Abstract
Importin β-type transport receptors mediate the vast majority of transport pathways between cell nucleus and cytoplasm. We identify here the translation elongation factor 1A (eEF1A) as the predominant nuclear export substrate of RanBP21/exportin 5 (Exp5). This cargo–exportin interaction is rather un usual in that eEF1A binds the exportin not directly, but instead via aminoacylated tRNAs. Exp5 thus represents the second directly RNA-binding exportin and mediates tRNA export in parallel with exportin-t. It was suggested recently that 10–15% of the cellular translation would occur in the nucleus. Our data rule out such a scenario and instead suggest that nuclear translation is actively suppressed by the nuclear export machinery. We found that the vast majority of translation initiation factors (eIF2, eIF2B, eIF3, eIF4A1, eIF5 and eIF5B), all three elongation factors (eEF1A, eEF1B and eEF2) and the termination factor eRF1 are strictly excluded from nuclei. Besides Exp5 and importin 13, CRM1 and as yet unidentified exportins also contribute to the depletion of translation factors from nuclei.
Keywords: eEF1A/exportin/nuclear transport/translation/tRNA
Introduction
Translation in bacteria, such as Escherichia coli, is coupled intimately to transcription and already initiated on nascent transcripts (Das et al., 1967; Miller et al., 1970). The situation in eukaryotes, however, is complicated by the presence of introns within the primary mRNA transcripts. These introns need to be removed by a splicing reaction, before translation can occur (Berget et al., 1977; Hastings and Krainer, 2001). Translating unspliced or incompletely spliced transcripts would produce truncated proteins that are, in the best case, just non-functional, but in the worst case could compete with the functional gene products in a dominant-negative fashion. Eukaryotes elegantly avoid such problems by confining transcription and translation to distinct compartments.
Transcription is a nuclear process, and the spliceosome retains pre-mRNAs in the nuclear compartment until splicing has been completed (Legrain and Rosbash, 1989; Custodio et al., 1999). This ensures that normally only fully spliced mRNAs become exported for cytoplasmic translation. Compartmentation thus forces translation to occur subsequent to splicing, and this principle obviously relies on strict exclusion of translation from the nuclei. However, key components of translation, namely ribosomes, mRNAs and tRNAs, are produced in the nuclear compartment, and even aminoacylated tRNAs (aa-tRNAs) can be detected in the nucleus (Lund and Dahlberg, 1998). This poses the interesting question as to how nuclear translation is prevented. The problem is the focus of this study and we will return to it after a brief introduction to the nuclear transport machinery.
The nuclear envelope (NE) separates the nuclear from the cytoplasmic compartment and thereby necessitates nucleocytoplasmic transport (for reviews, see Dahlberg and Lund, 1998; Mattaj and Englmeier, 1998; Görlich and Kutay, 1999; Nakielny and Dreyfuss, 1999). All nuclear proteins originate from the cytoplasm and need to be imported. Conversely, mRNAs, ribosomes and tRNAs must be exported to the cytoplasm, where they function in translation. All nucleocytoplasmic trafficking occurs through nuclear pore complexes (NPCs), which allow passage of material in two modes, referred to as passive diffusion and facilitated translocation. Passive diffusion is efficient for small molecules, such as metabolites, but becomes increasingly restricted as the size of the transported species approaches a limit of ∼20–40 kDa. In contrast, facilitated translocation can accommodate the transport of even very large objects up to diameters of nearly 40 nm and masses of several MDa. It is typically receptor mediated and coupled to an input of metabolic energy, which in turn allows substrate (cargo) accumulation against gradients of chemical activity.
Transport receptors of the importin β (Impβ) family account for most, but not all, nuclear transport pathways (Mattaj and Englmeier, 1998; Görlich and Kutay, 1999; Nakielny and Dreyfuss, 1999; Conti and Izaurralde, 2001). They circulate between the nucleus and cytoplasm, recognize cargo molecules and transfer them from one side of the NE to the other. Substrate loading and release are guided by a concentration gradient of RanGTP across the NE, which is sensed through the RanGTP-binding domains present in the transport receptors. Nuclear export mediators (exportins) preferentially bind their export substrates at high nuclear RanGTP levels and exit the nucleus as trimeric cargo–exportin–RanGTP complexes. These complexes disassemble in the cytoplasm upon hydrolysis of the Ran-bound GTP. The exportins can then re-enter nuclei, leaving their cargoes behind, while Ran returns to the nucleus via NTF2. Importins operate in an exactly converse manner to exportins. They recruit cargo molecules at low RanGTP levels in the cytoplasm and release them upon RanGTP binding in the nucleus.
The superfamily of Impβ-type transport receptors comprises >20 family members in higher eukaryotes, and the elucidation of their cellular functions has been a major goal in the field. At least 10 family members (importins β, 4, 5, 7, 8, 9, 11 and 13 as well as transportins 1 and SR) function in import (for reviews, see Mattaj and Englmeier, 1998; Strom and Weis, 2001; Görlich and Jäkel, 2002). Most importins bind their cargoes directly. Impβ, however, can combine with other import receptors or a variety of adaptor molecules and thereby expand its range of cargoes considerably. The most well characterized examples of such adaptors are the members of the Impα family, which recognize cargoes with a classical nuclear localization signal (NLS).
In higher eukaryotes, six transport receptors have been reported to function in nuclear export, namely CAS, CRM1, exportin-t (Exp-t), Exp4, Exp5 and Imp13. CAS mediates the retrieval of Impαs back to the cytoplasm (Kutay et al., 1997). CRM1 is the exportin with the broadest range of substrates; it exports a large variety of proteins with leucine-rich nuclear export signals (NESs), the import adaptor snurportin 1, U snRNAs (via the PHAX-CBC adaptor system) and even the small and large ribosomal subunits (Fornerod et al., 1997; Moy and Silver, 1999; Paraskeva et al., 1999; Ohno et al., 2000; Gadal et al., 2001). Exp-t mediates the export of mature tRNAs (Arts et al., 1998a,b; Kutay et al., 1998; Lipowsky et al., 1999), while Exp4 so far has only a single substrate, eIF5A (Lipowsky et al., 2000), a factor with an unclear role in translation. Exp5 has been reported to export double-stranded RNA (dsRNA)-binding proteins (Brownawell and Macara, 2002). Finally, Imp13 is quite unique in that it moves different cargoes in opposite directions. It imports several proteins, but also exports the translation initiation factor eIF1A from nuclei (Mingot et al., 2001).
Here, we identify the elongation factor 1A (eEF1A)–aa-tRNA complex as a major export substrate of Exp5 and show that this cargo is recognized via the aa-tRNA. Efficient Exp5-mediated export, combined with a very low nuclear influx rate, keep the nuclear eEF1A levels down to ≤1/100 of the cytoplasmic concentration. eEF1A delivers aa-tRNAs to the ribosome and is one of the two GTPases that drive translational elongation. The observed nuclear exclusion of eEF1A alone would already be sufficient to prevent nuclear translation. However, to address the issue systematically, we determined the nucleocytoplasmic distribution of a larger number of additional, essential translation factors. These were: eIF2, eIF2B, eIF4A1, eIF5, eIF5B, eEF1B, eEF2 and eRF1 (for reviews, see Merrick and Nyborg, 2000; Welch et al., 2000; Pestova et al., 2001). eEF1B functions as the guanine nucleotide exchange factor (GEF) for eEF1A. The GTPase eEF2 drives the codon-wise movement of the ribosome along the mRNA (translational translocation). eIF4A1 is an essential RNA helicase; it unwinds the 5′ end of the mRNA and thereby helps the pre-initiation complex to reach the start codon (scanning). eIF2 delivers the tRNAiMet to the pre-initiation complex; it is a GTPase and cooperates with eIF2B and eIF5, which in turn function as the eIF2-specific GEF and GTPase-activating protein (GAP), respectively. eIF5B, the homologue of bacterial IF2, is a GTPase implicated in joining the ribosomal subunits. Finally, the release factor eRF1 recognizes stop codons and is essential for termination. Strikingly, all the translation factors just mentioned appear to be actively depleted from nuclei and show strict nuclear exclusion. Eukaryotic cells might employ several strategies to prevent translation in the nucleus, but the expulsion of translation factors from the nuclei is certainly a major mechanism. A considerable share of the nuclear transport capacity and several export receptors appear to be allocated to accomplish this task.
