Abstract
c-MOS, a MAP kinase kinase kinase, is a regulator of oocyte maturation. The concentration of c-MOS is controlled in part through its conditional degradation. Previous studies proposed the ‘second-codon rule’, according to which the N-terminal proline (Pro) of c-MOS is a destabilizing residue that targets c-MOS for degradation. We analyzed the degradation signal (degron) of c-MOS in Xenopus oocytes, found it to be a portable degron, and demonstrated that, contrary to the model above, the N-terminal Pro residue of c-MOS is entirely dispensable for its degradation if Ser-2 (encoded Ser-3) of c-MOS is replaced by a small non-phosphorylatable residue such as Gly. The dependence of c-MOS degradation on N-terminal Pro is shown to be caused by a Pro-mediated downregulation of the net phosphorylation of Ser-2, a modification that halts c-MOS degradation in oocytes. Thus, the N-terminal Pro residue of c-MOS is not a recognition determinant for a ubiquitin ligase, in agreement with earlier evidence that Pro is a stabilizing residue in the N-end rule.
Keywords: c-MOS/N-end rule/phosphorylation/proteolysis/ubiquitin
Introduction
c-MOS (called MOS below) is a serine/threonine protein kinase that is normally expressed largely, if not exclusively, in oocytes and spermatocytes. MOS functions as a regulator of oocyte maturation (Sagata, 1997; Ferrell, 1999, 2002; Nebreda and Ferby, 2000; Duesbery and Vande Woude, 2002; Dupré et al., 2002). A fully grown (stage VI) Xenopus laevis oocyte is arrested in the G2 phase of the first meiotic cell cycle. Oocyte maturation, initiated by progesterone, involves the completion of meiosis I, followed by meiosis II and the arrest at metaphase of meiosis II. At this stage, the mature oocyte, called an egg, is ready for fertilization (Ferrell, 1999; Hochegger et al., 2001; Reimann and Jackson, 2002).
MOS phosphorylates, and thereby activates, the kinase MEK1. In the MAP cascade’s terminology, MOS is a MAP kinase kinase kinase (Nebreda and Ferby, 2000). The synthesis of MOS begins shortly after progesterone stimulation, and ceases near the end of oocyte maturation (Mendez et al., 2000; Charlesworth et al., 2002). The concentration of MOS is controlled both through its synthesis and through its conditional degradation by the ubiquitin (Ub) system (Nishizawa et al., 1992, 1993; Castro et al., 2001).
The Ub-dependent N-end rule pathway targets proteins bearing destabilizing N-terminal residues. This pathway also targets proteins bearing a distinct class of internal (non-N-terminal) degradation signals (degrons) (Bachmair et al., 1986; Varshavsky, 1996; Turner et al., 2000; Kwon et al., 2001, 2002; Rao et al., 2001). The N-degron, an N-terminal degron recognized by the N-end rule pathway, consists of two determinants: a substrate’s destabilizing N-terminal residue and an internal Lys residue (Bachmair and Varshavsky, 1989; Suzuki and Varshavsky, 1999). A primary destabilizing N-terminal residue (Arg, Lys, His, Phe, Tyr, Trp, Leu or Ile) is recognized (bound) by the Ub ligases (complexes of E3 and E2 enzymes) of the N-end rule pathway. Several other N-terminal residues function as tertiary (Asn, Gln, Cys) and secondary (Asp, Glu) destabilizing residues in that they are recognized after their enzymatic conjugation to Arg, a primary destabilizing residue; in the case of N-terminal Asn, Gln and Cys, this conjugation is preceded by other enzymatic modifications (Varshavsky, 1996; Davydov and Varshavsky, 2000; Kwon et al., 2001, 2002).
Studies by Sagata and colleagues (Nishizawa et al., 1992, 1993) have shown that wild-type MOS, bearing the N-terminal Pro residue, is short-lived upon its expression in oocytes, whereas the otherwise identical MOS derivative bearing N-terminal Gly is long-lived. These and related findings led Nishizawa et al. (1992, 1993) to propose the ‘second-codon rule’, in which the N-terminal Pro residue of MOS targets MOS for degradation. In the N-end rule terminology (Varshavsky, 1996), the above conjecture meant that the MOS protein contains an N-degron whose N-terminal Pro is recognized by a specific Ub ligase, as suggested by Nishizawa et al. (1992). One difficulty with this model was that N-terminal Pro, in the context of previously characterized N-end rule reporters, was clearly a stabilizing residue (Bachmair et al., 1986; Gonda et al., 1989; F.Lévy and A.Varshavsky, unpublished data).
To address this problem, we analyzed the MOS degron in greater detail, and discovered that, contrary to the model above, the N-terminal Pro residue of MOS was entirely dispensable for its degradation if Ser-2 (encoded Ser-3) of MOS was replaced by a small non-phosphorylatable residue such as Gly. We also found that the previ ously observed dependence of MOS degradation on its N-terminal Pro (Nishizawa et al., 1992, 1993) is caused by a Pro-mediated downregulation of the net phosphorylation of Ser-2, a modification that inhibits MOS degradation.
Results
To increase the accuracy of pulse–chase assays, we employed the UPR (Ub-protein-reference) technique, which yields (initially) equimolar amounts of a Ub-containing reference protein and a test protein bearing a predetermined N-terminal residue (Figure 1A and B). The UPR technique compensates for data scatter in a pulse– chase assay, and in addition makes it possible to measure the degradation of a test protein during the pulse (Suzuki and Varshavsky, 1999; Turner and Varshavsky, 2000; Turner et al., 2000; Varshavsky, 2000; Kwon et al., 2001).
