima et al., 1995; Garcia et al., 2001; Nakaseko et al., 2001). Similar mitotic defects were observed in asynchronously dividing cells, albeit less frequently (2%, data not shown). It is of note that anaphase spindles in alp14dis1 mutants are apparently shorter than those in dis1klp5 mutants (middle panels in Figure 7B and C). Taken together, we conclude that Alp14/Dis1 and Klp5/Klp6 play a cooperative role in vivo in the establishment of bivalent attachment of the kinetochores to the mitotic spindle.
Fig. 7. Klp5/Klp6 and Dis1/Alp14 MAPs share an essential function in mitotic progression. (A–C) Defective mitotic phenotypes of alp14dis1, alp14klp5 or dis1klp5 mutants. Cells that were deleted simultaneously for alp14+ and dis1+, alp14+ and klp5+ or dis1+ and klp5+, but kept viable by episomal multicopy plasmids carrying the dis1+ or alp14+ gene were starved of nitrogen for 12 h, washed, resuspended in fresh rich medium and incubated further. At each time point, aliquots were taken for immunofluorescence microscopy with anti-tubulin antibody. Nuclear DNA was stained with DAPI. The frequency of cells that displayed defects in chromosome segregation was scored [(A), at least 100 cells were counted at each time point]. Representatives of cells displaying mis-segregation defects are shown in (B) (dis1klp5) and (C) (alp14dis1). DAPI (left), anti-tubulin (middle) and merged images (right, green for tubulin and red for DAPI) are shown. (D) Kinetochore localization of Alp14 in klp5 mutants. Alp14–GFP signals are shown in mitotic klp5 mutants. Images were taken from live time-lapse movies (0, 150 and 200 s). Arrows (red) and arrowheads (white) show the SPBs and the centromeres, respectively. (E) Localization of Klp5–GFP in alp14 mutants. Wild-type and alp14 deletion strains containing Klp5–GFP were shifted up to 36°C and samples were taken hourly. The percentage of cells that contain Klp5–GFP at the spindle and the kinetochore was determined. The bar indicates 10 µm in (B), (C) and (E), and 5 µm in (D).
Kinetochore localization of Klp5 is reduced in alp14 mutants
We next sought to determine the requirement for Klp5 for Alp14 localization to the kinetochore and, vice versa, the requirement for Alp14 for Klp5 kinetochore localization. As shown in Figure 7D, Alp14 (tagged with GFP) was clearly capable of localizing to the kinetochores (arrowheads) as well as the SPB (arrows) in the absence of Klp5. In sharp contrast, the percentage of cells that contained Klp5–GFP at the kinetochores was substantially decreased in alp14 mutants incubated at 36°C (Figure 7E). Although 10% of alp14 mutant cells were still capable of retaining Klp5–GFP at the spindle and the kinetochore, this value might be deceptive, as >50% of alp14 mutant cells display mitotic phenotypes under these conditions (Garcia et al., 2001). This means that nearly 80% of mitotic alp14 cells are unable to retain Klp5 at their kinetochores. This suggests, therefore, that Alp14/Dis1 MAPs play a role in targeting of Klp5/Klp6 to the kinetochores during mitosis.
Discussion
In this study, we have presented evidence that the major role of two fission yeast Kin I homologues (Klp5 and Klp6) lies in the establishment of bivalent attachment of the kinetochores to the mitotic spindle. We and others have shown previously that the Dis1/TOG MAP homologues, Alp14 and Dis1, are required for an interaction between the spindles and the kinetochores (Garcia et al., 2001; Nakaseko et al., 2001), which prompted us to explore the functional relationship between Klp5/Klp6 and Alp14/Dis1. We show that these two families share an essential function, in which lethal phenotypes are characteristic of the failure in attachment. Our analysis, therefore, establishes an in vivo collaborative role for Kin I and Dis1/TOG in the formation of bipolar spindles.