Results
Molecular cloning of mouse and Drosophila Exp5
Based on sequence similarity to established members of the Impβ family, we identified several expressed sequence tags (ESTs) that ultimately allowed the cloning of mouse and Drosophila RanBP21/Exp5. Mouse and Drosophila Exp5 share 87 and 34% sequence identity, respectively, with the previously published human protein sequence (Brownawell and Macara, 2002). We next generated a monospecific antibody that recognizes both human and rodent Exp5. By immunoblotting, we found abundant expression of Exp5 in all mammalian cell lines tested, including human HeLa cells, mouse 3T3 and baby hamster kidney (BHK) cells (see Supplementary data available at The EMBO Journal Online), suggesting that Exp5 mediates a constitutive nuclear transport pathway.
Identification of eEF1A as an Exp5 cargo
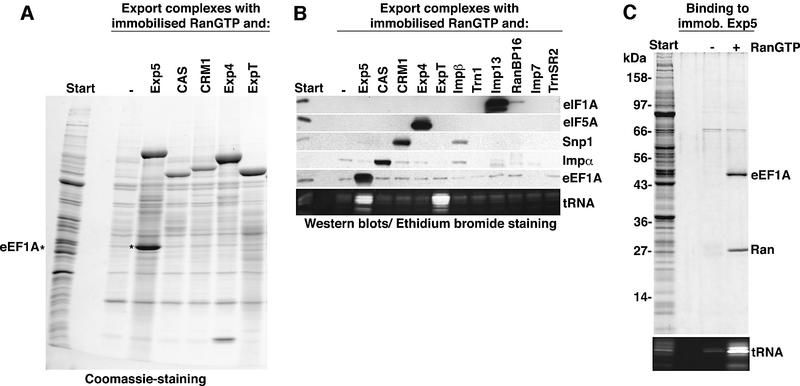
Typical export complexes contain the exportin, its cargo and RanGTP (Fornerod et al., 1997; Kutay et al., 1997). Therefore, exportin–cargo complexes can be purified with immobilized RanGTP. To identify cargoes of a given exportin by this method, no other nuclear transport receptor should be present in the system. To achieve that, we first depleted all nuclear transport receptors from a HeLa cytosolic extract by the phenyl-Sepharose method (Ribbeck and Görlich, 2002). Aliquots of the resulting extract were then replenished with single recombinant receptors and subjected to RanGTP binding. As seen in Figure 1A, a stoichiometric 50 kDa band was retrieved along with Exp5. The protein was identified by peptide fingerprinting as eEF1A, and western blotting confirmed that eEF1A assembled into export complexes specifically with Exp5, but not with any of the other nuclear transport receptors tested (Figure 1B).
Fig. 1. (A) Identification of the eEF1A–tRNA complex as a putative export substrate for Exp5. A cytosolic extract from HeLa cells was depleted of endogenous nuclear transport receptors and 500 µl aliquots were then each supplemented with a single nuclear transport receptor (1 µM) as indicated. Export complexes were formed with 2 µM zz-tagged RanGTP (GTPase-deficient RanQ69L mutant) and purified with IgG–Sepharose. Analysis was by SDS–PAGE followed by Coomassie staining. The load of the bound fractions corresponds to 40 times that of the starting material. With Exp5, a stoichiometric 50 kDa band was recovered and identified by peptide fingerprinting as eEF1A. (B) Export complexes were formed with the indicated transport receptors as in (A), but analysis was by immunoblotting with antibodies against the indicated nuclear export substrates or by ethidium bromide staining for tRNA (10% polyacrylamide gel). Note, eEF1A interacts with Exp5, but not with any of the other established export mediators. tRNA was retrieved specifically with both, Exp-t (ExpT) and Exp5. ‘Imp’, importin; ‘Exp’, exportin; ‘Trn’, transportin; and ‘Snp’, snurportin 1. (C) Binding of a HeLa extract to zz-tagged Exp5 in the absence or presence of 5 µM RanQ69L (GTP).
Exp5 recognized eEF1A very selectively and did not interact with Impα, snurportin 1, eIF5A or eIF1A, which represent export cargoes of CAS, CRM1, Exp4 and Imp13, respectively (Kutay et al., 1997; Paraskeva et al., 1999; Lipowsky et al., 2000; Mingot et al., 2001). However, ethidium bromide staining revealed that tRNAs were recruited by Exp5 as efficiently as by Exp-t (Arts et al., 1998a; Kutay et al., 1998). eEF1A recognizes aa-tRNAs with exquisite specificity (see Dreher et al., 1999), and the data suggest that eEF1A binds to Exp5 as the ternary GTP–eEF1A–aa-tRNA complex. We will return to this issue later.
We have demonstrated so far that a stable RanGTP– Exp5–eEF1A complex could form. In order to test if Ran regulates the interaction between Exp5 and its putative export cargo(es), we performed the assay the other way round and bound the cytosolic extract to immobilized Exp5. Strikingly, the addition of RanGTP greatly enhanced Exp5 binding of tRNA and eEF1A (Figure 1C), suggesting that the formed Exp5 complexes represent bona fide nuclear export intermediates.
Both isoforms of eEF1A are strictly excluded from nuclei
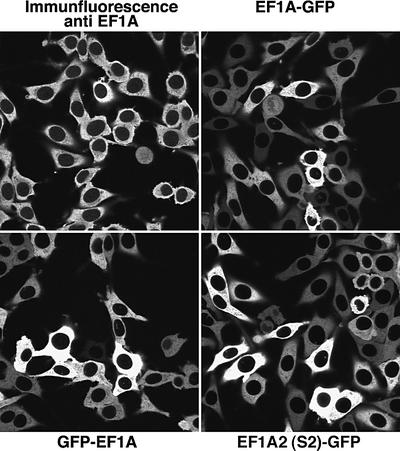
We next analysed the cellular distribution of eEF1A by two complementary methods. First, we performed immunofluorescence with highly specific antibodies raised against an exposed loop of the protein (see Materials and methods and Supplementary data). Secondly, we generated stably transfected BHK cell lines expressing three different green fluorescent protein (GFP)-tagged eEF1A fusions, namely GFP–eEF1A, eEF1A–GFP and GFP-tagged S2 (the second isoform of eEF1A). Confocal sections through these samples revealed in all cases an exclusive cytoplasmic localization and complete nuclear exclusion of the elongation factor (Figure 2). Quantitation of the data showed that the nuclear signal was at least 100 times weaker than that of the cytoplasm (see Table I). We found little variation in the eEF1A distribution between cell types. Our antibody cross-reacts perfectly with eEF1A from other mammals and even Drosophila, and we observed the same nearly complete nuclear exclusion of eEF1A in human HeLa cells, mouse 3T3 cells and Drosophila Schneider cells (data not shown). Likewise, the eEF1A–GFP fusions were also excluded from nuclei in CHO and 3T3 cells (not shown).