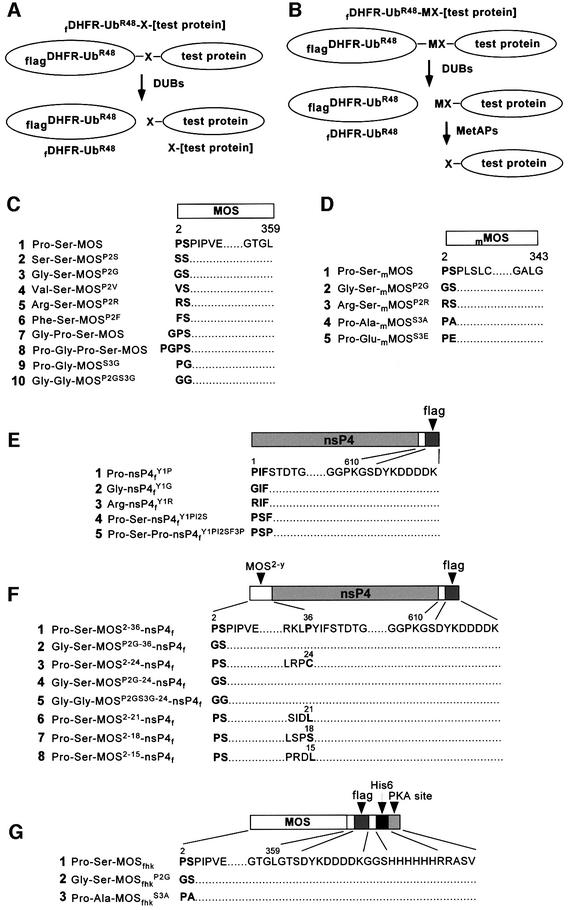
Fig. 1. Test proteins. The fusions were fDHFR-UbR48–X-[test protein] (A) or fDHFR-UbR48–MX-[test protein] (B). f is the flag epitope; DHFR is the mouse dihydrofolate reductase moiety; X in (A) is a variable residue at the Ub–protein junction that became the N-terminal residue after cleavage at the junction; and MX in (B) is the Met-X sequence of another set of test proteins (see Results). (C) Wild-type and mutant Xenopus MOS proteins, expressed as either A- or B-type Ub fusions. (D) Wild-type and mutant mouse MOS (mMOS) proteins, expressed as either A- or B-type Ub fusions. (E and F) Variants of the C-terminally flag-tagged nsP4f and MOS-nsP4f fusions, expressed as either A- or B-type Ub fusions in Xenopus oocytes. (G) MOSfhk and its derivatives, which were expressed directly (not as Ub fusions) in E.coli. The numbers above amino acid sequences indicate positions in the wild-type MOS or nsP4 ORFs.
Replacement of N-terminal Pro with Gly, Ser or Val abolishes degradation of Xenopus MOS in oocytes
We injected stage VI oocytes with in vitro synthesized mRNA encoding wild-type MOS or its derivatives, and carried out pulse–chase of proteins labeled with exogenously supplied [35S]methionine in the presence of progesterone (see Materials and methods). Xenopus MOS and its derivatives bearing predetermined N-terminal residues were produced through the co-translational cleavage, by deubiquitylating enzymes (DUBs), of fDHFR-UbR48–X-MOS fusions at the UbR48–X junction, yielding X-MOS and the long-lived reference fDHFR- UbR48 (Figure 1A). Since Pro is the only N-terminal residue of MOS that cannot be produced through a fusion of this kind (Varshavsky, 2000), a different set of fusions, fDHFR-UbR48–Met-X-MOS, was also employed (Figure 1B). The DUB-mediated cleavage of fDHFR-UbR48– Met-X-MOS at the UbR48–Met junction is followed by the cleavage at either Met-Pro or other N-terminal Met-X bonds by Met-aminopeptidases (MetAPs) (Bradshaw et al., 1998).
In the terminology below, made necessary by the range of constructs produced and examined in this work (Figure 1), the wild-type Xenopus MOS is denoted as Pro-Ser-MOS. An otherwise identical protein bearing, for example, N-terminal Gly is denoted as Gly-Ser-MOSP2G. The superscript indicates an underlying alteration (in this case, Pro→Gly), and cites the residue number in the ORF of wild-type MOS, in which Pro is the second residue after (initially present) Met.
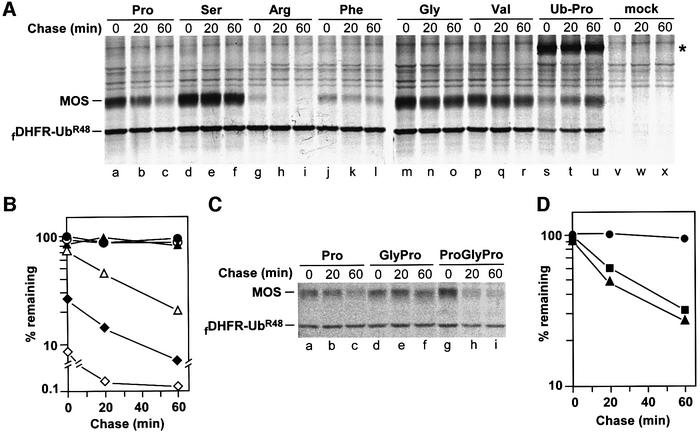
The wild-type Pro-Ser-MOS (Figure 1C1) was a short-lived protein in oocytes (t1/2 ≈ 25 min) (Figure 2A, lanes a–c, and B). In contrast, Ser-Ser-MOSP2S, Gly-Ser-MOSP2G and Val-Ser-MOSP2V (Figure 1C2–C4) were long-lived (Figure 2A, lanes d–f and m–r, and B). It was largely their finding that Pro-Ser-MOS was short-lived in oocytes, whereas Ser-Ser-MOSP2S and Gly-Ser-MOSP2G were long-lived, that led Nishizawa et al. (1992, 1993) to propose the ‘second-codon rule’ (see Introduction). Nishizawa et al. (1992) also reported that wild-type Pro-Ser-MOS was more unstable in vivo than MOS derivatives bearing N-terminal Met-Arg or Met-Lys, instead of wild-type Met-Pro. (Arg and Lys are destabilizing residues in the N-end rule.) However, MetAPs, which readily cleave the N-terminal Met-Pro bond, do not cleave Met-Arg, Met-Lys, or other Met-X bonds where X is a residue with the side chain’s radius of gyration larger than that of Val (1.29 Å) (Bradshaw et al., 1998). Thus, the comparisons by Nishizawa et al. (1992) were of Pro-Ser-MOS with proteins such as Met-Arg-Ser- MOSP2R, rather than Arg-Ser-MOSP2R.
