Abstract
Transport of proteins and RNA into and out of the cell nucleus is mediated largely by a family of RanGTP-binding transport receptors. Export receptors (exportins) need to bind RanGTP for efficient loading of their export cargo. We have identified eukaryotic elongation factor 1A (eEF1A) and tRNA as RanGTP-dependent binding partners of exportin-5 (Exp5). Exp5 stimulates nuclear export of eEF1A when microinjected into the nucleus of Xenopus laevis oocytes. Surprisingly, the interaction between eEF1A and Exp5 is dependent on tRNA that can interact directly with Exp5 and, if aminoacylated, recruits eEF1A into the export complex. These data suggested to us that Exp5 might support tRNA export. Indeed, not only the canonical tRNA export receptor, exportin-t, but also Exp5 can drive nuclear export of tRNA. Taken together, we show that there exists an alternative tRNA export pathway which can be exploited to keep eEF1A out of the cell nucleus.
Keywords: eEF1A/export receptor/exportin-5/nucleo-cytoplasmic transport/tRNA
Introduction
Exchange of proteins and RNAs between the cytoplasm and the nucleus occurs through nuclear pore complexes (NPCs). Selectivity in transport is achieved by nuclear transport receptors that specifically recognize their transport substrates and facilitate the translocation of the cargo through the NPC by interacting with NPC constituents, the nucleoporins. Transport receptors may recognize and bind their cargo directly or associate with adaptor proteins that function in substrate selection. Most of these transport receptors belong to a protein family that, in higher eukaryotes, comprises >20 different importins and exportins, driving import into or export out of the nucleus and, in some cases, transport in both directions. To function in multiple rounds of transport, these receptors shuttle continuously between the cytoplasm and the nucleus (for reviews, see Mattaj and Englmeier, 1998; Görlich and Kutay, 1999; Kuersten et al., 2001; Macara, 2001).
All members of this transport receptor family share a similar molecular architecture and can interact specifically with the GTP-bound form of the small GTPase Ran. Binding of RanGTP to importins and exportins controls their compartment-specific interaction with the transport cargo and thereby confers directionality to the transport reaction. RanGTP is distributed asymmetrically in the cell. In the nucleus, Ran is maintained in its GTP-bound form by the chromatin-associated nucleotide exchange factor RCC1. RanGTP binding to importins in the nucleus causes release of the substrate after the import reaction. In contrast, RanGTP binding to exportins in the nucleus facilitates cargo loading on to export receptors, resulting in the formation of export complexes consisting of the exportin, RanGTP and the export substrate. In the cytoplasm, RanGTP is not stable because it is actively converted to RanGDP by the cytoplasmic RanGTPase-activating protein (RanGAP). Therefore, import complexes can form efficiently in this compartment. Con versely, disassembly of export complexes in the cytoplasm is triggered by the action of the RanGAP and additional factors (for reviews, see Mattaj and Englmeier, 1998; Görlich and Kutay, 1999).
In higher eukaryotes, six exportins involved in protein and RNA export from the cell nucleus have been described (for a review, see Macara, 2001 and references therein). Some are specialized to transport one particular type of cargo, such as CAS, which exports the import adaptor protein importin α (Kutay et al., 1997). In contrast, CRM1 (exportin-1) is a general exportin and functions in nuclear export of a large variety of different proteins, many of which contain a leucine-rich nuclear export signal (NES) (Fornerod et al., 1997; Fukuda et al., 1997). CRM1 can also export RNA, since certain NES-containing proteins serve as adaptors for the export of specific RNAs out of the nucleus (Fischer et al., 1995; Ho et al., 2000; Ohno et al., 2000; Gadal et al., 2001).
Nuclear export of tRNAs in higher eukaryotes is driven by a specialized exportin, exportin-t (Exp-t/Xpo-t) (Arts et al., 1998a; Kutay et al., 1998). Exp-t is the only exportin known so far that can interact directly with an RNA substrate. tRNA binding to Exp-t occurs in a RanGTP-stimulated fashion in the cell nucleus, leading to the formation of a trimeric tRNA/Exp-t/RanGTP export complex that subsequently translocates to the cytoplasm where it is finally dissociated. Then, Exp-t re-enters the nucleus to export the next tRNA molecule. Many different tRNA isoacceptors and tRNAs from different species can bind to Exp-t (Arts et al., 1998a; Lund and Dahlberg, 1998; Lipowsky et al., 1999). Therefore, it is accepted presently that Exp-t has no preference for specific tRNAs but rather recognizes features common to all of them. Both correct tertiary folding and matured 5′ and 3′ ends have been shown to be prerequisites for high affinity binding of tRNA to Exp-t. Consequently, Exp-t contributes to the selective export of matured versus premature tRNAs from the cell nucleus (Arts et al., 1998b; Lund and Dahlberg, 1998; Lipowsky et al., 1999). In yeast, tRNA export occurs according to the same general principle and is mediated by Los1p, the yeast homolog of Exp-t (Hellmuth et al., 1998; Sarkar and Hopper, 1998). However, LOS1 is dispensable for viability of yeast (Hurt et al., 1987). As nuclear exit of tRNA is certainly an essential process, dispensability of LOS1 suggests either that tRNA can leave the cell nucleus by diffusion or that there exists an alternative, receptor-mediated export pathway. Indeed, several mechanisms and factors have been implicated in alternative tRNA export pathways in yeast, although no alternative exportin has yet been described. One such factor, the CCA-adding enzyme Cca1p, has been demonstrated to shuttle between the cytoplasm and nucleus, and it has therefore been suggested that it might function as either an export adaptor or a receptor (Feng and Hopper, 2002). Another screen identified the translation elongation factor 1A (eEF1A) as a suppressor of a synthetically lethal los1 mutant (Grosshans et al., 2000a). eEF1A is a GTP-binding protein that targets aminoacylated tRNAs (aa-tRNAs) to the ribosome. Mutations in eEF1A led to the nuclear accumulation of tRNAs (Grosshans et al., 2000a). Furthermore, it has been demonstrated that aminoacyl ation of specific tRNAs can occur in the cell nucleus (Lund and Dahlberg, 1998). Interference with aminoacylation decreases the rate of cytoplasmic appearance of specific tRNAs in Xenopus oocytes or leads to their nuclear accumulation in yeast (Lund and Dahlberg, 1998; Sarkar et al., 1999; Grosshans et al., 2000a). It is unclear presently, however, which alternative export pathway aa-tRNAs would enter.