Fig. 2. eEF1A is strictly excluded from nuclei. The cellular distribution of eEF1A was studied by several methods. First, Alexa 488-labelled anti-eEF1A antibodies were used to localize the endogenous protein within cultured BHK cells. The indicated panels show the distribution of eEF1A–GFP or GFP–eEF1A fusion proteins in stably transfected BHK cell lines. Finally, GFP-tagged S2, the second isoform of eEF1A was also studied. All images represent confocal sections through the equators of the nuclei.
Table I. Quantitation of the cellular distribution of translation factors.
| Translation factor | Nuclear:cytoplasmic concentration ratio |
|---|---|
| eIF1 | ∼2.6 |
| eIF6 | ∼2.8 |
| eIF2β | ∼0.013 |
| eIF2γ | ∼0.013 |
| eIF2B-ε | ∼0.015 |
| eIF3-p44 | ∼0.073 |
| eIF4A1 | ∼0.018 |
| eIF5 | <0.01 |
| eEF1A | ∼0.01 |
| eEF1A-2 (S2) | <0.01 |
| eEF1Bβ | <0.01 |
| eEF2 | <0.01 |
| eRF1 | <0.014 |
Analysis is based on the cell lines shown in Figure 6. Nuclear and cytoplasmic fluorescence signals were integrated, the background (i.e. signal outside cells) was subtracted and the nuclear signal was normalized to the cytoplasmic one. The nuclear signal of the nuclear-excluded factors had to be given as an upper limit for the actual nuclear concentration, because some signal inevitably spills over from the very bright cytoplasm into the dark nuclear areas.
Exp5 is the functional export receptor for eEF1A
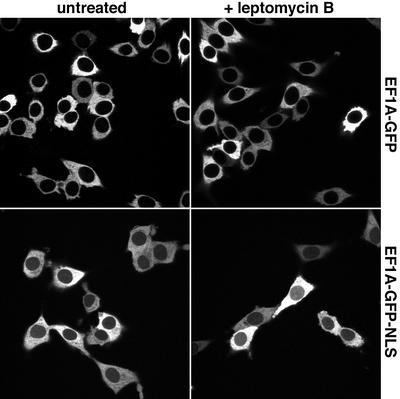
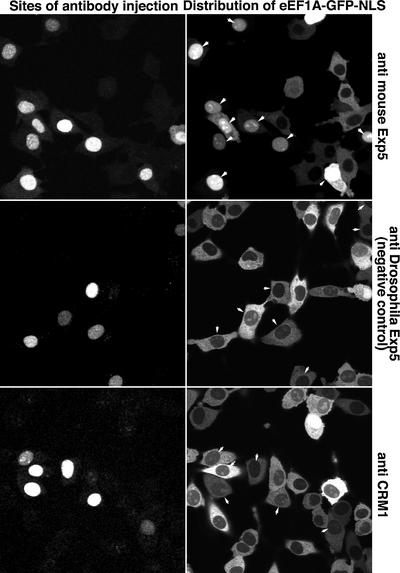
We next added the NLS from the SV40 large T antigen (Kalderon et al., 1984) to the reporter construct to generate an eEF1A–GFP–NLS fusion. Even though this NLS is a strong import signal, only a faint nuclear signal was observed (Figure 3), pointing to a very efficient nuclear export of the protein. Inhibiting CRM1 with leptomycin B (LMB; Nishi et al., 1994; Wolff et al., 1997) had no effect on eEF1A localization (Figure 3), which is consistent with the fact that eEF1A does not assemble with CRM1 into export complexes (Figure 1). However, injecting an affinity-purified anti-Exp5 antibody into nuclei caused a strong or even complete nuclear accumulation of the eEF1A–GFP–NLS fusion (Figure 4). The effect was highly specific and not observed after injection of antibodies raised against CRM1 or against Drosophila Exp5, which do not cross-react with the mammalian protein (Figure 4). Conversely, the anti-mouse Exp5 antibodies caused nuclear accumulation of eEF1A– GFP–NLS, but not of CRM1 substrates (not shown). Thus, we can conclude that eEF1A is actively depleted from nuclei and that Exp5 mediates this process.
Fig. 3. Neither an ectopic import signal nor a block of CRM1 export is sufficient for nuclear accumulation of eEF1A. Panels show the distribution of eEF1A–GFP and eEF1A–GFP–NLS fusions in stably transfected BHK cells. The addition of the NLS only produces a faint nuclear signal that is not enhanced further by a 30 min block of CRM1 by 5 ng/ml LMB.
Fig. 4. Exp5 is a functional nuclear export receptor for eEF1A. Monospecific antibodies raised against mouse Exp5 were injected into nuclei of BHK cells expressing the eEF1A–GFP–NLS fusion. Cells were fixed 90 min post-injection. The injection marker Texas red dextran and the eEF1A–GFP–NLS fusion protein were detected in separate fluorescence channels of the confocal laser scanning microscope. Arrows indicate injected cells. Note that anti-mouse Exp5-injected cells showed strong nuclear accumulation of the eEF1A–GFP–NLS fusion. Antibodies against CRM1 or Drosophila Exp5, which do not cross-react with mammalian Exp5, had no effect on the localization of the eEF1A–GFP–NLS fusion.
Exp5 binds eEF1A via tRNA
Figure 1 shows that not only eEF1A assembled into Exp5 complexes, but also low molecular weight RNAs, which could indicate that eEF1A is recovered as a complex with aa-tRNA. To clarify this issue and to ask whether Exp5 recognizes eEF1A, the tRNA or both, we performed binding experiments with fractionated HeLa extracts (Figure 5). eEF1A binding to Exp5 was completely abolished if the tRNA had been depleted with Q-Sepharose. Readdition of tRNA restored the binding. Binding was not restored if the tRNA had been deacylated prior to addition (Derwenskus et al., 1984), which is known to prevent the tRNA–eEF1A interaction (Dreher et al., 1999). We can thus conclude that Exp5 recognizes eEF1A only in complex with aa-tRNA. Interestingly, tRNA was recovered with Exp5 independently of the eEF1A, which clearly demonstrates that Exp5 is a tRNA-binding protein and that eEF1A is recovered in the export complex via the tRNA.
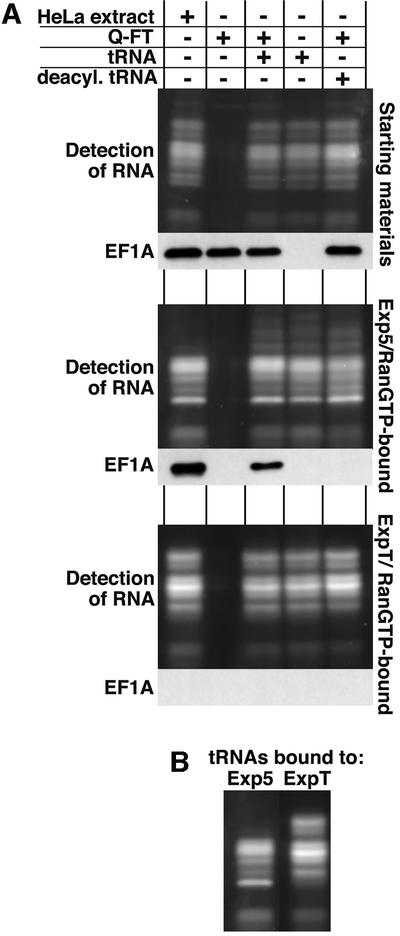
Fig. 5. The interaction between Exp5 and eEF1A is mediated by tRNA. (A) A cytosolic HeLa extract was depleted of nuclear transport receptors and fractionated further to separate eEF1A from tRNA: the flowthrough from a Q-Sepharose column contained eEF1A, but lacked tRNA, while the phenol/chloroform extract contained tRNA, but no protein. Where indicated, tRNA was used after deacylation. These fractions were tested singly or in combination for binding to Exp5/RanGTP or ExpT/RanGTP as in Figure 1. The figure shows starting materials (upper parts), Exp5-bound materials (middle parts) and ExpT-bound materials (lower parts). Detection of eEF1A was by western blotting. RNA was detected after Sybr-green staining of the 12% polyacrylamide gel. The load of the bound fractions corresponds to 10 times the starting materials. Note that eEF1A binding to Exp5 was strictly RNA dependent, but tRNA binding to Exp5 occurred independently of eEF1A. Deacylation of the tRNA (which prevents the tRNA–EF1A interaction) only abolished recruitment of eEF1A into Exp5 export complexes, but still allowed efficient tRNA binding to either Exp5 or ExpT. (B) A comparison of RNA patterns when bound to Exp5 and Exp-t.