Role of Klp5 and Klp6 in the formation of bivalent spindles: capture and tension
We have found that in the klp5 mutant, Bub1 and Mad2 remain at the mitotic kinetochores for a prolonged period, corresponding to prometaphase. The kinetochores, to which both Bub1 and Mad2 localize, most probably represent the unattached state, indicating that Klp5 (and Klp6) plays a role in capturing the kinetochores (Figure 8). We noticed that the kinetochore tends to localize in close proximity to the SPBs. As in budding yeast (Guacci et al., 1997), the centromeres cluster in the vicinity of the SPB during fission yeast interphase (Chikashige et al., 1997). It is likely that SPB-proximal kinetochores are incapable of being detached from the SPB even upon the onset of mitosis, and as a result retain the interphase spatial configuration.
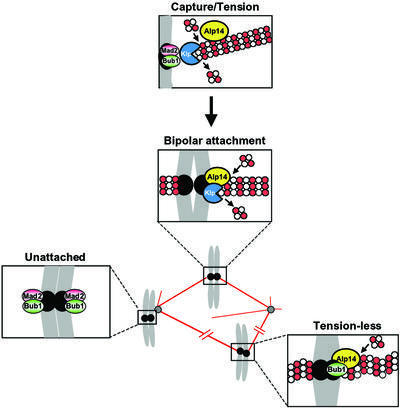
Fig. 8. A role for Klp5/Klp6 and Alp14/Dis1 in the formation of bipolar mitotic spindles. Alp14 and Dis1 (shown as Alp14 in the figure) localize to both the mitotic spindles [shown by filaments consisting of tubulin dimers (white and red circles)] and the mitotic kinetochores (closed black circles). Klp5 and Klp6 (shown as Klp) also localize to the mitotic kinetochores. In their absence, the major defect is failure in the attachment of the kinetochores to which Bub1 and Mad2 localize. Furthermore, at least Klp5 and Klp6 have an additional role in generation of tension at the kinetochores upon attachment. In the absence of Klp5 and Klp6, the kinetochores fail to produce tension, as the spindles tend only to polymerize without the poleward force (depicted by wavy spindles). These tension-less kinetochores recruit Bub1, but not Mad2.
In addition, unlike wild-type cells, the klp5 mutant displays multiple split signals of Bub1 and Mad2. The existence of the kinetochores, to which only Bub1, but not Mad2, localizes, is of particular significance. We envisage that these kinetochores are attached, as Mad2 is detached, but lack tension. This may mimic the situation in animal culture cells, which were treated with a low dose of vinblastine; Bub1 but not Mad2 was recruited (Skoufias et al., 2001). The absence of microtubule-destabilizing Klp5/Klp6 leads to spindle stabilization and, as a result, to the loss of the poleward force at the kinetochores. Forces both away from and towards the pole are vital for chromosome congression (Nicklas, 1997). The defect in the poleward force would lead to imbalance at the centromeres, which results in a tension-less state at these kinetochores (Figure 8). In summary, we propose that the Kin I homologues play a crucial role in mitotic progression by contributing to bipolar spindle formation at dual steps, the first is attachment and the second is generation of tension upon capture.
Functional collaboration between Klp5/Klp6 and Alp14/Dis1
As shown here, kinetochore localization of Klp5 is substantially dependent upon Alp14 function. Our previous analysis indicates that Alp14 is required for bipolar spindle formation and plays an essential role in this process as a structural linker between the spindle and the kinetochore (Garcia et al., 2001). It is likely, therefore, that Klp5 (and Klp6) is one of the molecules that traverse through the spindles towards the mitotic kinetochores in an Alp14/Dis1-dependent fashion.
In Xenopus and budding yeast, XMAP215 (Stu2) and XKCM1 (Kip3) have been proposed to play an antagonistic role. Although these results and those in our study appear to represent a reverse relationship for the two families, they may not be contradictory. Whilst Klp5 and Klp6 also act as microtubule-destabilizing factors in fission yeast (West et al., 2001; Garcia et al., 2002; this study), these two molecules, as shown here, play a vital role in capturing the kinetochores. Importantly, this role is shared by Alp14 and Dis1, which promote microtubule assembly (Garcia et al., 2001). The apparent opposing role of Kin I and Dis1/TOG may not be analogous to that between protein kinase and phosphatase, nor that between histone acetyl transferase and deacetylase, which act genetically and biologically in an antagonistic fashion. Instead, these two families function in harmony towards the regulation of in vivo microtubule dynamics, in which the essential role is the establishment of bivalent attachment of the kinetochores to the mitotic spindle (Figure 8). In fact, a recent in vitro study (Kinoshita et al., 2001) has shown that XMAP215 and XKCM1 are necessary and sufficient to reconstitute physiological microtubule dynamics, indicating that these two molecules also act in a coordinated manner in vertebrates.