Fig. 2. Replacement of N-terminal Pro with Gly, Ser or Val abolishes degradation of Xenopus MOS in oocytes. (A) mRNAs encoding either fDHFR-UbR48–MP-MOS (lanes a–c, Pro-Ser-MOS), or fDHFR-UbR48–X-MOS fusions, the residue X being Ser (lanes d–f, Ser-Ser-MOSP2S), Arg (lanes g–i, Arg-Ser-MOS P2R), Phe (lanes j–l, Phe-Ser-MOSP2F), Gly (lanes m–o, Gly-Ser-MOSP2G), Val (lanes p–r, Val-Ser-MOSP2V) or Pro (lanes s–u, Ub-Pro-Ser-MOS), were injected into stage VI Xenopus oocytes, followed by pulse–chase. Oocytes injected with H2O were used as a control (lanes v– x). The positions of X-MOS and the reference protein fDHFR-UbR48 are indicated. An asterisk denotes the position of slowly cleaved fDHFR-UbR48–Pro-MOS fusion. (B) Quantitation of pulse–chase patterns in (A). The ratio of 35S in wild-type or mutant MOS to 35S in the reference protein fDHFR-UbR48 in the same lane was determined using a PhosphorImager and plotted as a percentage of this ratio for MOSP2V (one of the metabolically stable MOS mutants) at time zero. Open triangles, Pro-Ser-MOS (wild-type MOS); closed triangles, Ser-Ser-MOSP2S; open diamonds, Arg-Ser-MOSP2R; closed diamonds, Phe-Ser-MOSP2F; open circles, Gly-Ser-MOSP2G; closed circles, Val-Ser-MOSP2V. (C) Comparison of Pro-Ser-MOS, Gly-Pro-Ser-MOS and Pro-Gly-Pro-Ser-MOS. (D) Quantitation of pulse–chase patterns in (C). At each time point, the ratio of wild-type or mutant MOS to fDHFR-UbR48 in the same lane was plotted as a percentage of this ratio for the long-lived Gly-Pro-Ser-MOS at time zero. Closed circles, Gly-Pro-Ser-MOS; closed squares, wild-type Pro-Ser-MOS; closed triangles, Pro-Gly-Pro-Ser-MOS.
We used the UPR technique to produce Arg-Ser- MOSP2R and Phe-Ser-MOSP2F (Figure 1C5 and C6), both of which, being bona fide N-end rule substrates, were highly unstable in vivo, significantly more so than Pro-Ser-MOS (Figure 2A and B). The relative amounts of test proteins (determined through the use of reference protein) in Figure 2B differed strongly even at time zero (the end of pulse). For example, ∼20% of wild-type Pro-Ser-MOS and >90% of Arg-Ser-MOSP2R were destroyed before the beginning of chase (Figure 2B). Since the cleavage of the Ub-Pro bond by DUBs is slow (Bachmair et al., 1986) (Figure 2A, lanes s–u), a modified UPR-type fusion, fDHFR-UbR48–Met-Pro-MOS, was used to produce Pro-Ser-MOS and its derivatives bearing N-terminal Pro (Figures 1B and 2A, lanes a–c).
Mouse MOS is ∼55% identical to Xenopus MOS, and the first four encoded residues, Met-Pro-Ser-Pro, are identical between the two proteins. We injected UPR-based mRNAs encoding mouse MOS and its derivatives into Xenopus oocytes; the results were essentially the same as those with Xenopus MOS. We also compared wild-type Pro-Ser-MOS with either Gly-Pro-Ser-MOS (with Gly upstream of wild-type Pro; Figure 1C7) or Pro-Gly-Pro-Ser-MOS (with Pro-Gly upstream of wild-type Pro; Figure 1C8). Whereas Gly-Pro-Ser-MOS was long-lived, similarly to Gly-Ser-MOSP2G (Figure 2C, lanes d–f, and A, lanes m–o), Pro-Gly-Pro-Ser-MOS was unstable, similarly to wild-type Pro-Ser-MOS (Figure 2C, lanes g–i, A and C, lanes a–c, and D). Thus, Pro must remain N-terminal, in the above sequence contexts, to enhance the degradation of MOS, while the distance of N-terminal Pro from Ser-2 (encoded Ser-3) is a flexible parameter.
The 20-residue N-terminal region of MOS contains a portable degron
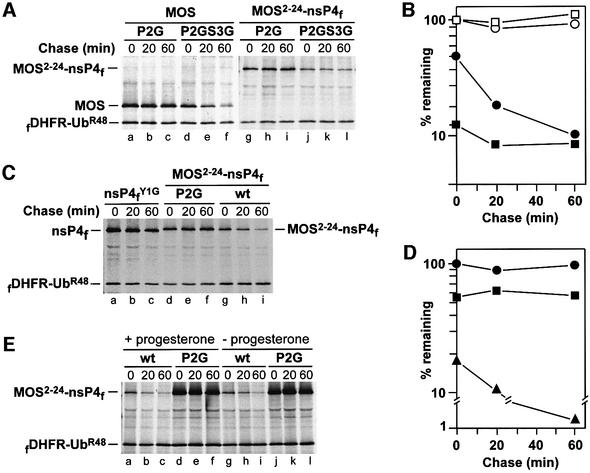
The Sindbis virus RNA polymerase, called nsP4, is a 70 kDa protein produced through cleavage of a polyprotein precursor. Wild-type nsP4 bears N-terminal Tyr, a destabilizing residue, and is a substrate of the N-end rule pathway (deGroot et al., 1991). We used C-terminally flag-tagged derivatives of nsP4 (X-nsP4f) to map the MOS degron. Oocytes were injected with mRNAs encoding fDHFR-UbR48–X-nsP4f or fDHFR-UbR48–Met-X-nsP4f, followed by pulse–chase (Figure 3). In agreement with earlier evidence that Pro and Gly are stabilizing residues in the N-end rule (Varshavsky, 1996), Pro-nsP4fY1P and Gly-nsP4fY1G (Figure 1E1 and E2) were long-lived proteins in oocytes, in contrast to Arg-nsP4fY1R (Figure 3A and B). Thus, N-terminal Pro, in the context of Pro-nsP4fY1P, was, in fact, a stabilizing residue in oocytes. Pro-Ser-nsP4fY1PI2S and Pro-Ser-Pro-nsP4fY1PI2SF3P, in which either two or three N-terminal residues of nsP4 were replaced by either Pro-Ser or Pro-Ser-Pro (the N-terminal sequences of MOS), were constructed (Figure 1E4 and E5) and expressed in oocytes as UPR fusions. Similarly to Pro-nsP4fY1P, these proteins were also long-lived in oocytes (Figure 3C and D).