Thus, Exp5 is the second Impβ-type transport receptor, besides Exp-t, that binds tRNA directly. Interestingly, a comparison of the pattern of bound RNA indicates that Exp5 and Exp-t differ in their preference for individual tRNA species (see Figure 5B). We are in the process of systematically testing the selectivity of Exp5 for different RNAs.
The vast majority of translation initiation, elongation and release factors show strict nuclear exclusion
It has been claimed recently that a significant proportion (10–15%) of cellular protein synthesis takes place in the nucleus, and occasional reports of nuclear pools for some translation factors were proposed to support that claim (Iborra et al., 2001). Nuclear translation, however, would require the complete set of essential translation factors at non-limiting concentrations in the nucleus. The striking nuclear exclusion of eEF1A already strongly argues against this scenario, but we wanted to address this issue systematically and obtain a comprehensive overview of the nuclear/cytoplasmic distribution of the key translation factors. For this purpose, we generated stably transduced cell lines expressing GFP fusions of initiation and elongation factors as well as a termination factor. Many of these translation factors are part of larger complexes; therefore, we had to avoid an overexpression situation, where an excess of the GFP-tagged subunit becomes only partly integrated into the respective complex. Thus, the fusions were expressed under control of the ‘Tet on’ system (Urlinger et al., 2000), and the inducer doxycycline was titrated down until the cellular distribution of the fusion proteins no longer changed (extrapolation to zero expression; for details see Materials and methods).
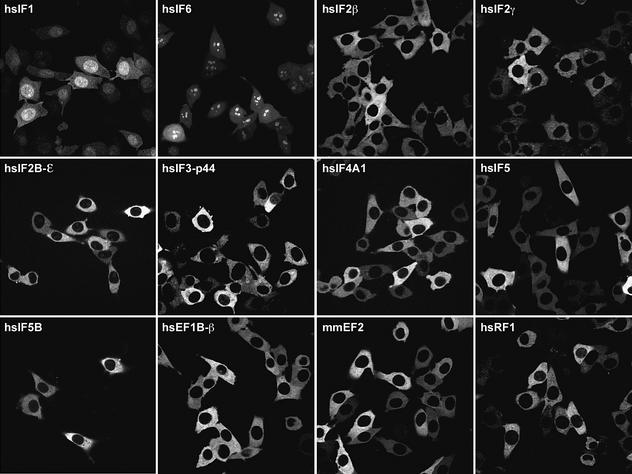
Of 12 translation factors tested, only three showed a clear nuclear pool, namely eIF1, eIF6 (Figure 6) and eIF1A (not shown). eIF6 functions, at least in yeast, in the biogenesis of the 60S ribosomal subunit and not in initiation. The cellular distribution of the human protein is consistent with this. eIF1 and eIF1A might also have as yet unidentified nuclear functions. In any case, they possess extended, rather basic mRNA-binding domains (Fletcher et al., 1999; Battiste et al., 2000) that bind several importins and appear to function as cryptic nuclear import signals (K.Regener, unpublished observation). Constant export of eIF1 and eIF1A is probably required to prevent complete nuclear accumulation and to ensure that cytoplasmic levels suffice for efficient translation. In the case of eIF1A, this export is mediated by Imp13 (Mingot et al., 2001). The export pathway for eIF1 currently is under investigation.
Fig. 6. The majority of translation factors show strict nuclear exclusion. Panels show confocal sections through BHK cells expressing the indicated GFP-tagged translation factors. eIF5B–GFP was introduced by transient transfection; all other translation factor–GFP fusions were expressed weakly in stable cell lines under tet control. For quantitation, see Table I; for details, see Materials and methods and main text.
All other translation factors, however, showed a clear nuclear exclusion (see Figure 6 for images and Table I for quantitation). These are the eIF2 complex (tested for the β and γ subunits), the eIF2B complex (tested for the ε subunit), eIF3 (tested for the p44 subunit), eIF4AI, eIF5 and eIF5B. The three elongation factors, i.e. eEF1A (see above), its nucleotide exchange factor eEF1B and elongation factor eEF2, are also firmly excluded from nuclei, and even the termination factor eRF1 has only a negligible nuclear pool.
CRM1 mediates nuclear exclusion of several translation factors
Although NPCs restrict the flux of inert macromolecules into nuclei, they do not represent absolute barriers. Even macromolecules above the ‘passive diffusion limit’ of ∼40 kDa will enter nuclei, though equilibration might take hours. The very steep nucleocytoplasmic gradient in translation factor concentration can therefore not be explained just by a very slow entry rate. Instead, active export must also be required to maintain such gradients. In addition, active export of translation factors will be part of the cellular efforts to separate cytoplasmic from nuclear contents after the mixing of the two compartments during open mitosis.
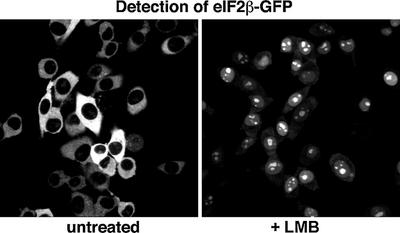
CRM1 is the exportin with the broadest substrate specificity. We therefore wanted to test if this exportin also exports translation factors and so used the CRM1-specific inhibitor LMB for this purpose. The eIF2β subunit shifted, after 30 min of LMB treatment, from its exclusively cytoplasmic to a predominantly nuclear localization (Figure 7). The protein might contain a weak import signal that is counteracted by very efficient CRM1-mediated export. In contrast, LMB treatment was insufficient to cause nuclear accumulation of the eIF2γ subunit and, even in combination with a fused NLS, only a weak nuclear signal was observed (Figure 8). This indicates that an exportin distinct from CRM1 recognizes eIF2γ. It thus appears that at least two export pathways synergize to expel the active eIF2 complex from the nucleus.
Fig. 7. CRM1 excludes eIF2β from nuclei. Panels show confocal sections through stably transduced BHK cells expressing an eIF2β–GFP fusion. Where indicated, cells had been treated with 5 ng/ml LMB before fixation. Note that LMB treatment changed the eIF2β localization from completely cytoplasmic to predominantly nuclear. The duration of LMB treatment was kept short (30 min) to rule out non-specific side effects of the drug.