Evolutionary divergence between fission yeast and vertebrates
It has been shown that the mammalian Kin I homologue MCAK is not involved in chromosome alignment, instead it is required for chromosome segregation at anaphase (Maney et al., 1998). Although the Kin I family plays an essential role in bipolar spindle formation in animal cells (Walczak et al., 1996), at present it appears premature to conclude a degree of functional conservation with regard to the mitotic role of the Kin I family between fission yeast and animal cells (West et al., 2002). It is possible that the requirement for Kin I-mediated microtubule-depolymerizing activity during mitosis has diverged throughout evolution, such that in fission yeast it is required during prometaphase, whilst in animal cells it might be essential at anaphase instead. Nonetheless, it is worth stressing that in both fission yeast and animals, the Kin I family of proteins localize specifically to the centromeres/kinetochores in a mitosis-specific fashion (Wordeman and Mitchison, 1995; Garcia et al., 2002), which suggests that their mitotic role is conserved through evolution. Further analysis is required to clarify this important point.
Spindle bipolarity and chromosome bi-orientation
As Klp5 and Klp6 function independently of Rad21 and Ark1, it is unlikely that these two Kin I homologues are involved in sister chromatid bi-orientation, in which cohesion molecules and the aurora kinase play a pivotal role (Tanaka et al., 2000, 2002; Sonoda et al., 2001; Tanaka, 2002; Toyoda et al., 2002). Functional parallelism between Klp5/Klp6 and cohesin (and the aurora kinase) indicates that at least two independent pathways operate for the establishment of bivalent mitotic spindles. One is the spindle-based pathway, which ensures bipolarity of the mitotic spindles, whilst the other is the chromosome-mediated pathway, which establishes sister chromatid bi-orientation. It should be noted that despite parallelism, these pathways merge at the kinetochores, as mutations in either Klp5/Klp6 or cohesin activate the common Mad2 checkpoint (Garcia et al., 2002; Toyoda et al., 2002). How spindle-based and chromosome-mediated pathways interact structurally and functionally is an interesting challenge for future study.
Materials and methods
Strains
Strains used in this study are listed in the Supplementary data available at The EMBO Journal Online.
Immunochemical assays
Affinity-purified rabbit polyclonal anti-Sad1 antibody was obtained from Dr Mizuki Shimanuki. Mouse monoclonal anti-α-tubulin (TAT-1, provided by Dr Keith Gull; or T-5168, Sigma) and anti-HA (16B12, BAbCO) antibodies were also used.
Indirect immunofluorescence microscopy
Cells were fixed with methanol or formaldehyde, and primary antibodies (TAT-1 1/50, anti-Sad1 antibody 1/10 000 or anti-HA antibody 1/1000) were applied, followed by Cy3-conjugated goat anti-rabbit or anti-mouse IgG (Sigma) and fluorescein-linked sheep anti-mouse IgG (Amersham).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Drs Tony Carr, Keith Gull, Yasushi Hiraoka, Osami Niwa, Mizuki Shimanuki, Ayumu Yamamoto and Mitsuhiro Yanagida for providing materials used in this study, Dr Jesús Vázquez for encouragement, and Dr Jacqueline Hayles and Frank Uhlmann for critical reading of the manuscript and useful suggestions. M.A.G. was supported by an EMBO long-term fellowship. This work is supported by Cancer Research UK and an HFSP research grant.