Fig. 3. The 20-residue N-terminal region of MOS contains a portable degron. (A) N-terminal Pro alone does not render nsP4 short-lived in Xenopus oocytes. Positions of fDHFR-UbR48, MOS and nsP4f are indicated on the left. Lanes a–c, Pro-Ser-MOS; lanes d–f, Gly-Ser-MOSP2G; lanes g–i, Pro-nsP4fY1P; lanes j–l, Gly-nsP4fY1G; lanes m–o, Arg-nsP4fY1R. (B) Quantitation of pulse–chase patterns in lanes g–o of (A). Closed circles, Pro-nsP4fY1P; closed squares, Gly-nsP4fY1G; closed triangles, Arg-nsP4fY1R. At each time point, the ratio of X-nsP4f to fDHFR-UbR48 was plotted as a percentage of the same ratio for Pro-nsP4fY1P at time zero. (C) N-terminal Pro-Ser-Pro of MOS does not render nsP4 short-lived in oocytes. Lanes a–c, Pro-nsP4fY1P; lanes d–f, Pro-Ser-nsP4fY1PI2S; lanes g–i, Pro-Ser-Pro-nsP4fY1PI2SF3P. (D) Quantitation of pulse–chase patterns in (C). Closed circles, Pro-nsP4fY1P; closed squares, Pro-Ser-nsP4fY1PI2S; closed triangles, Pro-Ser-Pro-nsP4fY1PI2SF3P. (E) The 20-residue or longer N-terminal regions of MOS confer instability on nsP4f. Lanes a–c, Pro-Ser-MOS2–36-nsP4f; lanes d–f, Pro-Ser-MOS2–24-nsP4f; lanes g–i, Pro-Ser-MOS2–21-nsP4f; lanes j–l, Pro-Ser-MOS2–18-nsP4f; lanes m–o, Pro-Ser-MOS2–15-nsP4f. (F) Quantitation of pulse–chase patterns in (E). Closed circles, Pro-Ser-MOS2–36-nsP4f; closed squares, Pro-Ser-MOS2–24-nsP4f; closed upright triangles, Pro-Ser-MOS2–21-nsP4f; closed diamonds, Pro-Ser-MOS2–18-nsP4f; closed inverted triangles, Pro-Ser-MOS2–15-nsP4f. (G) Replacement of N-terminal Pro with Gly results in long-lived Gly-Ser-MOS2–y-nsP4f. Lanes a–c, Pro-Ser-MOS2–36-nsP4f; lanes d–f, Gly-Ser-MOSP2G-36-nsP4f; lanes g–i, Pro-Ser-MOS2–24-nsP4f; lanes j–l, Gly-Ser-MOSP2G-24-nsP4f. (H) Quantitation of pulse–chase patterns in (G). Open squares, Pro-Ser-MOS2–36-nsP4f; closed squares, Gly-Ser-MOSP2G-36-nsP4f; open circles, Pro-Ser-MOS2–24-nsP4f; closed circles, Gly-Ser-MOSP2G-24-nsP4f.
We also constructed Pro-Ser-MOS2–15-nsP4f, Pro-Ser- MOS2–18-nsP4f, Pro-Ser-MOS2–21-nsP4f, Pro-Ser-MOS2–24- nsP4f and Pro-Ser-MOS2–36-nsP4f (Figure 1F). Whereas the N-terminal sequences of MOS shorter than 20 residues did not significantly destabilize nsP4f in oocytes, longer sequences did: the degradation of Pro-Ser-MOS2–21-nsP4f, Pro-Ser-MOS2–24-nsP4f and Pro-Ser-MOS2–36-nsP4f was comparable to, or faster than, the degradation of full-length Pro-Ser-MOS (Figure 3E and F). To determine whether the MOS-derived portable degron still exhibited the (operationally defined) requirement for N-terminal Pro, we examined Gly-Ser-MOSP2G-24-nsP4 and Gly- Ser-MOSP2G-36-nsP4f, which bore Gly instead of Pro at their N-termini (Figure 1F2 and F4). In striking contrast to instability of their Pro-bearing counterparts, both Gly-Ser-MOSP2G-24-nsP4f and Gly-Ser-MOSP2G-36-nsP4f were long-lived in vivo (Figure 3G and H).
N-terminal proline affects the net phosphorylation of Ser-2
To assay the degradation of MOS in extracts from eggs or oocytes, MOS bearing C-terminal tags (denoted as Pro-Ser-MOSfhk), and its derivatives Gly-Ser-MOSfhkP2G and Pro-Ala-MOSfhkS3A (Figure 1G), were expressed in Escherichia coli and purified by affinity chromatography. The in vivo degradation of Pro-Ser-MOSfhk in intact oocytes was indistinguishable from that of its untagged Pro-Ser-MOS counterpart (data not shown). The expected in vivo removal of N-terminal Met from E.coli-expressed Met-Pro-Ser-MOSfhk was verified and confirmed through N-terminal sequencing, by Edman degradation, of the purified protein (data not shown).