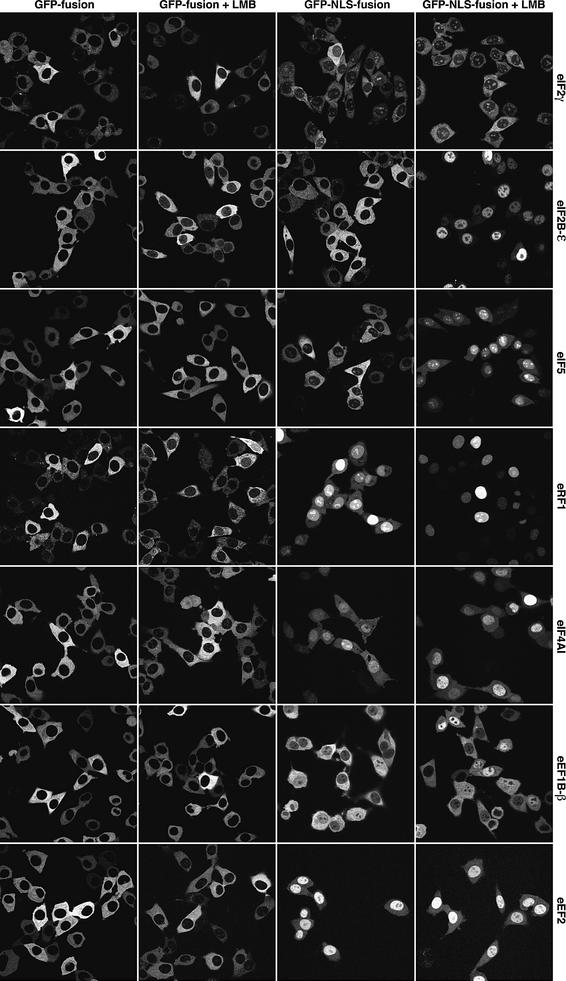
Fig. 8. CRM1 contributes to nuclear exclusion of several translation factors. Stably transduced BHK cells expressing the indicated translation factors were analysed. The translation factors were tagged with either GFP alone or with GFP–NLS (to increase the nuclear influx rate). The distribution of the fusion proteins was analysed by laser scanning microscopy without or after LMB treatment (5 ng/ml for 30 min).
Elongation factors eEF2 and eEF1B are examples of a fully CRM1-independent nuclear exclusion (Figure 8); LMB treatment had no effect on the localization of the corresponding GFP or GFP–NLS fusions. The fact that the ectopic NLS was insufficient for complete nuclear accumulation points to CRM1-independent export of the fusions. We are currently seeking the corresponding exportin(s).
CRM1 might also be involved in export of eIF4A1, because LMB treatment shifted the eIF4A1–GFP fusion from a complete nuclear exclusion to a weak nucleolar signal (Figure 8). The eIF4A1–GFP–NLS fusion showed a heterogeneous pattern. In ∼50% of the cells, a predominant nuclear signal was observed. This number increased to ∼70% after LMB treatment. Nevertheless, the accumulation was incomplete, and ∼30% of the cells only showed a nucleocytoplasmic equilibration of the fusion protein. CRM1 is therefore probably not the only exportin that takes eIF4A1 to the cytoplasm.
The initiation factors eIF2Bε and eIF5 as well as the termination factor eRF1 are clearly CRM1 export substrates (Figure 8). Like many other translation factors, however, they enter nuclei only very reluctantly, and a clear nuclear accumulation only occurred when the LMB treatment was combined with the fusion of an ectopic NLS to the translation factors.
Discussion
Is there such a thing as nuclear translation?
Translation in prokaryotes is coupled to transcription and already initiated on nascent transcripts. Typical eukaryotic genes, however, contain introns and so the pre-mRNA must be fully spliced before the message can be translated into a functional gene product. Eukaryotes elegantly enforce this order of events with the help of the NE; they confine translation to the cytoplasm and thus keep it separate from the nuclear processes of transcription and RNA processing.
Incorrectly spliced or mutant mRNAs with premature stop codons become rapidly degraded through the nonsense-mediated decay mechanism (NMD), which detects nonsense codons during a pioneer round of translation (Hentze and Kulozik, 1999; Lykke-Andersen, 2001; Maquat and Carmichael, 2001; Schell et al., 2002). NMD in yeast is clearly cytoplasmic (Gonzalez et al., 2001). In higher eukaryotes, NMD also appears to occur largely in the cytoplasm, but nuclear translation has been suggested as a proof-reading mechanism for mRNAs prior to export (see, for example, Buhler et al., 2002). Before we discuss the firm experimental evidence against nuclear translation, several conceptual problems with the nuclear translational proof-reading should be mentioned. First, such proof-reading of each and every message would require an unreasonably large translation capacity in the nucleus. Secondly, there is no explanation as to how this proof-reading could be suppressed before completion of splicing attempts and yet be enforced before export occurs. Thirdly, the proof-reading would have to occur between release of the mRNA from the spliceosomes and export; this time window is very short (Custodio et al., 1999) and in fact probably far shorter than the time required for a full translation of the message (with, at best, trace amounts of elongation factors). Finally, the positioning of splice signatures, such as the exon junction complex, plausibly explains how the first round of cytoplasmic translation can detect premature stop codons and initiate NMD (reviewed in Gonzalez et al., 2001); there is simply no need to implicate a nuclear proof-reading event.
Nevertheless, it was claimed recently that 10–15% of cellular protein synthesis would occur in the nucleus (Iborra et al., 2001). In that study, nascent polypeptides had been labelled by incorporation of a fluorescent dye (BODIPY) from an ε-modified lysyl tRNALys, and some nuclear signal was taken as proof of nuclear translation. The fundamental problems are that nuclear import can occur in the time scale of seconds (Ribbeck and Görlich, 2001) and that the BODIPY detection cannot distinguish translation intermediates from biotinylated polypeptides that entered nuclei post-translationally. These authors dismissed any nuclear import of translation products and argued that their assay would operate without nucleo cytoplasmic exchange. They ignored the fact that their assay strictly depends on nucleocytoplasmic exchange, as the (cytoplasmically added) ε-modified lysyl tRNALys would have to reach the ‘nuclear translation sites’. If a charged tRNA (25 kDa) can enter nuclei during the assay, then small translation products can too, and it is simply impossible to tell whether the BODIPY moiety arrived in the nuclear compartment as an aa-tRNA or after cytoplasmic incorporation into a polypeptide. The approach is thus unsuitable for identifying translation sites, and the data provide no support for nuclear translation.
A second study (Brogna et al., 2002), claiming the association of ribosomes and translation factors with nascent transcripts at Drosophila chromosomes, must be viewed equally critically. The technical problem there is that the specimens were first treated with a high concentration of detergent before the chromosomes were isolated. The treatment dissolves the NE, and no control excluded the (probable) scenario of translation components reaching the transcription sites after the detergent treatment. In fact, we observed that nuclei of cultured Drosophila Schneider cells lack eEFIA (not shown) and thus probably the capacity of translation.
Ultrastructural evidence clearly stands against nuclear translation. For example, membrane-bound ribosomes (which synthesize proteins destined for the secretory pathway) are highly abundant at the outer nuclear membrane, but absent from the inner one (see, for example, Sabatini and Kreibich, 1975). Likewise, the study of the giant Balbiani ring mRNAs has clearly demonstrated that mRNAs do not assemble into polysomes until their 5′ end has reached the cytoplasm (reviewed in Daneholt, 1997). Finally, it has to be emphasized that nuclear translation would be a rather wasteful and even dangerous process: translation of incompletely spliced messages would yield truncated proteins that are not only non-functional, but potentially even act as dominant-negative inhibitors. An additional problem is the lack of functional rough endoplasmic reticulum within the nuclei, which implies that nuclear proteins with hydrophobic signal sequences would miss the secretory pathway and instead end up in aggregates.