The export receptor exportin-5 (Exp5) was identified based on its homology to CRM1 but, in fact, if compared with the yeast nuclear transport receptors, it is more closely related to the exportin Msn5p than to Crm1p (Brownawell and Macara, 2002). Similarly to CRM1, yeast Msn5p is involved in nuclear export of various proteins, including the transcription factor Pho4p (Kaffman et al., 1998), the cell cycle regulator Far1p (Blondel et al., 1999) and the transcriptional repressor Mig1p (DeVit and Johnston, 1999). These export substrates do not contain a consensus NES but in all cases phosphorylation of the export substrate seems to be required for the interaction with the export receptor Msn5p. In contrast, the only export cargo of mammalian Exp5 identified so far, a double-stranded RNA-binding protein (interleukin enhancer-binding protein 3, ILF3), binds to Exp5 independently of phosphorylation (Brownawell and Macara, 2002).
Here, we report on the identification of novel export substrates of Exp5. Both eEF1A and tRNA are shown to associate with Exp5 in a RanGTP-dependent manner. Exp5 can mediate nuclear export of eEF1A. However, the binding of eEF1A to Exp5 is dependent on the presence of tRNA. In fact, Exp5 can bind tRNA directly in the absence of eEF1A and stimulates tRNA export.
Results
Identification of Exp5-binding proteins
In order to characterize the function of a vertebrate ortholog of yeast Msn5p, we cloned mammalian Msn5 based on the sequence information of homologous expressed sequence tags (ESTs) present in the database (see Materials and methods). The deduced amino acid sequence matches the sequence of Exp5 that has been published meanwhile (Brownawell and Macara, 2002).
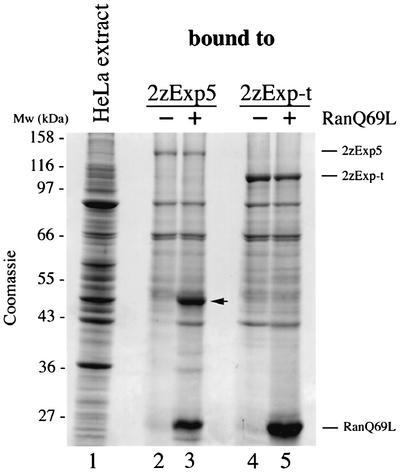
Exportins stably associate with their export substrates only when simultaneously bound to RanGTP. To identify potential export substrates of Exp5, we subjected HeLa cell extracts to affinity chromatography on immobilized Exp5 in the presence of RanQ69L(GTP), a GTPase-deficient mutant of Ran. As a specificity control, we performed binding to another exportin, Exp-t. The binding of a 50 kDa protein to Exp5, but not to Exp-t, was greatly promoted in the presence of RanQ69L (Figure 1). Mass spectrometric analysis revealed the protein to be identical to eEF1A.
Fig. 1. Binding of proteins from HeLa cell extract to Exp5. HeLa cell extract (lane 1) was incubated with immobilized 2zExp5 (lanes 2 and 3) or 2zExp-t (lanes 4 and 5) in the absence or presence of RanQ69L(GTP). Bound proteins were separated by SDS–PAGE and detected by Coomassie Blue staining. The arrow indicates the 50 kDa protein that was excised, subjected to mass spectrometry and identified as eEF1A.
Exp5 promotes nuclear export of eEF1A
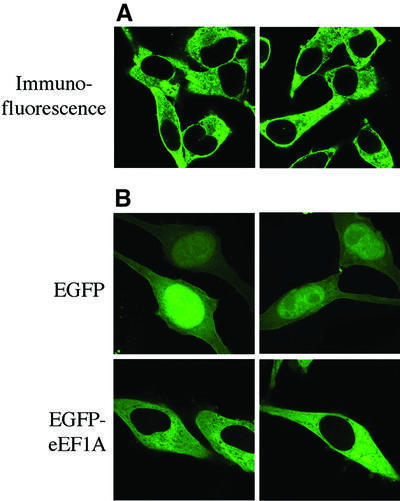
eEF1A catalyzes the GTP-dependent binding of aa-tRNA to the ribosome. Besides this well-established role in cytoplasmic protein synthesis, eEF1A has also been implicated in other cellular processes including organization of the cytoskeleton, oncogenic transformation, signal transduction and nuclear export of tRNA (for reviews, see Negrutskii and El’skaya, 1998; Ejiri, 2002). Immunolocalization studies have demonstrated convincingly that eEF1A is a cytoplasmic protein (Kjaer et al., 2001). However, nuclear localization of a minor fraction of eEF1A has also been reported, for instance after mitogenic stimulation of cells (Gangwani et al., 1998). To analyze the localization of eEF1A in HeLa cells, we imaged the steady-state distribution of eEF1A by confocal laser scanning microscopy. We could detect eEF1A in the cell nucleus neither by immunolocalization of the endogeneous protein (Figure 2A) nor after expression of a green fluorescent protein (GFP)-tagged version of eEF1A in transiently transfected HeLa cells (Figure 2B), even though the cells were grown in the presence of 10% fetal calf serum. Cytoplasmic localization of eEF1A could be achieved by cytoplasmic retention, nuclear export or both.
Fig. 2. Cytoplasmic localization of eEF1A in HeLa cells. (A) Immunostaining of eEF1A in HeLa cells. HeLa cells were grown on coverslips to ∼30% confluency and fixed with 4% paraformaldehyde. For immunostaining, a mouse monoclonal antibody to eEF1A was used in conjunction with a fluorescein-conjugated secondary antibody. (B) Localization of EGFP–eEF1A. Detection of EGFP–eEF1A (left panel) and EGFP alone was ∼36 h after transfection and fixation of the cells with 4% paraformaldehyde. While EGFP is found in both the nucleus and the cytoplasm, EGFP–eEF1A is completely excluded from the cell nucleus. All images were taken by confocal fluorescence microscopy. Two representative images are shown for each experiment.
The identification of eEF1A as a RanGTP-dependent binding partner of Exp5 suggested that it might be an export substrate of this exportin. To investigate whether eEF1A can be exported actively from the nucleus by Exp5, we performed microinjection experiments. eEF1A was translated in vitro in rabbit reticulocyte lysate in the presence of [35S]methionine and injected into the nuclei of Xenopus laevis oocytes. Radiolabeled GST served as an injection control. After 2 h, only very little eEF1A could be found in the cytoplasmic fraction. If, however, Exp5 and RanQ69L(GTP) were co-injected, the export of eEF1A was greatly stimulated such that ∼50% of the injected eEF1A was exported (Figure 3A). Co-injection of RanQ69L on its own also slightly enhanced eEF1A export, suggesting that an export receptor for eEF1A exists either in Xenopus oocytes or in the co-injected reticulocyte lysate. Importantly, Exp5 alone significantly stimulated eEF1A export to ∼40%. Export stimulation by Exp5 was abolished upon nuclear co-injection of the normally cytoplasmic RanGAP, thereby depleting the nuclear pool of RanGTP. Taken together, these data show that nuclear export of eEF1A requires the presence of nuclear RanGTP and that the concentration of Exp5 in the oocyte is rate limiting for eEF1A export. Further control experiments demonstrated the specificity of export stimulation by Exp5 since neither CRM1 nor Exp-t could stimulate nuclear export of the translation factor (Figure 3B).