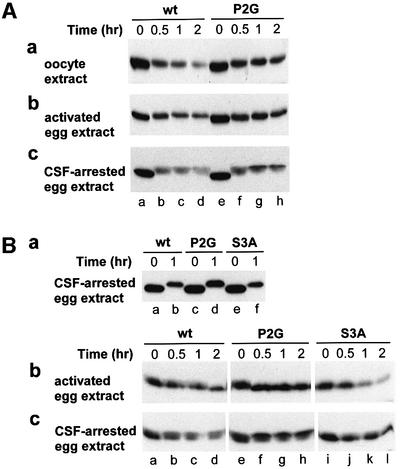
Pro-Ser-MOS is short-lived before germinal vesicle breakdown (GVBD), but becomes stable at the metaphase of meiosis II, thereby increasing in concentration and acting to maintain unfertilized egg at metaphase II. Upon fertilization, Pro-Ser-MOS becomes short-lived again (see Introduction). Extracts that represented these three stages—oocyte extract, cytostatic factor (CSF)-arrested egg extract and activated egg extract—were prepared and used to analyze, by immunoblotting with anti-flag antibody, the degradation of purified Pro-Ser-MOSfhk and its derivatives. As expected, Pro-Ser-MOSfhk was degraded in oocyte extract (Figure 4Aa, lanes a–d). Interestingly, Gly-Ser-MOSfhkP2G, which was stable in vivo (Figure 2A and B), was also degraded during the first 30 min of incubation in the extract, but became stable over the next 90 min of incubation (Figure 4Aa, lanes e–h versus a–d). The minor upshift in electrophoretic mobility of both Pro-Ser-MOSfhk and Gly-Ser-MOSfhkP2G was presumably caused by phosphorylation.
Fig. 4. Degradation and phosphorylation of E.coli-derived Xenopus MOS and its derivatives in extracts from Xenopus oocytes, activated eggs and CSF-arrested eggs. (A) Pro-Ser-MOSfhk and Gly-Ser- MOSfhkP2G in oocyte extract (a), activated egg extract (b) and CSF-arrested egg extract (c). Degradation (monitored through immunoblotting) and phosphorylation (detected through shifts in electrophoretic mobility) as a function of time after the addition of MOS to an extract. (B) Pro-Ser-MOSfhk, Gly-Ser-MOSfhkP2G and Pro-Ala-MOSfhkS3A in activated egg extract and CSF-arrested egg extract. In these experiments, a MOS test protein was pre-incubated, at 100 µg/ml, in 30 µl of the CSF-arrested egg extract for 1 h (a), followed by the addition of a resulting sample to either the activated egg extract (b) or CSF-arrested egg extract (c), to a final total volume of 25 µl and a final MOS concentration of ∼25 µg/ml.
The results of otherwise identical tests with extract from activated eggs were similar to those above (Figure 4Ab). When Pro-Ser-MOSfhk and Gly-Ser-MOSfhkP2G were incubated in CSF-arrested egg extract, both proteins were much more unstable than they were in either the oocyte extract or the activated egg extract (Figure 4Ac). This result seemed to be in contradiction to the known stability of MOS in CSF-arrested eggs (Nishizawa et al., 1992, 1993). Note, however, that in CSF-arrested eggs, the endogenously produced Pro-Ser-MOS is largely phosphorylated at Ser-2 (encoded Ser-3) and thus protected from degradation (Nishizawa et al., 1992, 1993), whereas in our in vitro setting, the E.coli-derived Pro-Ser-MOSfhk and its derivatives were, at least initially, unphosphorylated and therefore susceptible to degradation. However, the differences between Pro-Ser-MOSfhk and Gly-Ser-MOSfhkP2G persisted in CSF-arrested extract as well. Specifically, the mutant Gly-Ser-MOSfhkP2G was significantly more stable after 30 min, with a persisting upper (shifted) band at 1 and 2 h time points; this species may be a version of MOS fully stabilized through phosphorylation at Ser-2 (Figure 4Ac, lanes g and h versus c and d).
Thus, in either of the three extracts, the (initially) unphosphorylated Gly-Ser-MOSfhkP2G, which bore N-terminal Gly (a stabilizing residue in the N-end rule), was as short-lived during the first 30 min of incubation as was its Pro-bearing wild-type counterpart, Pro-Ser- MOSfhk (Figure 4A). However, Gly-Ser-MOSfhkP2G became stable (and more phosphorylated) after 30 min of incubation, in contrast to Pro-Ser-MOSfhk (Figure 4A). These findings suggested that the strong stabilizing effect of N-terminal Gly on MOS in vivo (Figures 2A, B and 3A) stemmed from faster phosphorylation (and/or slower dephosphorylation) of MOS at Ser-2 in the presence of N-terminal Gly than in the presence of N-terminal Pro. In this interpretation, Pro-MOS is targeted for degradation through an N-terminus-proximal degron whose activity requires the absence of phosphorylation of Ser-2. If so, the degron of Pro-Ser-MOS is not an N-degron, in which, by definition, the substrate’s N-terminal residue is an essential determinant recognized by a Ub ligase. One prediction of this model is that MOS phosphorylated at Ser-2 (encoded Ser-3) would be resistant to degradation, and also, conversely, that MOS that could not be phosphorylated at that position would be short-lived, irrespective of the identity of its N-terminal residue.