References
- Amon A. (1999) The spindle checkpoint. Curr. Opin. Genet. Dev., 9, 69–75. [DOI] [PubMed] [Google Scholar]
- Bernard P., Hardwick,K. and Javerzat,J.-P. (1998) Fission yeast Bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis. J. Cell Biol., 143, 1775–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge A.J., Morphew,M., Bartlett,R. and Hagan,I.M. (1998) The fission yeast SPB component Cut12 links bipolar spindle formation to mitotic control. Genes Dev., 12, 927–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill D.P., Lengauer,C., Yu,J., Riggins,G.J., Willson,J.K.V., Markowitz,S.D., Kinzler,K.W. and Vogelstein,B. (1998) Mutations of mitotic checkpoint genes in human cancers. Nature, 392, 300–303. [DOI] [PubMed] [Google Scholar]
- Chen R.-H., Waters,J.C., Salmon,E.D. and Murray,A.W. (1996) Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science, 274, 242–246. [DOI] [PubMed] [Google Scholar]
- Chikashige Y., Ding,D.-Q., Imai,Y., Yamamoto,M., Haraguchi,T. and Hiraoka,Y. (1997) Meiotic nuclear reorganization: switching the position of centromeres and telomeres in the fission yeast Schizosaccharomyces pombe. EMBO J., 16, 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Verma,S., Mitchison,T.J. and Walczak,C.E. (1999) Kin I kinesins are microtubule-destabilizing enzymes. Cell, 96, 69–78. [DOI] [PubMed] [Google Scholar]
- Garcia M.A., Vardy,L., Koonrugsa,N. and Toda,T. (2001) Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO J., 20, 3389–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M.A., Koonrugsa,N. and Toda,T. (2002) Two kinesin-like Kin I family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr. Biol., 12, 610–621. [DOI] [PubMed] [Google Scholar]
- Guacci V., Hogan,E. and Koshland,D. (1997) Centromere position in budding yeast: evidence for anaphase A. Mol. Biol. Cell, 8, 957–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I. and Yanagida,M. (1995) The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol., 129, 1033–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikui A.E., Furuya,K., Yanagida,M. and Matsumoto,T. (2002) Control of localization of a spindle checkpoint protein, Mad2, in fission yeast. J. Cell Sci., 115, 1603–1610. [DOI] [PubMed] [Google Scholar]
- Kinoshita K., Arnal,I., Desai,A., Drechsel,D.N. and Hyman,A.A. (2001) Reconstitution of physiological microtubule dynamics using purified components. Science, 294, 1340–1343. [DOI] [PubMed] [Google Scholar]
- Kinoshita K., Habermann,B. and Hyman,A.A. (2002) XMAP215: a key component of the dynamic microtubule cytoskeleton. Trends Cell Biol., 6, 267–273. [DOI] [PubMed] [Google Scholar]
- Kline-Smith S.L. and Walczak,C.E. (2002) The microtubule-destabilizing kinesin XKCM1 regulates microtubule dynamic instability in cells. Mol. Biol. Cell, 13, 2718–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverson J.D., Huang,H.-K., Forsburg,S.L. and Hunter,T. (2002) The Schizosaccharomyces pombe Aurora-related kinase Ark1 interacts with the inner centromere protein Pic1 and mediates chromosome segregation and cytokinesis. Mol. Biol. Cell, 13, 1132–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. and Nicklas,R.B. (1995) Mitotic forces control a cell-cycle checkpoint. Nature, 373, 630–632. [DOI] [PubMed] [Google Scholar]
- Li Y. and Benezra,R. (1996) Identification of a human mitotic checkpoint gene: hsMAD2. Science, 274, 246–248. [DOI] [PubMed] [Google Scholar]
- Mallavarapu A., Sawin,K. and Mitchison,T. (1999) A switch in microtubule dynamics at the onset of anaphase B in the mitotic spindle of Schizosaccharomyces pombe. Curr. Biol., 9, 1423–1426. [DOI] [PubMed] [Google Scholar]