Fig. 3. Exp5 stimulates nuclear export of eEF1A. A mixture of in vitro translated, radiolabeled eEF1A and GST was injected into the oocyte nuclei. After 2 h or at the indicated time, protein was extracted from either total oocytes (T), or, after dissection of the oocytes, from the cytoplasmic (C) and nuclear fractions (N). Proteins were separated by 10% SDS–PAGE and detected after phosphoimaging (for quantification) and autoradiography. (A) Export of eEF1A is promoted by Exp5 and requires nuclear RanGTP. Where indicated, purified 2zExp5, RanQ69L(GTP) or RanGAP (Rna1p) were co-injected. The concentrations in the injected samples were: 5 µM 2zExp5, 15 µM RanQ69L and 20 µM RanGAP. Note that only full-length eEF1A molecules were exported, while eEF1A fragments derived from the synthesis in the reticulocyte lysate stayed nuclear. (B) Nuclear export of eEF1A is not stimulated by other exportins. Co-injection of 2zExp-t (5 µM) or 2zCRM1 (5 µM) did not enhance the nuclear export of eEF1A. Both factors were fully functional, as they supported the export of GST–NES (not shown) and tRNA (Figure 6), respectively.
Characterization of the eEF1A–Exp5 interaction
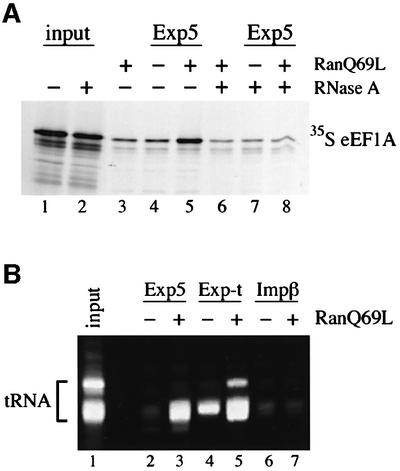
It has been shown for other transport receptor–substrate complexes that cargo binding can be either direct or mediated by adaptor molecules. Therefore, we next asked whether eEF1A is able to bind its export receptor directly. However, all our attempts to demonstrate the formation of a complex between either purified recombinant eEF1A or eEF1A purified from HeLa cell extracts and Exp5/RanGTP failed (data not shown). This could be explained by the lack of an essential mediator of the interaction. Since we could not detect another protein in stoichiometric amounts to eEF1A on immobilized Exp5 in pull-down experiments (e.g. Figure 1), we reasoned that the eEF1A/Exp5/RanGTP interaction might be mediated by an RNA. Consequently, we tested whether eEF1A binding to Exp5/RanGTP could occur at all in the absence of RNA. eEF1A was translated in vitro in a reticulocyte lysate in the presence of [35S]methionine and allowed to bind to immobilized Exp5. Comparable with the binding from HeLa cell extracts, eEF1A was recruited to Exp5 in a RanGTP-dependent manner (Figure 4A). Strikingly, if the lysate was treated with RNase A prior to binding, the recruitment of eEF1A was reduced to background levels, supporting our assumption that RNA is necessary for the RanGTP-dependent interaction between eEF1A and Exp5.
Fig. 4. Characterization of the eEF1A–Exp5 interaction. (A) Binding of eEF1A to Exp5 is RNase A sensitive. eEF1A was translated in vitro in the presence of [35S]methionine and either mock (lanes 1, 3, 4 and 5) or RNase A treated (lanes 2, 6, 7 and 8). Binding of radiolabeled eEF1A to 2zExp5 was performed in either the absence (lanes 4 and 7) or presence (lanes 5 and 8) of RanQ69L(GTP). Then, 2zExp5 and associated factors were retrieved from the reaction mixtures with IgG–Sepharose. Control reactions for unspecific binding without 2zExp5 were performed in the presence of RanQ69L(GTP) (lanes 3 and 6). After washing, bound proteins were eluted from the beads and separated by 8% SDS–PAGE. The gel was first stained with Coomassie Blue to confirm equal retrieval of 2zExp5 and, where applicable, RanQ69L (not shown). Radiolabeled eEF1A was analyzed subsequently by phosphoimaging and autoradiography. Lanes 1 and 2 show the mock- and RNase A-treated starting lysates, respectively. The load corresponds to 15% of the total for the starting lysates and to 20% for the bound fractions. (B) tRNA is bound to Exp5 from HeLa cell extracts. HeLa cell extract was incubated with immobilized 2zExp5, 2zExp-t or 2zImpβ in the absence or presence of RanQ69L(GTP) as described in Materials and methods. Bound RNAs were separated by 8% urea–PAGE and visualized by SYBR®Gold (Molecular Probes) staining. HeLa cell tRNA is shown in the ‘input’ lane.
The fact that eEF1A is a tRNA-binding protein prompted us to investigate whether tRNA might be required for eEF1A binding to Exp5. To determine whether tRNA is indeed part of the eEF1A/Exp5/ RanGTP complex, we analyzed whether tRNA can also be found on immobilized Exp5 in pull-down experiments performed with HeLa cell extracts. Therefore, we repeated the binding experiment presented in Figure 1 and this time also inspected the RNAs in the bound fractions (Figure 4B). Indeed, tRNA was recruited to Exp5 in the presence of RanGTP, albeit with reduced efficiency compared with its binding to Exp-t. The observation that tRNA can interact with Exp5 suggested to us that tRNA might be necessary for the binding of eEF1A to Exp5.