Since MOSfhk purified from E.coli could be phosphorylated in CSF-arrested egg extract (Figure 4Ac), we incubated the E.coli-derived Pro-Ser-MOSfhk, Gly-Ser- MOSfhkP2G and Pro-Ala-MOSfhkS3A in that extract for 1 h (Figure 4Ba), and the resulting samples were added, separately, to 4 vols of either activated egg extract or CSF-arrested (the same) egg extract (Figure 4Bb and c). As indicated by the upward shifts in mobility, Pro-Ser- MOSfhk, Gly-Ser-MOSfhkP2G and Pro-Ala-MOSfhkS3A were phosphorylated (in addition to their initial degradation) after 1 h in CSF-arrested extract (Figure 4Ba). Significantly more of the added Gly-Ser-MOSfhkP2G was left intact after 1 h incubation than in the case of the other two proteins (Figure 4Ba), in agreement with other data (Figure 4A).
Upon fertilization of CSF-arrested eggs, the fully phosphorylated (and stable) MOS is rapidly degraded, owing to its fertilization-induced dephosphorylation (Nishizawa et al., 1993). The addition of phosphorylated MOS to activated egg extract would be an analog of this event. Indeed, the addition of pre-incubated, phosphorylated Pro-Ser-MOSfhk to activated egg extract resulted in its rapid dephosphorylation (Figure 4Bb, lanes a–d). The lower mobility of Gly-Ser-MOSfhkP2G at time zero (Figure 4Bb, lanes e–h) indicated that dephosphoryl ation of MOS bearing N-terminal Gly was substantially slower than dephosphorylation of MOS bearing N-terminal Pro. In addition, Gly-Ser-MOSfhkP2G was more stable than either Pro-Ser-MOSfhk or Pro-Ala-MOSfhkS3A (Figure 4Bb).
When pre-incubated, phosphorylated Pro-Ser-MOSfhk, Gly-Ser-MOSfhkP2G and Pro-Ala-MOSfhkS3A were added to a fresh CSF-arrested egg extract, Pro-Ser-MOSfhk was initially dephosphorylated, but then the undegraded Pro-Ser-MOSfhk was phosphorylated again, as indicated by the shifts in mobility (Figure 4Bc, lanes a–d). Pro-Ser-MOSfhk was unstable throughout the incubation. In contrast, Gly-Ser-MOSfhkP2G was degraded much more slowly, and also, as predicted by the above model, was phosphorylated faster and more extensively than Pro-Ser-MOSfhk (Figure 4Bc, lanes e–h versus a–d). As for Pro-Ala-MOSfhkS3A, it was unstable throughout the incubation, and could not be fully phosphorylated, as indicated by the absence of a strong upper band, characteristic of Gly-Ser-MOSfhkP2G (Figure 4Bc, lanes i–l versus e–h).
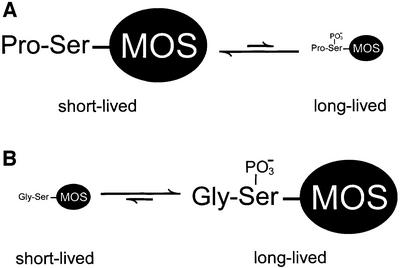
The above results (Figure 4) suggested that the presence of Gly (or another stabilizing residue) instead of wild-type Pro at the N-terminus of MOS may inhibit dephosphorylation of Ser-2 (encoded Ser-3), and that the net difference in the level of phosphorylation of Ser-2 determines the rate of MOS degradation in this setting. In other words, the stabilizing effect of N-terminal Gly, relative to Pro, would be through an increase in the level of phosphorylated Ser-2 in MOS (Figure 5).
Fig. 5. N-terminal proline of MOS influences net phosphorylation of Ser-2, a modification that halts MOS degradation in oocytes. This model, based on the findings of the present work, is described in Results and Discussion.
N-terminus-proximal, conditional degron of MOS
The rate of degradation of Pro-Gly-MOSS3G (Figure 1C9) was similar to that of Pro-Ser-MOS (data not shown). The above model (Figure 5) predicted that Gly-Ser-MOSP2G, which was long-lived in vivo (Figures 2A, B and 3A), would become short-lived, similarly to wild-type Pro-Ser-MOS, if phosphorylatable Ser-2 would be replaced by an unphosphorylatable residue such as Gly. Oocytes were injected with mRNAs encoding UPR-based fusions that yielded either Gly-Ser-MOSP2G or Gly-Gly-MOSP2GS3G (Figure 1C10). As predicted, whereas Gly-Ser-MOSP2G was long-lived in oocytes, Gly-Gly-MOSP2GS3G was strikingly more unstable, with a large (∼50%) fraction of its degradation taking place during the pulse (Figure 6A, lanes a–c versus d–f, and B). These results, in conjunction with the data above (Figures 2–4), directly proved that the N-terminal Pro residue of wild-type Pro-Ser-MOS was not an essential determinant of the MOS degron, since N-terminal Gly (a stabilizing residue in the N-end rule) could function just as well, provided that position 2 was not occupied by a phosphorylatable residue such as Ser. Thus, the N-terminus-proximal degron of MOS is not, mechanistically, an N-degron (see Introduction for the definition of N-degrons).