Mechanisms to suppress nuclear translation
As tRNAs, mRNAs and ribosomes originate from the nucleus, eukaryotic cells have to employ efficient mechanisms to suppress premature translation of mRNA in the nucleus. This suppression might already occur at the level of ribosome biogenesis. For example, the 20S rRNA of the small ribosomal subunit is processed to the mature 18S form only after export to the cytoplasm (Hannon et al., 1989; Stevens et al., 1991; Moy and Silver, 1999; Vanrobays et al., 2001). Likewise, the ribosomal proteins P1 and P2 are added to pre-ribosomal particles after export to the cytoplasm (see Zurdo et al., 2000, and references therein). Ribosomes might therefore acquire translation competence only after their arrival in the cytoplasmic compartment. We have investigated here another mechanism and demonstrated that translation factors are actively depleted from nuclei, typically down to 1/100 of their cytoplasmic concentrations. This applies to all elongation factors (eEF1B, eEF2 and both isoforms of eEF1A), to the termination factor eRF1 and the initiation factors eIF2 (β and γ subunits), eIF2B, eIF5, eIF5B and eIF4A1 (see Figures 2 and 6). In addition, available published data indicate that the nuclear steady-state levels of other subunits of the eIF3 and eIF4 complexes as well as of the small and large ribosomal subunits correspond to, at most, a tiny fraction of the cytoplasmic one (see, for example, Valasek et al., 2001; Kedersha et al., 2002).
Translation is a multimolecular reaction. Its efficiency will therefore decrease not just linearly with dilution of the active components, but rather with a power >1. If several essential translation factors are depleted to ∼1% of their optimal concentration, the translation efficiency will drop to a far smaller percentage and might even go down to zero. eEF1A is a good example to address synergistic effects of multiple depletions. eEF1A is not only a rare species in the nucleus (Figure 2), but the nuclear exclusion of its GEF (eEF1B, Figure 6) would prevent its multi-round action: even if an eEF1A molecule could deliver an aa-tRNA to an elongating ribosome, it would persist thereafter in the GDP-bound form, unable to recruit another aa-tRNA. Likewise, eIF2 (the carrier of the initiator tRNA) not only is depleted in the nucleus to very low concentrations, but the nuclear pool will be largely blocked in the GTPase cycle, because the eIF2-specific GEF (eIF2B) and GAP (eIF5) are also excluded.
Several export pathways synergize to keep translation cytoplasmic. First, there is export of eIF1A by Imp13 (Mingot et al., 2001). Secondly, CRM1 mediates export of eIF2 (via the β-subunit), eIF2B, eIF5 and eRF1 (Figures 7 and 8). CRM1 also makes a small contribution to export of eIF4A1, but additional (as yet unidentified) exportins appear to be involved. Finally, Exp5 exports eEF1A (via aa-tRNA), while the export mechanism for the elongation factors eEF1B and eEF2 remains elusive. An interesting point is that exportins might not only interfere with nuclear translation by physically removing translation factors from the nucleus, but they might act as nucleus-specific inhibitors of translation, because the binding of a bulky exportin–RanGTP complex to a translation factor, to a ribosomal subunit or (via CBC and PHAX; Ohno et al., 2000) to the 7mG cap of mRNAs is likely to interfere with translation-relevant interactions. We are testing this at present.
Exp5/RanBP21: a second RNA-binding exportin
The export of eEF1A by Exp5 is a highly unusual example of protein export in that the protein is not recognized directly, but via aa-tRNA instead. To our knowledge, this is the first example of protein export through an RNA adaptor.
Exp5 might also contribute to the export of other proteins, but so far we have found no indications for a direct interaction between Exp5 and any proteinaceous cargo. This contrasts with the view of Brownawell and Macara (2002), who observed that dsRNA competes the interaction between Exp5 and a dsRNA-binding protein and concluded that Exp5 recognizes dsRNA-binding domains. We found that Exp5 directly binds dsRNAs (not shown) and we would therefore give a different interpretation of the Brownawell experiment: the dsRNA-binding protein is recruited to Exp5 indirectly via RNA (that is already present in the extract), and excess of free RNA therefore competes the Exp5–RNPs interaction.
Exp5 obviously mediates tRNA export in parallel with Exp-t. Such a parallel pathway could already be anticipated from published data. For example, anti-Exp-t antibodies do not block tRNA export completely (Arts et al., 1998b; Lipowsky et al., 1999) and we would assume that the residual export was mediated by Exp5. Clear indications also came from studies in the yeast Saccharomyces cerevisiae (Sarkar and Hopper, 1998; Grosshans et al., 2000), where inactivation of LOS1 (coding for the Exp-t orthologue) only causes a mild block in tRNA export. A far stronger effect was observed with mutations in RNA1 (coding for the RanGAP), which should disrupt not only Los1p-dependent export, but also transport by other exportins. The yeast homologue of Exp5 is called Msn5p. It exports several phosphorylated transcription factors (Kaffman et al., 1998) and would be an excellent candidate for the suspected second exporter for tRNA in yeast and perhaps for the yeast eEF1A–aa-tRNA complex.
Even though both Exp-t and Exp5 act in tRNA export, there are also clear differences between the two. First, they bind to distinct sites on the tRNA. This is evident from the fact that eEF1A can bind to the same tRNA molecule together with Exp5, but not with Exp-t (Figures 1 and 5). Secondly, the two exportins appear to enrich complementary sets of tRNA (see Figure 5B). Thirdly, while Exp-t has an exquisite specificity for mature tRNA, Exp5 appears less ‘choosy’ and also recognizes, for example, an U1 snRNA, 7S RNA from the signal recognition particle or naked mRNAs (not shown). Exp5 might thus synergize with other RNA exporters, such as with the CBC– PHAX–CRM1 complex in U snRNA export (Ohno et al., 2000) or with TAP/Mex67 in mRNA export (see Segref et al., 1997; Grüter et al., 1998) whenever high-affinity RNA binders spare suitable RNA motifs for Exp5.
Materials and methods
Molecular cloning of mouse exportin 5
We identified several human and mouse ESTs of, at that time, an uncharacterized mammalian cDNA which displayed similarity to members of the Impβ family, in particular to MSN5 from yeast. Based on this sequence information, we cloned full-length Exp5 cDNA from human and mouse by RT–PCR and also identified two ESTs coding for the Drosophila protein.
Antibodies
Antibodies against human CRM1, and mouse and Drosophila Exp5 were raised against the C-termini of the proteins. Anti-eEF1A antibodies were raised against residues 48–57 of the human protein. Antibodies against snurportin 1, eIF1A and Impα were raised against the full-length proteins. All antibodies were raised in rabbits and used after affinity purification on the immobilized antigens. For microinjections, affinity-purified antibodies were concentrated by ammonium sulfate precipitation to 15 mg/ml and dialysed extensively against phosphate-buffered saline (PBS). Test blots with whole-cell lysates verified their monospecificity.
Recombinant protein expression and purification
Expression and purification of the following proteins has been described previously: zzRanQ69L, CAS, CRM1, Exp4, Exp-t, Impβ, transportin 1, Imp13 and Imp7 (see Mingot et al., 2001, and references therein). RanBP16, transportin SR2 (human Mtr10a) and Exp5 were expressed from pQE derivatives with histidine tags and purified on nickel-NTA–agarose, followed by gel filtration on Superdex 200.
Binding assays
The selective depletion of nuclear transport receptors from a cytosolic HeLa extract with low substitution phenyl-Sepharose has been described previously (Ribbeck and Görlich, 2002). To remove RNA from a cytosolic HeLa extract, the extract was adjusted to 50 mM NaCl and passed through a Q-Sepharose FF column. eEF1A was recovered in the flowthrough fractions, while RNA bound tightly to the matrix. As the nucleotide-free form of mammalian eEF1A appears highly unstable, the partially purified preparation was supplemented immediately with GTP and a GTP-regenerating system.
Low molecular weight RNA was prepared from cytosolic HeLa extract by extraction with phenol (pH 5.0) and chloroform and finally precipitated with ethanol. Deacylation of tRNA was performed as described by Derwenskus et al. (1984).
Binding assays with zz-tagged components immobilized to IgG–Sepharose were performed at 50 mM NaCl essentially as described (Mingot et al., 2001). Specific details are given in the figure legends.