Exp5 is a tRNA-binding protein
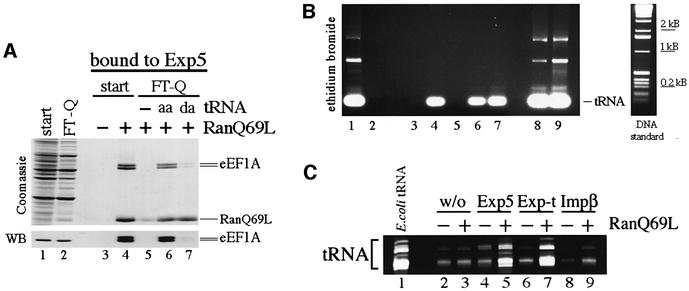
Based on these data, we wanted to test directly whether tRNA can facilitate binding of eEF1A to Exp5. For that reason, HeLa cell extracts were passed over an anion exchange column to deplete tRNA from the extract. While eEF1A was found in the flowthrough fraction, tRNA was retained on the column efficiently. When the flowthrough fraction, the tRNA-depleted extract, was used in a pull-down experiment with Exp5, eEF1A was unable to bind Exp5 as expected (Figure 5A, lane 5). Next, we tested whether the addition of purified tRNA could restore the binding of eEF1A to Exp5. The binding of eEF1A was reconstituted efficiently upon addition of tRNA to the depleted extract but only if the added tRNA was aminoacylated (Figure 5A, lane 6). This is explained by the fact that eEF1A/GTP has a strong binding preference for aa-tRNA (Roobol and Moller, 1978). Consistently, the addition of deacylated tRNA (da-tRNA) could not restore eEF1A binding, demonstrating that aminoacylation of the tRNA is required for efficient complex formation between eEF1A and Exp5. When we analyzed the RNA in the Exp5-bound fractions, we noticed, to our surprise, that both da- and aa-tRNA had been recruited to Exp5 with the same efficiency (see Figure 5B, lanes 6 and 7). This suggests that the binding of tRNA is direct and independent of eEF1A. Obviously, it is the other way round for eEF1A, i.e. eEF1A needs to bind aa-tRNA for its interaction with Exp5. This is supported further by our observation that only eEF1A containing both the tRNA- and GTP-binding domains, together indispensable for binding of aa-tRNA, binds to Exp5 (data not shown). Moreover, only the export of full-length eEF1A was stimulated by Exp5 in the oocyte injection experiments (see Figure 3A). Taken together, these results suggest that aa-tRNA facilitates the interaction between eEF1A and Exp5.
Fig. 5. tRNA confers binding of eEF1A to Exp5. (A) aa-tRNA stimulates binding of eEF1A to Exp5. tRNA was depleted from HeLa cell extracts by passing the extract over Q-Sepharose as described in Materials and methods. The depleted extract was supplemented with 1 mM GMPPNP to compensate for the loss of nucleotides and to allow for the efficient association of eEF1A with aa-tRNA. The starting (start) and depleted extracts (FT-Q) were subjected to binding to 2zExp5 in the presence of the factors indicated at the top of the figure. tRNA was supplemented to the FT-Q in either the aminoacylated (aa) or deacylated (da) form. 2zExp5 and bound factors were isolated from the reaction mixtures as described in Materials and methods. Then, each sample was divided into two equal parts. From one part, bound proteins were eluted and separated by 8% SDS–PAGE followed by Coomassie Blue staining or immunoblotting (WB). The load in the extract lanes (lanes 1 and 2) is 1/300 of the input, and the load from the bound fractions (lanes 3–7) corresponds to 1/12 of the total. The recruitment of eEF1A to 2zExp5 could only be detected in the presence of RanGTP and if the depleted extract had been replenished with aa-tRNA (lane 6). In comparison, the addition of tRNA that had been deacylated by treatment at high pH did not restore eEF1A binding (lane 7). Note that eEF1A migrates here as a double band of 50 and 52 kDa. Protein from both bands was subjected to mass spectrometric analysis and identified as eEF1A. (B) Exp5 binds aminoacylated and deacylated tRNA. The experiment was performed as in (A). Bound RNAs were extracted from the second part of each sample and analyzed by agarose gel electrophoresis (1.5%) and ethidium bromide staining. To demonstrate that tRNA was removed efficiently in the depleted extract, RNA was also extracted from the starting extract and the FT-Q (lanes 1 and 2). The load equals 1/30 of the total RNA retrieved from these extracts. The load in the bound fractions corresponds to 2/5 of the total. In addition, 1/15 of the aa-tRNA and da-tRNA (lanes 8 and 9) that were used to supplement the Q-Sepharose flowthrough are also shown. Note that tRNA can bind to Exp5 independently of its aminoacylation status. (C) tRNA can bind directly to Exp5. Escherichia coli tRNA (lane 1) was subjected to binding to 2zExp5 (lanes 4 and 5), 2zExp-t (lanes 6 and 7) or 2zImpβ (lanes 8 and 9) in the absence or presence of RanQ69L(GTP). 2z-tagged proteins were collected from the samples with IgG–Sepharose. The amount of unspecific binding of tRNA to IgG–Sepharose was also analyzed (lanes 2 and 3). RNA was extracted from the bound fractions, separated by 8% urea–PAGE and visualized by SYBR®Gold staining. Lane 1 shows 5% of the input RNA. The load in the bound fractions corresponds to 25% of the totally bound RNA.
It remained to be tested whether tRNA could interact directly with Exp5. tRNA from Escherichia coli was subjected to Exp5 binding. Indeed, not only Exp-t but also Exp5 was able to bind tRNA directly in the presence of RanGTP (Figure 5C). To exclude the possibility that tRNA binding was to RanGTP, we also used importin β as a control, as it binds to RanGTP with high affinity. We were unable to detect recruitment of tRNA to importin β–RanGTP over background levels.
Exp5 promotes nuclear export of tRNA
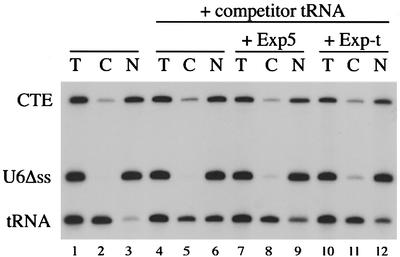
The experiments presented so far hinted that Exp5 might be able to drive tRNA export: Exp5 stimulates eEF1A export, eEF1A binding to Exp5 is dependent on tRNA, and tRNA can interact directly with Exp5. To determine whether Exp5 can mediate tRNA export, we injected a mixture of radiolabeled RNAs including tRNAiMet into Xenopus oocyte nuclei. U6Δss RNA served as an injection control as it marks the injection site since it can neither enter nor leave the cell nucleus. In addition, we added an RNA known to utilize an unrelated export pathway, namely the constitutive transport element (CTE) derived from the mRNA of Mason Pfitzer monkey virus (for a review, see Cullen, 1998). CTE binds to the mRNA export receptor TAP and by doing so accesses the cellular mRNA export pathway (Grüter et al., 1998).
As shown in Figure 6, tRNAiMet was exported efficiently to the cytoplasm 30 min after injection (95% export). This export was probably mediated by Exp-t, as Exp-t is known to be the major tRNA export receptor in Xenopus oocytes (Arts et al., 1998b; Lipowsky et al., 1999). In order to analyze the role of Exp5 in tRNA export, we competed tRNAiMet export by co-injection of unlabeled total tRNA from E.coli. Injection of the competitor reduced tRNAiMet export to 45%. Co-injection of either Exp-t or Exp5 relieved the export block and promoted export of up to 80% of tRNAiMet as quantified by phosphoimaging. Importantly, the export of CTE was not affected by the co-injection of either export receptor, demonstrating that the enhancement of tRNA export by Exp5 was specific. This result shows that not only Exp-t but also Exp5 can drive nuclear export of tRNA.