- Maney T., Hunter,A.W., Wagenbach,M. and Wordeman,L. (1998) Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J. Cell Biol., 142, 787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millband D.N. and Hardwick,K.G. (2002) Fission yeast Mad3p is required for Mad2p to inhibit the anaphase-promoting complex and localizes to kinetochores in a Bub1p-, Bub3p- and Mph1p-dependent manner. Mol. Cell. Biol., 22, 2728–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moores C.A., Yu,M., Guo,J., Beraud,C., Sakowicz,R. and Milligan,R.A. (2002) A mechanism for microtubule depolymerization by KinI kinesins. Mol. Cell, 9, 903–909. [DOI] [PubMed] [Google Scholar]
- Morishita J., Matsusaka,T., Goshima,G., Nakamura,T., Tatebe,H. and Yanagida,M. (2001) Bir1/Cut17 moving from chromosome to spindle upon the loss of cohesion is required for condensation, spindle elongation and repair. Genes Cells, 6, 743–763. [DOI] [PubMed] [Google Scholar]
- Nabeshima K., Kurooka,H., Takeuchi,M., Kinoshita,K., Nakaseko,Y. and Yanagida,M. (1995) p93dis1, which is required for sister chromatid separation, is a novel microtubule and spindle pole body-associating protein phosphorylated at the Cdc2 target sites. Genes Dev., 9, 1572–1585. [DOI] [PubMed] [Google Scholar]
- Nabeshima K., Nakagawa,T., Straight,A.F., Murray,A., Chikashige,Y., Yamashita,Y.M., Hiraoka,Y. and Yanagida,M. (1998) Dynamics of centromeres during metaphase–anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell, 9, 3211–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaseko Y., Goshima,G., Morishita,J. and Yanagida,M. (2001) M phase-specific kinetochore proteins in fission yeast microtubule-associating Dis1 and Mtc1 display rapid separation and segregation during anaphase. Curr. Biol., 11, 537–549. [DOI] [PubMed] [Google Scholar]
- Nicklas R.B. (1997) How cells get the right chromosomes. Science, 275, 632–637. [DOI] [PubMed] [Google Scholar]
- Niederstrasser H., Salehi-Had,H., Gan,E.C., Walczak,C. and Nogales,E. (2002) XKCM1 acts on a single protofilament and requires the C terminus of tubulin. J. Mol. Biol., 316, 817–828. [DOI] [PubMed] [Google Scholar]
- Niwa O., Matsumoto,T., Chikashige,Y. and Yanagida,M. (1989) Characterization of Schizosaccharomyces pombe minichromosome deletion derivatives and a functional allocation of their centromere. EMBO J., 8, 3045–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H. and Yanagida,M. (1991) S.pombe gene sds22+ essential for a mid-mitotic transition encodes a leucine-rich repeat protein that positively modulates protein phosphatase-I. Cell, 64, 149–157. [DOI] [PubMed] [Google Scholar]
- Ohkura H., Adachi,Y., Kinoshita,N., Niwa,O., Toda,T. and Yanagida,M. (1988) Cold-sensitive and caffeine supersensitive mutants of the Schizosaccharomyces pombe dis genes implicated in sister chromatid separation during mitosis. EMBO J., 7, 1465–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H., Garcia,M.A. and Toda,T. (2001) Dis1/TOG universal microtubule adaptors—one MAP for all? J. Cell Sci., 114, 3805–3812. [DOI] [PubMed] [Google Scholar]
- Petersen J., Paris,J., Willer,M., Philippe,M. and Hagan,I.M. (2001) The S.pombe aurora-related kinase Ark1 associates with mitotic structures in a stage dependent manner and is required for chromosome segregation. J. Cell Sci., 114, 4371–4384. [DOI] [PubMed] [Google Scholar]
- Rajagopalan S. and Balasubramanian,M.K. (2002) Schizosaccharomyces pombe Bir1p, a nuclear protein that localizes to kinetochores and the spindle midzone, is essential for chromosome condensation and spindle elongation during mitosis. Genetics, 160, 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin F., Habermann,B., Huffaker,T. and Hyman,T. (2001) Stu2 promotes mitotic spindle elongation in anaphase. J. Cell Biol., 153, 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias D.A. andreassen,P.R., Lacroix,F.B., Wilson,L. and Margolis,R.L. (2001) Mammalian mad2 and bub1/bubR1 recognize distinct spindle-attachment and kinetochore-tension checkpoints. Proc. Natl Acad. Sci. USA, 98, 4492–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E. et al. (2001) Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev. Cell, 1, 759–770. [DOI] [PubMed] [Google Scholar]