Immunofluorescence
Cells were grown on coverslips, fixed for 7 min with 3% paraformaldehyde and 0.1% glutaraldehyde, quenched with 1 mg/ml NaBH4, permeabilized with 0.1% Triton X-100 and blocked with 1% bovine serum albumin (BSA) in 10% rabbit normal serum. The sample was then incubated with the affinity-purified Alexa488-labelled anti-EF1A antibody. The fluorescent signal was recorded with a confocal scanning laser microscope.
Stable cell lines expressing translation factor–GFP fusions under Tet control
The generation of such cell lines by retroviral transduction has been described (Engling et al., 2002). cDNA clones for human eIF1, eIF2β, eIF2γ, eIF2B-ε, eIF3-p44, eIF5, eIF6 and eEF1A-2 (S2), and mouse eEF2 were obtained from the Resource Centre/Primary Database (RZPD). eIF1A, eIF4A-1, eEF1A-1, eEF1B-β and eRF1 were cloned directly from human cDNA. Translation factors were cloned in front of an eGFP or eGFP–NLS (GPKKKRKVE) cassette of a pRevTRE2 vector (Clontech). eEF1A-1 was also expressed as a GFP–EF1A-1 construct. Expression of GFP fusions was induced for 24 h by the following optimized doxycycline concentrations: 100 ng/ml for eIF2B-ε and eEF1A; 50 ng/ml for eIF2γ and eIF5; 10 ng/ml for eEF1B-β and eEF2; 5 ng/ml for eIF6, eIF2β, eIF4A1 and eRF1; and 1 ng/ml for eIF3-p44, eIF1 and eIF1A.
Microinjection of antibodies
Nuclear injections into cells grown on coverslips were performed utilizing the Zeiss AIS system with an inverse Axiovert 35 microscope. Capillaries had a tip size of 0.7 µm. Injection was for 0.3 s at 100–400 hPa. After injection, cells were grown for 1.5 h, fixed and examined by confocal laser scanning microscopy.
DDBJ/EMBL/GenBank accession numbers
The nucleotide sequences reported in this study have been deposited under the following accession numbers: mouse RanBP21/ Exp5, ACC AF343581; human RanBP21/Exp5, ACC AF271159; and Drosophila RanBP21/Exp5, ACC AF222746.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We wish to thank A.Kehlenbach for cell sorting, A.Bosserhoff and T.Ruppert for mass spectrometry, P.Rübmann and U.Jäkle for excellent technical help, M.Trendelenburg for access to his microinjection facility, and D.Mohr, J.M.Mingot and M.Pool for critical reading of the manuscript. We are grateful to the DFG (SFB 352 and Graduiertenkolleg Molekulare Zellbiologie) and the Alfried Krupp Foundation for financial support.
References
- Arts G.J., Fornerod,M. and Mattaj,I.W. (1998a) Identification of a nuclear export receptor for tRNA. Curr. Biol., 8, 305–314. [DOI] [PubMed] [Google Scholar]
- Arts G.J., Kuersten,S., Romby,P., Ehresmann,B. and Mattaj,I.W. (1998b) The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J., 17, 7430–7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battiste J.L., Pestova,T.V., Hellen,C.U. and Wagner,G. (2000) The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol. Cell, 5, 109–119. [DOI] [PubMed] [Google Scholar]
- Berget S.M., Moore,C. and Sharp,P.A. (1977) Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc. Natl Acad. Sci. USA, 74, 3171–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogna S., Sato,T.A. and Rosbash,M. (2002) Ribosome components are associated with sites of transcription. Mol. Cell, 10, 93–104. [PubMed] [Google Scholar]
- Brownawell A.M. and Macara,I.G. (2002) Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins. J. Cell Biol., 156, 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M., Wilkinson,M.F. and Muhlemann,O. (2002) Intranuclear degradation of nonsense codon-containing mRNA. EMBO rep., 3, 646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E. and Izaurralde,E. (2001) Nucleocytoplasmic transport enters the atomic age. Curr. Opin. Cell Biol., 13, 310–319. [DOI] [PubMed] [Google Scholar]
- Custodio N., Carmo-Fonseca,M., Geraghty,F., Pereira,H.S., Grosveld,F. and Antoniou,M. (1999) Inefficient processing impairs release of RNA from the site of transcription. EMBO J., 18, 2855–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg J.E. and Lund,E. (1998) Functions of the GTPase Ran in RNA export from the nucleus. Curr. Opin. Cell Biol., 10, 400–408. [DOI] [PubMed] [Google Scholar]
- Daneholt B. (1997) A look at messenger RNP moving through the nuclear pore. Cell, 88, 585–588. [DOI] [PubMed] [Google Scholar]
- Das H.K., Goldstein,A. and Lowney,L.I. (1967) Attachment of ribosomes to nascent messenger RNA in Escherichia coli. J. Mol. Biol., 24, 231–245. [DOI] [PubMed] [Google Scholar]
- Derwenskus K.H., Fischer,W. and Sprinzl,M. (1984) Isolation of tRNA isoacceptors by affinity chromatography on immobilized bacterial elongation factor Tu. Anal. Biochem., 136, 161–167. [DOI] [PubMed] [Google Scholar]
- Dreher T.W., Uhlenbeck,O.C. and Browning,K.S. (1999) Quantitative assessment of EF-1α·GTP binding to aminoacyl-tRNAs, aminoacyl-viral RNA and tRNA shows close correspondence to the RNA binding properties of EF-Tu. J. Biol. Chem., 274, 666–672. [DOI] [PubMed] [Google Scholar]
- Engling A., Backhaus,R., Stegmayer,C., Zehe,C., Seelenmeyer,C., Kehlenbach,A., Schwappach,B., Wegehingel,S. and Nickel,W. (2002) Biosynthetic FGF-2 is targeted to non-lipid raft micro domains following translocation to the extracellular surface of CHO cells. J. Cell Sci., 115, 3619–3631. [DOI] [PubMed] [Google Scholar]
- Fletcher C.M., Pestova,T.V., Hellen,C.U. and Wagner,G. (1999) Structure and interactions of the translation initiation factor eIF1. EMBO J., 18, 2631–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M., Ohno,M., Yoshida,M. and Mattaj,I.W. (1997) Crm1 is an export receptor for leucine rich nuclear export signals. Cell, 90, 1051–1060. [DOI] [PubMed] [Google Scholar]
- Gadal O., Strauss,D., Kessl,J., Trumpower,B., Tollervey,D. and Hurt,E. (2001) Nuclear export of 60S ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol., 21, 3405–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C.I., Bhattacharya,A., Wang,W. and Peltz,S.W. (2001) Nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Gene, 274, 15–25. [DOI] [PubMed] [Google Scholar]
- Görlich D. and Jäkel,S. (2002) Nucleocytoplasmic transport. In Dalbey,R.E. and von Heijne,G. (eds), Protein Targeting, Transport and Translocation. Academic Press, New York, pp. 293–321.