Fig. 6. Exp5 stimulates nuclear export of tRNA. A mixture of radio labeled tRNAiMet, U6Δss and constitutive transport element (CTE) RNAs was injected into the nuclei of Xenopus oocytes, either alone (lanes 1–3) or in the presence of unlabeled competitor tRNA (0.65 µg/µl in the injection mixture) derived from E.coli (lanes 4–12). Injection mixtures were supplemented with either recombinant 2zExp5 (lanes 7–9), recombinant 2zExp-t (lanes 10–12) or the corresponding buffer (lanes 1–6). Note that the concentration of 2zExp5 in the injected sample was 40 µM while that of 2zExp-t was only 10 µM. Export was analyzed 30 min after injection. T, C and N are defined as in Figure 3. Isolated RNAs were separated by 8% urea–PAGE and analyzed first by phosphoimaging for quantification and thereafter by autoradiography.
Discussion
In this study, we undertook a biochemical search for interaction partners of the human transport receptor Exp5. This led to the identification of eEF1A as the main proteinaceous binding partner of Exp5 present in HeLa cell extracts (Figure 1). Complex formation between eEF1A and Exp5 was shown to be dependent on the presence of RanGTP, suggestive of a function for Exp5 in eEF1A export from the cell nucleus. Indeed, eEF1A nuclear export was stimulated specifically upon co-injection of Exp5 into Xenopus oocyte nuclei, demonstrating that Exp5 can drive eEF1A export (Figure 3). Analysis of the Exp5/eEF1A interaction revealed that tRNA is not only another Exp5 binding partner but, rather, is essential for Exp5/eEF1A complex formation. While eEF1A binding to Exp5 was strictly dependent on tRNA, tRNA could interact with Exp5 even in the absence of eEF1A, demonstrating that tRNA recruits the elongation factor into an Exp5 complex (Figure 5). Hence, tRNA can bridge the interaction between Exp5 and eEF1A, but only if it is aminoacylated. Exp5 can bind both charged and uncharged tRNA molecules and drive tRNA export.
Nuclear export of eEF1A
Different models concerning the cellular role of Exp5-mediated export of eEF1A can be envisaged.
The first possibility would be that eEF1A plays a role in tRNA export. The major pathway for tRNA export is governed by the export receptor Exp-t in vertebrates and Los1p in yeast (for a review, see Grosshans et al., 2000b). However, yeast strains harboring a deletion of LOS1 are viable. This led to a search for redundant export pathways. Besides eEF1A, other factors involved in tRNA biogenesis or consumption have been proposed to be part of such alternative tRNA export pathways (see Introduction). If we assume that eEF1A plays a role in constitutive tRNA export, then eEF1A should shuttle rapidly between the nucleus and the cytoplasm. At steady state, in our hands, eEF1A is completely excluded from the cell nucleus (see Figure 2). This confirms a previous report in which two highly specific single-chain antibodies directed to eEF1A were used. These antibodies detected the protein exclusively in the cytoplasm (Kjaer et al., 2001). Although a few reports described a nuclear localization of eEF1A (Billaut-Mulot et al., 1996; Sanders et al., 1996; Gangwani et al., 1998), we have been unable to demonstrate shuttling or nuclear localization of eEF1A. One study suggested that eEF1A binds to zinc finger protein R (ZPR) upon serum or epidermal growth factor (EGF) stimulation of serum-starved cells, and this complex then translocates into the nucleus (Gangwani et al., 1998). Our cells were grown in the presence of serum and yet did not show nuclear eEF1A staining. In addition, when, for instance, eEF1A was microinjected into the cytoplasm of Xenopus oocytes, it never appeared in the nucleus even after prolonged incubation times (data not shown). This is noteworthy given the slow rate of export observed after nuclear injection of eEF1A in the absence of exogenous Exp5 (Figure 3). Since eEF1A shuttling was also not detectable in yeast, it has been suggested that eEF1A might act from the cytoplasm to sustain tRNA export (Grosshans et al., 2000a,b). Although the molecular mechanism is not yet understood, it is possible that the tRNA equilibrium in the cell is shifted towards the cytoplasm by binding of aa-tRNAs to eEF1A. For these reasons, and consistent with data obtained in yeast, we currently have no reason to assume that shuttling of eEF1A contributes to nuclear export of tRNA. Most importantly, tRNA can bind to Exp5 independently of eEF1A. Therefore, a nuclear contribution of eEF1A to tRNA export is unlikely to be required. On the contrary, our data suggest that eEF1A nuclear export is a consequence of tRNA export driven by Exp5. To serve efficiently as a bridge between Exp5 and eEF1A, tRNAs have to be aminoacylated (Figure 5A). Consequently, the nuclear presence of aa-tRNA would be a prerequisite for eEF1A export. Because the eEF1A used for our export experiments was generated in a reticulocyte lysate, aa-tRNA was inherently a part of the injected material. Notably, aa-tRNAs have also been shown to exist normally in the nuclei of Xenopus oocytes and yeast (Lund and Dahlberg, 1998; Azad et al., 2001).
Recently, it has been proposed that translation occurs in the cell nucleus with unforeseen high efficiencies (Iborra et al., 2001). While indeed many components needed for translation can be found in the cell nucleus, eEF1A and the other constituents of the EF1 holoenzyme, namely eEF1Bα, eEF1Bβ and eEF1Bγ, show striking absence in this compartment (Sanders et al., 1996). In addition, as we demonstrate here, Exp5 can drive the nuclear export of the tRNA-binding subunit of this translation factor. Therefore, it might be reasonable to assume that it is a function of Exp5 to keep eEF1A out of the cell nucleus in case eEF1A would ever enter nuclei by passive diffusion or upon reformation of the nuclear envelope in organisms with an open mitosis. If nuclear translation should indeed take place with the reported effectiveness, then either an as yet undiscovered elongation factor might account for this activity or nuclear translation might only occur under specific circumstances.