- Straight A.F., Belmont,A.S., Robinett,C.C. and Murray,A.W. (1996) GFP tagging of budding yeast chromosomes reveals that protein–protein interactions can mediate sister chromatid cohesion. Curr. Biol., 6, 1599–1608. [DOI] [PubMed] [Google Scholar]
- Tatebayashi K., Kato,J. and Ikeda,H. (1998) Isolation of a Schizosaccharomyces pombe rad21ts mutant that is aberrant in chromosome segregation, microtubule function, DNA repair and sensitive to hydroxyurea: possible involvement of Rad21 in ubiquitin-mediated proteolysis. Genetics, 148, 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T.U. (2002) Bi-orientating chromosomes on the mitotic spindles. Curr. Opin. Cell Biol., 14, 365–371. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Fuchs,J., Loidl,J. and Nasmyth,K. (2000) Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nature Cell Biol., 2, 492–499. [DOI] [PubMed] [Google Scholar]
- Tanaka T.U., Rachidi,N., Janke,C., Pereira,G., Galova,M., Schiebel,E., Stark,M.J.R. and Nasmyth,K. (2002) Evidence that the Ipl1–Sli15 (aurora kinase–INCENP) complex promotes chromosome bi-orientation by altering kinetochore–spindle pole connections. Cell, 108, 317–329. [DOI] [PubMed] [Google Scholar]
- Taylor S.S. and McKeon,F. (1997) Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell, 89, 727–735. [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Hussein,D., Wang,Y., Elderkin,S. and Morrow,C.J. (2001) Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J. Cell Sci., 114, 4385–4395. [DOI] [PubMed] [Google Scholar]
- Tournebize R. et al. (2000) Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat. Cell Biol., 2, 13–19. [DOI] [PubMed] [Google Scholar]
- Toyoda Y., Furuya,K., Goshima,G., Nagao,K., Takahashi,K. and Yanagida,M. (2002) Requirement of chromatid cohesion proteins Rad21/Scc1 and Mis4/Scc2 for normal spindle–kinetochore interaction in fission yeast. Curr. Biol., 12, 347–358. [DOI] [PubMed] [Google Scholar]
- Vardy L., Fujita,A. and Toda,T. (2002) The γ-tubulin complex protein Alp4 provides a link between the metaphase checkpoint and cytokinesis in fission yeast. Genes Cells, 7, 365–373. [DOI] [PubMed] [Google Scholar]
- Walczak C.E., Mitchison,T.J. and Desai,A. (1996) XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell, 84, 37–47. [DOI] [PubMed] [Google Scholar]
- Waters J.C., Chen,R.-H., Murray,A.W. and Salmon,E.D. (1998) Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol., 141, 1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R.R., Malmstrom,T., Troxell,C.L. and McIntosh,J.R. (2001) Two related kinesins, klp5+ and klp6+, foster microtubule disassembly and are required for meiosis in fission yeast. Mol. Biol. Cell, 12, 3919–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R.R., Malmstrom,T. and McIntosh,J.R. (2002) Kinesins klp5+ and klp6+ are required for normal chromosome movement in mitosis. J. Cell Sci., 115, 931–940. [DOI] [PubMed] [Google Scholar]
- Wordeman L. and Mitchison,T.J. (1995) Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J. Cell Biol., 128, 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Panda,D., Landen,J.W., Wilson,L. and Joshi,H.C. (2002a) Minor alteration of microtubule dynamics causes loss of tension across kinetochore pairs and activates the spindle checkpoint. J. Biol. Chem., 277, 17200–17208. [DOI] [PubMed] [Google Scholar]
- Zhou J., Yao,J. and Joshi,H.C. (2002b) Attachment and tension in the spindle assembly checkpoint. J. Cell Sci., 115, 3547–3555. [DOI] [PubMed] [Google Scholar]