Fig. 6. Identity of the N-terminal residue of MOS determines the rate of phosphorylation of Ser-2 that inhibits MOS proteolysis. (A) If Ser-2 (encoded Ser-3) of MOS is replaced by Gly, the degradation of MOS or MOS2–24-nsP4f in oocytes does not require N-terminal Pro. Lanes a–c, Gly-Ser-MOSP2G; lanes d–f, Gly-Gly-MOSP2GS3G; lanes g–i, Gly-Ser-MOSP2G-24-nsP4f; lanes j–l, Gly-Gly-MOSP2GS3G-24-nsP4f. (B) Quantitation of pulse–chase patterns in (A). Open circles, Gly-Ser-MOSP2G; closed circles, Gly-Gly-MOSP2GS3G; open squares, Gly-Ser-MOSP2G-24-nsP4f; closed squares, Gly-Gly-MOSP2GS3G-24-nsP4f. (C) Degradation of MOS-nsP4f during labeling in oocytes. Lanes a–c, Gly-nsP4fYIG; lanes d–f, Gly-Ser-MOSP2G-24-nsP4f; lanes g–i, Pro-Ser-MOS2–24-nsP4f. (D) Quantitation of pulse–chase patterns in (C). Closed circles, Gly-nsP4fY1G; closed squares, Gly-Ser-MOSP2G-24-nsP4f; closed triangles, Pro-Ser-MOS2–24-nsP4f. At each time point, the ratio of 35S in a test protein to 35S in fDHFR-UbR48 (the reference protein) was plotted as a percentage of this ratio for Gly-nsP4fY1G at time zero. (E) The patterns of degradation- suppressing phosphorylation of Ser-2 in Pro-Ser-MOS in stage VI Xenopus oocytes are similar in the presence or absence of progesterone. Stage VI oocytes were injected with mRNAs encoding either Pro-Ser-MOS2–24-nsP4f or Gly-Ser-MOSP2G-24-nsP4f, and were incubated for 30 min in MBS containing progesterone, followed by pulse–chase in the presence of progesterone. Lanes a–c, Pro-Ser-MOS2––24-nsP4f; lanes d–f, Gly-Ser-MOSP2G-24-nsP4f. In a parallel test, the injected oocytes were incubated for 30 min in the absence of progesterone, followed by pulse–chase also in the absence of progesterone. Lanes g–i, Pro-Ser-MOS2–24-nsP4f; lanes j–l, Gly-Ser-MOSP2G-24-nsP4f.
To determine whether these findings could be reproduced with the MOS-derived 20-residue portable degron linked to nsP4, similar microinjection experiments were carried out with mRNAs encoding UPR-based fusions that yielded either Gly-Ser-MOSP2G-24-nsP4f (Figure 1F4) or Gly-Gly-MOSP2GS3G-24-nsP4f (Figure 1F5). Whereas Gly-Ser-MOSP2G-24-nsP4f was long-lived (see also Figure 3G and H), the otherwise identical Gly-Gly-MOSP2GS3G-24-nsP4f, in which Ser-2 was converted to Gly, was strikingly unstable (Figure 6A, lanes g–i versus j–l, and B). Thus, the 20-residue N-terminal region of MOS faithfully mimics the entire range of tested properties of the N-terminus-proximal MOS degron.
Although Gly-Ser-MOSP2G was long-lived in oocytes, in contrast to Pro-Ser-MOS (Figure 2A and B), the model of Figure 5 predicted that a newly formed Gly-Ser- MOSP2G that was not yet phosphorylated at Ser-2 would be, initially, as short-lived as Pro-Ser-MOS. An initial burst of degradation can be missed in a pulse–chase assay, especially if the bulk of degradation occurs during the pulse. Thus, if the model of Figure 5 is correct, one would expect an initial degradation of Gly-Ser-MOSP2G, followed by phosphorylation-mediated stabilization of Gly-Ser-MOSP2G. The latter process would be relatively slow with Pro-Ser-MOS, thus accounting for its continued degradation. We addressed this prediction by comparing the UPR-based in vivo decay curves, in oocytes, of a bona fide long-lived protein such as Gly-nsP4fY1G (bearing a stabilizing N-terminal residue and lacking MOS degron; Figure 1E2) with that of Gly-Ser-MOSP2G-24-nsP4f (Figure 1F4). The overtly short-lived Pro-Ser-MOS2–24-nsP4f was assayed as well, the latter two proteins being only 23 residues longer than Gly-nsP4f Y1G, and otherwise identical to it. Both Gly-nsP4fY1G and Gly-Ser-MOSP2G-24-nsP4f were long-lived during the chase (Figure 6C, lanes a–c and d–f, and D), in contrast to Pro-Ser- MOS2–24-nsP4f (Figure 6C, lanes g–i, and D). However, the relative amount of Gly-Ser-MOSP2G-24-nsP4f (calibrated against the reference protein; Figure 1A) at time zero (the beginning of chase) was <60% of Gly-nsP4fY1G, indicating that >40% of the initially labeled Gly- Ser-MOSP2G-24-nsP4f was degraded during the pulse (Figure 6D). By comparison, >80% of initially labeled Pro-Ser-MOS2–24-nsP4f was degraded under the same conditions (Figure 6D). These results indicated that newly formed Gly-Ser-MOSP2G-24-nsP4f was transiently unstable, relative to Gly-nsP4fY1G, in agreement with a prediction of the model (Figure 5).
If Ser-2 (encoded Ser-3) of Pro-Ser-MOS was normally phosphorylated by MOS itself (Nishizawa et al., 1992) or cyclin B/cdc2 (Castro et al., 2001), one could predict that Gly-Ser-MOSP2G-24-nsP4f, which lacked kinase activity, would be short-lived in oocytes that had not been exposed to progesterone, because virtually no endogenous MOS is produced in such oocytes (Sagata et al., 1988), and their cyclin B/cdc2 complexes are held at low activity by inactivating phosphorylation (Ferrell, 1999). However, pulse–chases with Gly-Ser-MOSP2G-24-nsP4f and Pro- Ser-MOS2–24-nsP4f in progesterone-treated versus untreated oocytes indicated that Gly-Ser-MOSP2G-24-nsP4f remained stable during the chase in both settings, in contrast to Pro-Ser-MOS2–24-nsP4f (Figure 6E). Thus, a kinase that phosphorylates Ser-2 of MOS during oocyte maturation is not (exclusively) MOS itself or cyclin B/cdc2, and the relevant kinase must be present in both progesterone-treated and untreated stage VI oocytes, in agreement with earlier data (Freeman et al., 1992). In addition, the fact that Pro-Ser-MOS2–24-nsP4f was short-lived in either progesterone-treated or untreated oocytes implied slow net phosphorylation of Ser-2 under both conditions, and therefore suggested that a phosphatase that dephosphorylates phosphoserine-2, and/or an inhibitor of the relevant kinase, were already present in resting stage VI oocytes, before the induction of Pro-Ser-MOS synthesis by progesterone.