Exp5 and RNA export
We have shown that Exp5 mediates nuclear export of tRNA in Xenopus oocytes (Figure 6). This is a surprising observation because antibody injection experiments had previously demonstrated that Exp-t is the main tRNA export receptor in this system (Arts et al., 1998b; Lipowsky et al., 1999). However, the inhibition of tRNA export by anti-Exp-t antibodies had not been complete, raising the possibility that an alternative tRNA export pathway might exist. As demonstrated here, Exp5 is the second member of the importin β superfamily that can bind and export tRNA directly. Although Exp5 can function in tRNA export, we do not intend to claim that the tRNA export capability of Exp5 is comparable with that of Exp-t. First, the binding of tRNA to Exp5 is less efficient when directly compared with Exp-t (Figures 4B and 5C). Secondly, at a higher concentration of competitor tRNA, Exp-t was more efficient in rescuing tRNA export than Exp5 (not shown). Thirdly, a higher concentration of Exp5 was needed to observe the same stimulation of export if compared with Exp-t (Figure 6). Our data also suggest that Exp-t and Exp5 bind tRNA in a different manner. tRNA binding to Exp5 did not exclude the association of eEF1A. In contrast, eEF1A was neither bound to nor exported by Exp-t, suggesting that tRNA binding to Exp-t and eEF1A is mutually exclusive. Because Exp-t binds the acceptor arm and the CCA end of tRNA (Arts et al., 1998b), it thereby presumably blocks the access of eEF1A to the 3′ end of the tRNA. Exp5 must recognize tRNA differently, still allowing the association of eEF1A with Exp5-bound tRNA.
While Exp-t is a transport receptor highly specific for tRNA, Exp5 has been shown to act as a nuclear export receptor for the dsRNA-binding protein ILF3. We have observed that the binding of ILF3 to Exp5 is also RNase sensitive, suggesting that this interaction might also be facilitated by a cellular RNA (see Supplementary figure available at The EMBO Journal Online). This raises the possibility that Exp5 can interact not only with tRNA but perhaps also with other structured RNAs. Interestingly, Exp5 supports the nuclear export of another polymerase III transcript, namely adenoviral VA1 RNA (C.Gwidzek and C.Dargemont, personal communication). It will be interesting to investigate whether additional cellular RNAs can be identified that take Exp5 as an export carrier.
Exp5 is homologous to Saccharomyces cerevisiae Msn5p. Unlike Msn5p, Exp5 has not yet been demonstrated to interact with phosphorylated proteins. Con versely, it is unknown whether yeast Msn5p can contribute to tRNA export. It remains to be seen whether in this case similarity of sequence is truly a reflection of a common function. At least, a role for Msn5p in tRNA export would provide another explanation as to why LOS1 is unessential.
Materials and methods
Antibodies
The mouse monoclonal anti-eEF1A antibody (clone CBP-KK1) was obtained from Upstate Biotechnology and is monospecific as tested by immunoblotting of total HeLa cell extracts. The secondary antibody, fluorescein-labeled sheep anti-mouse antibody, was from Sigma.
Molecular cloning
The sequence information of the mouse EST (AAC AI663424) and human BAA86605 (KIAA1291) was used to design primers to amplify the complete coding region of mammalian Exp5 (5′-gggcccATG GAGATGGAGCAAGTGAACGCGC and 3′-gccgagatctGGGTTCAAA GATGGTGGCCAGGC) by PCR using HeLa cell cDNA as a template. Note that the use of the mouse 5′ primer changes the amino acids in positions 2 and 4 from the human to the mouse sequence. The PCR fragment was cloned into the NcoI–BamHI sites of pQE602z to yield pQE602z-Exp5. The complete coding region of eEF1A-1 was amplified by PCR from HeLa cell cDNA and cloned into the NcoI–BamHI sites of pQE60T7. pQE60T7-GST was generated in the same way using pGEX as a PCR template. EGFP–eEF1A was obtained by cloning a BamHI–XmaI-digested PCR fragment comprising the eEF1A coding region into the BglII–XmaI sites of pEGFP-C1 (Clontech).
Protein expression
Expression and purification of 2zExp-t, CRM1 and RanQ69L were performed as previously described (Izaurralde et al., 1997; Kutay et al., 1998; Mühlhäusser et al., 2001).
2zExp5 was expressed in Escherichia coli XL1(pBS161) at 21°C. Cells were lysed by sonication in 50 mM Tris–HCl pH 7.5, 700 mM NaCl, 10 mM MgCl2, 5% glycerol and 5 mM β-mercaptoethanol. Purification was performed by chromatography on nickel-NTA–agarose, followed by MonoQ and gel filtration in 50 mM Tris–HCl pH 7.5, 100 mM K acetate, 1 mM Mg acetate.
eEF1A was synthesized in the presence of [35S]methionine in rabbit reticulocyte lysate (TNT coupled reticulocyte lysate®; Promega) according to the manufacturer’s instructions, using pQE60T7-eEF1A as template DNA.
Protein identification by mass spectrometry
Protein identification by mass spectrometry was performed as described in Mühlhäusser et al. (2001). Identified eEF1A peptides included VETGVLKPGMVVTFAPVNVTTEVK, STTTGHLIYK, YYVTIIDAP GHR, LPLQDVYK, IGGIGTVPVGR and SGDAAIVDMVPGK.
Preparation of HeLa cell extracts and RNA
HeLa cell extracts for experiments presented in Figures 1 and 4B were prepared as described in Kutay et al. (2000). The extract used for the experiment presented in Figure 5A and B was prepared in essentially the same way, except that only 75 mM K acetate was used in the cell lysis buffer. Moreover, this extract was adjusted to 20% (w/v) glycerol before freezing since eEF1A activity is known to be preserved better at a high glycerol concentration (Negrutskii and El’skaya, 1998 and references therein). To deplete the extract of RNAs, 10 ml of extract were passed over a 4 ml column containing Q-Sepharose Fast Flow™ (Pharmacia) equilibrated with the same buffer. The flowthrough was collected. Prior to use in a binding experiment, these depleted extracts were passed over a PD10 column in the same buffer. Then, GMPPNP was added to a final concentration of 1 mM.
tRNA was prepared from high speed supernatants (90 min, 55 000 r.p.m., Ti 70). Extracts were diluted 1:3 with 100 mM Na acetate pH 5.0 and then extracted twice with acidic phenol and ethanol precipitated. To preserve the aminoacylation level of the extracted tRNAs, the RNA precipitate was resuspended in 10 mM Na acetate pH 5.0. For deacylation, the RNA was taken up in 0.1 M NH4OH/HCl pH 9.5 and incubated for 45 min at 37°C. The resulting RNA was reprecipitated and resuspended in 10 mM Tris–HCl pH 7.5. The quality of the tRNA was checked by denaturing urea–PAGE using commercial tRNA (Roche) from E.coli and calf liver as controls. Just prior to adding these tRNAs to the binding experiment, they were passed over a Sephadex G50 spin column.
In vitro binding assays
Pull-down from HeLa cell extracts.