- Görlich D. and Kutay,U. (1999) Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell. Dev. Biol., 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Grosshans H., Hurt,E. and Simos,G. (2000) An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes Dev., 14, 830–840. [PMC free article] [PubMed] [Google Scholar]
- Grüter P., Tabernero,C., von Kobbe,C., Schmitt,C., Saavedra,C., Bachi,A., Wilm,M., Felber,B.K. and Izaurralde,E. (1998) TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell, 1, 649–659. [DOI] [PubMed] [Google Scholar]
- Hannon G.J., Maroney,P.A., Branch,A., Benenfield,B.J., Robertson,H.D. and Nilsen,T.W. (1989) Accurate processing of human pre-rRNA in vitro. Mol. Cell. Biol., 9, 4422–4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings M.L. and Krainer,A.R. (2001) Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol., 13, 302–309. [DOI] [PubMed] [Google Scholar]
- Hentze M.W. and Kulozik,A.E. (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell, 96, 307–310. [DOI] [PubMed] [Google Scholar]
- Iborra F.J., Jackson,D.A. and Cook,P.R. (2001) Coupled transcription and translation within nuclei of mammalian cells. Science, 293, 1139–1142. [DOI] [PubMed] [Google Scholar]
- Kaffman A., Rank,N.M., O’Neill,E.M., Huang,L.S. and O’Shea,E.K. (1998) The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature, 396, 482–486. [DOI] [PubMed] [Google Scholar]
- Kalderon D., Richardson,W.D., Markham,A.F. and Smith,A.E. (1984) Sequence requirements for nuclear location of simian virus 40 large T antigen. Nature, 311, 33–38. [DOI] [PubMed] [Google Scholar]
- Kedersha N., Chen,S., Gilks,N., Li,W., Miller,I.J., Stahl,J. and Anderson,P. (2002) Evidence that ternary complex (eIF2– GTP–tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell, 13, 195–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U., Bischoff,F.R., Kostka,S., Kraft,R. and Görlich,D. (1997) Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell, 90, 1061–1071. [DOI] [PubMed] [Google Scholar]
- Kutay U., Lipowsky,G., Izaurralde,E., Bischoff,F.R., Schwarzmaier,P., Hartmann,E. and Görlich,D. (1998) Identification of a tRNA-specific nuclear export receptor. Mol. Cell, 1, 359–369. [DOI] [PubMed] [Google Scholar]
- Legrain P. and Rosbash,M. (1989) Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell, 57, 573–583. [DOI] [PubMed] [Google Scholar]
- Lipowsky G., Bischoff,F.R., Izaurralde,E., Kutay,U., Schäfer,S., Gross,H.J., Beier,H. and Görlich,D. (1999) Coordination of tRNA nuclear export with processing of tRNA. RNA, 5, 539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipowsky G., Bischoff,F.R., Schwarzmaier,P., Kraft,R., Kostka,S., Hartmann,E., Kutay,U. and Görlich,D. (2000) Exportin 4: a mediator of a novel nuclear export pathway in higher eukaryotes. EMBO J., 19, 4362–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E. and Dahlberg,J.E. (1998) Proofreading and aminoacylation of tRNAs before export from the nucleus. Science, 282, 2082–2085. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J. (2001) mRNA quality control: marking the message for life or death. Curr. Biol., 11, R88–R91. [DOI] [PubMed] [Google Scholar]
- Maquat L.E. and Carmichael,G.G. (2001) Quality control of mRNA function. Cell, 104, 173–176. [DOI] [PubMed] [Google Scholar]
- Mattaj I.W. and Englmeier,L. (1998) Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem., 67, 265–306. [DOI] [PubMed] [Google Scholar]
- Mehdi Q. and Yudkin,M.D. (1967) Coupling of transcription to translation in the induced synthesis of β-galactosidase. Biochim. Biophys. Acta., 149, 288–290. [DOI] [PubMed] [Google Scholar]
- Merrick C.W. and Nyborg,J. (2000) The protein biosynthesis elongation cycle. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 89–125.
- Miller O.L. Jr, Hamkalo,B.A. and Thomas,C.A.,Jr (1970) Visualization of bacterial genes in action. Science, 169, 392–395. [DOI] [PubMed] [Google Scholar]
- Mingot J.M., Kostka,S., Kraft,R., Hartmann,E. and Görlich,D. (2001) Importin 13: a novel mediator of nuclear import and export. EMBO J., 20, 3685–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy T.I. and Silver,P.A. (1999) Nuclear export of the small ribosomal subunit requires the ran-GTPase cycle and certain nucleoporins. Genes Dev., 13, 2118–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S. and Dreyfuss,G. (1999) Transport of proteins and RNAs in and out of the nucleus. Cell, 99, 677–690. [DOI] [PubMed] [Google Scholar]
- Nishi K., Yoshida,M., Fujiwara,D., Nishikawa,M., Horinouchi,S. and Beppu,T. (1994) Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J. Biol. Chem., 269, 6320–6324. [PubMed] [Google Scholar]
- Ohno M., Segref,A., Bachi,A., Wilm,M. and Mattaj,I.W. (2000) PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell, 101, 187–198. [DOI] [PubMed] [Google Scholar]
- Paraskeva E., Izaurralde,E., Bischoff,F.R., Huber,J., Kutay,U., Hartmann,E., Lührmann,R. and Görlich,D. (1999) CRM1-mediated recycling of snurportin 1 to the cytoplasm. J. Cell Biol., 145, 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Kolupaeva,V.G., Lomakin,I.B., Pilipenko,E.V., Shatsky,I.N., Agol,V.I. and Hellen,C.U. (2001) Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl Acad. Sci. USA, 98, 7029–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K. and Görlich,D. (2001) Kinetic analysis of translocation through nuclear pore complexes. EMBO J., 20, 1320–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K. and Görlich,D. (2002) The permeability barrier of nuclear pore complexes appears to operate through hydrophobic exclusion. EMBO J., 21, 2664–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini D.D. and Kreibich,G. (1975) Functional specialization of membrane-bound ribosomes in eukaryotic cells. In Martonosi,A.N. (ed.), Enzymes of Biological Membranes. Plenum Publishing Corp., NY, Vol. 2, pp. 531–579.
- Sarkar S. and Hopper,A.K. (1998) tRNA nuclear export in Saccharomyces cerevisiae: in situ hybridization analysis. Mol. Biol. Cell, 9, 3041–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell T., Kulozik,A.E. and Hentze,M.W. (2002) Integration of splicing, transport and translation to achieve mRNA quality control by the nonsense-mediated decay pathway. Genome Biol., 3, 1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A., Sharma,K., Doye,V., Hellwig,A., Huber,J., Lührmann,R. and Hurt,E. (1997) Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J., 16, 3256–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens A., Hsu,C.L., Isham,K.R. and Larimer,F.W. (1991) Fragments of the internal transcribed spacer 1 of pre-rRNA accumulate in Saccharomyces cerevisiae lacking 5′–3′ exoribonuclease 1. J. Bacteriol., 173, 7024–7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom A.C. and Weis,K. (2001) Importin-β-like nuclear transport receptors. Genome Biol., 2, 3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlinger S., Baron,U., Thellmann,M., Hasan,M.T., Bujard,H. and Hillen,W. (2000) Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc. Natl Acad. Sci. USA, 97, 7963–7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L., Hasek,J., Nielsen,K.H. and Hinnebusch,A.G. (2001) Dual function of eIF3j/Hcr1p in processing 20S pre-rRNA and translation initiation. J. Biol. Chem., 276, 43351–43360. [DOI] [PubMed] [Google Scholar]
- Vanrobays E., Gleizes,P.E., Bousquet-Antonelli,C., Noaillac-Depeyre,J., Caizergues-Ferrer,M. and Gelugne,J.P. (2001) Processing of 20S pre-rRNA to 18S ribosomal RNA in yeast requires Rrp10p, an essential non-ribosomal cytoplasmic protein. EMBO J., 20, 4204–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch E.M., Wang,W. and Peltz,S.W. (2000) Translation termination: it’s not the end of the story. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 467–485.
- Wolff B., Sanglier,J.J. and Wang,Y. (1997) Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol., 4, 139–147. [DOI] [PubMed] [Google Scholar]
- Zurdo J., Parada,P., van den Berg,A., Nusspaumer,G., Jimenez-Diaz,A., Remacha,M. and Ballesta,J.P. (2000) Assembly of Saccharomyces cerevisiae ribosomal stalk: binding of P1 proteins is required for the interaction of P2 proteins. Biochemistry, 39, 8929–8934. [DOI] [PubMed] [Google Scholar]