IgG–Sepharose beads were pre-saturated with the specified 2z-tagged transport receptors (100 pmol) and incubated with 750 µl of HeLa cell extract in a final volume of 1.5 ml for 4 h at 4°C, in the absence or presence of 1 µM RanQ69L(GTP) (Figures 1 and 4B). Beads were washed three times with binding buffer 50 mM Tris–HCl pH 7.5, 100 mM K acetate, 1 mM Mg acetate. Bound proteins were eluted with 1.5 M MgCl2, 50 mM Tris–HCl pH 7.5, precipitated with isopropanol and dissolved in SDS sample buffer. Bound RNAs were recovered from beads by treatment with proteinase K for 30 min at 37°C, followed by phenol extraction and ethanol precipitation.
Alternatively (Figure 5A and B), for each sample, 300 µl of HeLa cell extract were supplemented with purified 2zExp5 (200 pmol). RanQ69L was added to a final concentration of 2 µM. Purified tRNAs were supplemented to the same amount as present in the starting extract (∼660 pmol). The reactions were incubated in a final volume of 500 µl for 45 min on ice. Then, 20 µl of IgG–Sepharose beads (Pharmacia) were added and the samples mixed occasionally. After 30 min, beads were washed with 50 mM Tris–HCl pH 7.5, 100 mM K acetate, 1 mM Mg acetate, 20% glycerol, and treated as described above.
Binding reactions from reticulocyte lysate.
After in vitro synthesis of eEF1A, the corresponding reticulocyte lysate was either mock treated or incubated for 15 min at 37°C in the presence of 0.2 mg/ml RNase A. For each binding reaction, 12.5 µl of the lysate were incubated with 150 pmol of purified 2zExp5 in a final volume of 20 µl. Where indicated, 150 pmol of RanQ69L plus 1 mM additional GTP were added. After incubation for 20 min at 4°C, reactions were supplemented with 100 µl of binding buffer (50 mM Tris–HCl pH 7.5, 150 mM K acetate, 1 mM Mg acetate, 0.001% Triton X-100), spun for 10 min (13 000 r.p.m., 4°C, bench top centrifuge), and the 2z-tagged proteins retrieved from the supernatants by the addition of 15 µl of IgG–Sepharose as described above. After washing twice with 1.5 ml of binding buffer and once with buffer 50 mM Tris–HCl pH 7.5, 0.001% Triton X-100, bound proteins were eluted from the resin with sample buffer containing 4% SDS.
Binding of purified tRNA.
Binding reactions were assembled in 100 µl of binding buffer 50 mM Tris–HCl pH 7.5, 100 mM K acetate, 1 mM Mg acetate, 0.25 mg/ml bovine serum albumin (BSA), 0.001% Triton X-100. 2z-tagged transport receptors were added to a final concentration of 5 µM, RanQ69L(GTP) to 5 µM and E.coli tRNA to 800 pmol. The reactions were left on ice for 45 min. Then, 25 µl of IgG–Sepharose beads were added and the samples were mixed occasionally. After 30 min, beads were washed three times with 50 mM Tris–HCl pH 7.5, 100 mM K acetate, 1 mM Mg acetate, 0.001% Triton X-100, and bound RNAs were recovered after proteinase K treatment by phenol extraction and ethanol precipitation.
Transfection and immunolocalization
Immunolocalization and detection of GFP-tagged proteins after transfection were performed as described in Kutay et al. (2000).
Oocyte injections
Microinjection experiments of RNAs and proteins into X.laevis oocytes were performed as previously described (Jarmolowski et al., 1994; Izaurralde et al., 1997).
For the injection of radiolabeled proteins, synthesis was performed in rabbit reticulocyte lysate (see above). Unincorporated methionine was removed by G-50 gel filtration. 32P-labeled RNAs were generated by in vitro transcription in the presence of [α-32P]GTP as described previously (Jarmolowski et al., 1994). The templates for T7-tRNAiMet and T7-U6Δss have been described (Izaurralde et al., 1997). T7-CTE was a kind gift of Brian Cullen.
For co-injection of recombinant proteins, the injected sample was first adjusted with ‘intracellular buffer’ to a final concetration of 20 mM HEPES pH 7.4, 110 mM K acetate, 10 mM NaCl, 2 mM Mg acetate, and then the purified transport factors added to the indicated concentrations. Competitor tRNA was from E.coli (Roche) and used at a final concentration of 0.65 µg/µl in the injection mixture.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We wish to thank Howard Fried, Gerd Lipowsky, Judith Erkmann, Ludwig Englmeier, Hans Lutz and the members of the Kutay laboratory for critical reading of the manuscript and stimulating discussions, Franziska Thomas for initial tRNA export experiments, Brian Cullen for the T7-CTE construct, and Anton Lehmann for the tremendous support in optimizing our frog facility. This work was supported by a grant from the Swiss National Science Foundation to U.K.
References
- Arts G.J., Fornerod,M. and Mattaj,I.W. (1998a) Identification of a nuclear export receptor for tRNA. Curr. Biol., 8, 305–314. [DOI] [PubMed] [Google Scholar]
- Arts G.J., Kuersten,S., Romby,P., Ehresmann,B. and Mattaj,I.W. (1998b) The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J., 17, 7430–7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad A.K., Stanford,D.R., Sarkar,S. and Hopper,A.K. (2001) Role of nuclear pools of aminoacyl-tRNA synthetases in tRNA nuclear export. Mol. Biol. Cell, 12, 1381–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billaut-Mulot O., Fernandez-Gomez,R., Loyens,M. and Ouaissi,A. (1996) Trypanosoma cruzi elongation factor 1-α: nuclear localization in parasites undergoing apoptosis. Gene, 174, 19–26. [DOI] [PubMed] [Google Scholar]
- Blondel M., Alepuz,P.M., Huang,L.S., Shaham,S., Ammerer,G. and Peter,M. (1999) Nuclear export of Far1p in response to pheromones requires the export receptor Msn5p/Ste21p. Genes Dev., 13, 2284–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownawell A.M. and Macara,I.G. (2002) Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins. J. Cell Biol., 156, 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B.R. (1998) Retroviruses as model systems for the study of nuclear RNA export pathways. Virology, 249, 203–210. [DOI] [PubMed] [Google Scholar]
- DeVit M.J. and Johnston,M. (1999) The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr. Biol., 9, 1231–1241. [DOI] [PubMed] [Google Scholar]
- Ejiri S. (2002) Moonlighting functions of polypeptide elongation factor 1: from actin bundling to zinc finger protein R1-associated nuclear localization. Biosci. Biotechnol. Biochem., 66, 1–21. [DOI] [PubMed] [Google Scholar]
- Feng W. and Hopper,A.K. (2002) A Los1p-independent pathway for nuclear export of intronless tRNAs in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 99, 5412–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U., Huber,J., Boelens,W.C., Mattaj,I.W. and Lührmann,R. (1995) The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell, 82, 475–83. [DOI] [PubMed] [Google Scholar]
- Fornerod M., Ohno,M., Yoshida,M. and Mattaj,I.W. (1997) Crm1 is an export receptor for leucine rich nuclear export signals. Cell, 90, 1051–1060. [DOI] [PubMed] [Google Scholar]
- Fukuda M., Asano,S., Nakamura,T., Adachi,M., Yoshida,M., Yanagida,M. and Nishida,E. (1997) CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature, 390, 308–311. [DOI] [PubMed] [Google Scholar]
- Gadal O., Strauss,D., Kessl,J., Trumpower,B., Tollervey,D. and Hurt,E. (2001) Nuclear export of 60S ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol., 21, 3405–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangwani L., Mikrut,M., Galcheva-Gargova,Z. and Davis,R.J. (1998) Interaction of ZPR1 with translation elongation factor-1α in proliferating cells. J. Cell Biol., 143, 1471–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D. and Kutay,U. (1999) Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol., 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Grosshans H., Hurt,E. and Simos,G. (2000a) An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes Dev., 14, 830–840. [PMC free article] [PubMed] [Google Scholar]
- Grosshans H., Simos,G. and Hurt,E. (2000b) Review: transport of tRNA out of the nucleus—direct channeling to the ribosome? J. Struct. Biol., 129, 288–294. [DOI] [PubMed] [Google Scholar]
- Grüter P., Tabernero,C., von Kobbe,C., Schmitt,C., Saavedra,C., Bachi,A., Wilm,M., Felber,B.K. and Izaurralde,E. (1998) TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell, 1, 649–659. [DOI] [PubMed] [Google Scholar]
- Hellmuth K., Lau,D.M., Bischoff,F.R., Kunzler,M., Hurt,E. and Simos,G. (1998) Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol. Cell. Biol., 18, 6374–6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J.H., Kallstrom,G. and Johnson,A.W. (2000) Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol., 151, 1057–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt D.J., Wang,S.S., Lin,Y.H. and Hopper,A.K. (1987) Cloning and characterization of LOS1, a Saccharomyces cerevisiae gene that affects tRNA splicing. Mol. Cell. Biol., 7, 1208–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iborra F.J., Jackson,D.A. and Cook,P.R. (2001) Coupled transcription and translation within nuclei of mammalian cells. Science, 293, 1139–1142. [DOI] [PubMed] [Google Scholar]
- Izaurralde E., Kutay,U., von Kobbe,C., Mattaj,I.W. and Görlich,D. (1997) The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J., 16, 6535–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmolowski A., Boelens,W.C., Izaurralde,E. and Mattaj,I.W. (1994) Nuclear export of different classes of RNA is mediated by specific factors. J. Cell Biol., 124, 627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman A., Rank,N.M., O’Neill,E.M., Huang,L.S. and O’Shea,E.K. (1998) The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature, 396, 482–486. [DOI] [PubMed] [Google Scholar]
- Kjaer S., Wind,T., Ravn,P., Ostergaard,M., Clark,B.F. and Nissim,A. (2001) Generation and epitope mapping of high-affinity scFv to eukaryotic elongation factor 1A by dual application of phage display. Eur. J. Biochem., 268, 3407–3415. [DOI] [PubMed] [Google Scholar]
- Kuersten S., Ohno,M. and Mattaj,I.W. (2001) Nucleocytoplasmic transport: Ran, β and beyond. Trends Cell Biol., 11, 497–503. [DOI] [PubMed] [Google Scholar]
- Kutay U., Bischoff,F.R., Kostka,S., Kraft,R. and Görlich,D. (1997) Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell, 90, 1061–1071. [DOI] [PubMed] [Google Scholar]
- Kutay U., Lipowsky,G., Izaurralde,E., Bischoff,F.R., Schwarzmaier,P., Hartmann,E. and Görlich,D. (1998) Identification of a tRNA-specific nuclear export receptor. Mol. Cell, 1, 359–369. [DOI] [PubMed] [Google Scholar]
- Kutay U., Hartmann,E., Treichel,N., Calado,A., Carmo-Fonseca,M., Prehn,S., Kraft,R., Görlich,D. and Bischoff,F.R. (2000) Identifi cation of two novel RanGTP-binding proteins belonging to the importin β superfamily. J. Biol. Chem., 275, 40163–40168. [DOI] [PubMed] [Google Scholar]
- Lipowsky G., Bischoff,F.R., Izaurralde,E., Kutay,U., Schäfer,S., Gross, H.J., Beier,H. and Görlich,D. (1999) Coordination of tRNA nuclear export with processing of tRNA. RNA, 5, 539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E. and Dahlberg,J.E. (1998) Proofreading and aminoacylation of tRNAs before export from the nucleus. Science, 282, 2082–2085. [DOI] [PubMed] [Google Scholar]
- Macara I.G. (2001) Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev., 65, 570–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I.W. and Englmeier,L. (1998) Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem., 67, 265–306. [DOI] [PubMed] [Google Scholar]
- Mühlhäusser P., Müller,E.C., Otto,A. and Kutay,U. (2001) Multiple pathways contribute to nuclear import of core histones. EMBO rep., 2, 690–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrutskii B.S. and El’skaya,A.V. (1998) Eukaryotic translation elongation factor 1α: structure, expression, functions and possible role in aminoacyl-tRNA channeling. Prog. Nucleic Acid Res. Mol. Biol., 60, 47–78. [DOI] [PubMed] [Google Scholar]
- Ohno M., Segref,A., Bachi,A., Wilm,M. and Mattaj,I.W. (2000) PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell, 101, 187–198. [DOI] [PubMed] [Google Scholar]
- Roobol K. and Moller,W. (1978) The role of guanine nucleotides in the interaction between aminoacyl-tRNA and elongation factor 1 of Artemia salina. Eur. J. Biochem., 90, 471–7. [DOI] [PubMed] [Google Scholar]
- Sanders J., Brandsma,M., Janssen,G.M., Dijk,J. and Moller,W. (1996) Immunofluorescence studies of human fibroblasts demonstrate the presence of the complex of elongation factor-1 βγδ in the endoplasmic reticulum. J. Cell Sci., 109, 1113–1117. [DOI] [PubMed] [Google Scholar]
- Sarkar S. and Hopper,A.K. (1998) tRNA nuclear export in Saccharomyces cerevisiae: in situ hybridization analysis. Mol. Biol. Cell, 9, 3041–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Azad,A.K. and Hopper,A.K. (1999) Nuclear tRNA aminoacylation and its role in nuclear export of endogenous tRNAs in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 96, 14366–14371. [DOI] [PMC free article] [PubMed] [Google Scholar]