In mouse 3T3 cells, in Xenopus XTC cells and in rabbit reticulocyte extract the degradation of MOS does not require N-terminal proline
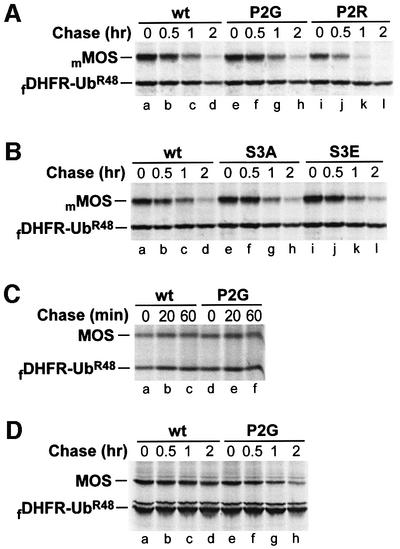
Whereas the major phosphorylation site of MOS in Xenopus oocytes is Ser-2 (encoded Ser-3) (Freeman et al., 1992; Nishizawa et al., 1992), it was reported that Ser-15 (encoded Ser-16) is the major phosphorylation site of MOS in transfected mammalian (green monkey) COS-1 cells (Pham et al., 1999). Either mouse MOS (mMOS) or Xenopus MOS was found to be short-lived (t1/2 ≈ 30 min) in mouse NIH 3T3 cells, irrespective of whether they bore the wild-type N-terminal Pro residue or the Gly residue (Figures 1D1 and D2, and 7A, lanes a–h; data not shown). Replacing N-terminal Pro of Pro-Ser-mMOS with Arg (Figure 1D3) yielded a highly unstable protein (Figure 7A, lanes i–l), confirming the presence of the N-end rule pathway in 3T3 cells. Furthermore, the degradation of Pro-Ala-mMOSS3A and Pro-Glu-mMOSS3E (Figure 1D4 and D5) in 3T3 cells was similar to that of wild-type Pro-Ser-MOS (Figure 7B), despite the absence of a phosphorylatable residue at the (encoded) position 3 of Pro-Ala-mMOSS3A and the presence of a negatively charged residue (a mimic of phosphorylated Ser) at that position in Pro-Glu-mMOSS3E. Nishizawa et al. (1992, 1993) have shown that Glu at position 2 of Pro-Glu-MOSS3E stabilized MOS in Xenopus oocytes. Thus, the degradation of MOS in mouse 3T3 cells does not involve the oocyte-specific degron of MOS analyzed in this study. (Oocytes are a physiologically relevant cell type with regard to MOS expression.)
Fig. 7. Degradation of either Xenopus or mouse MOS in mouse NIH 3T3 cells, Xenopus XTC cells and rabbit reticulocyte lysate does not depend on the N-terminal Pro residue of MOS. Positions of fDHFR-UbR48 and either Xenopus MOS (MOS) or mouse MOS (mMOS) are indicated on the left. (A) Pulse–chase in mouse NIH 3T3 cells. Lanes a–d, Pro-Ser-mMOS; lanes e–h, Gly-Ser-mMOSP2G; lanes i–l, Arg-Ser-mMOSP2R. (B) Mutations at Ser-2 (encoded Ser-3) have a negligible effect on degradation of mouse MOS in NIH 3T3 cells. Lanes a–d, Pro-Ser-mMOS; lanes e–h, Pro-Ala-mMOSS3A; lanes i–l, Pro-Glu-mMOSS3E. (C) Xenopus MOS is long-lived in Xenopus XTC cells. Lanes a–c, Pro-Ser-MOS; lanes d–f, Gly-Ser-MOSP2G. (D) N-terminal Pro of Xenopus MOS does not metabolically destabilize MOS in rabbit reticulocyte lysate. Lanes a–d, Pro-Ser-MOS; lanes e–h, Gly-Ser-MOSP2G.
In contrast to instability of MOS in mouse NIH 3T3 cells, both wild-type Pro-Ser-MOS and Gly-Ser-MOSP2G were long-lived in Xenopus fibroblast-like XTC cells (Figure 7C). However, Arg-Ser-MOSP2R was short-lived in XTC cells (data not shown), confirming the presence of the N-end rule pathway in these cells. Pro-Ser-MOS and Gly-Ser-MOSP2G were both moderately short-lived proteins in rabbit reticulocyte extract, with half-lives of ∼90 min (Figure 7D; data not shown), in contrast to the strikingly different rates of degradation of these proteins in Xenopus oocytes (Figure 2A and B).
Discussion
Studies by Sagata and colleagues have shown that wild-type MOS, bearing N-terminal Pro, is short-lived upon its expression in Xenopus oocytes, whereas the otherwise identical MOS derivative bearing N-terminal Gly is long-lived (Nishizawa et al., 1992, 1993). These and related findings led the authors to propose the ‘second-codon rule’, in which the N-terminal Pro residue of MOS targets MOS for degradation through the recognition of Pro by a Ub ligase (Nishizawa et al., 1992, 1993). In the N-end rule’s terminology (see Introduction), this conjecture meant that the MOS protein contains a distinct (Pro-specific) N-degron.
However, the N-terminal Pro residue was clearly a stabilizing residue in both the yeast Saccharomyces cerevisiae and mammalian cells, in the context of substrates other than MOS (Bachmair et al., 1986; Gonda et al., 1989; F.Lévy and A.Varshavsky, unpublished data). Given this conundrum, we reinvestigated the MOS degron in the present work, and demonstrated that MOS contains a portable, N-terminus-proximal degradation signal that is qualitatively distinct from an N-degron, in that the N-terminal Pro residue of MOS is entirely dispensable for the in vivo degradation of MOS in oocytes, if Ser-2 (encoded Ser-3) of MOS is replaced by a small non-phosphorylatable residue such as Gly. We also demonstrated that the previously observed dependence of MOS degradation on its N-terminal Pro residue (Nishizawa et al., 1992, 1993) is caused by a Pro-mediated downregulation of the net phosphorylation of Ser-2, a modification that inhibits MOS degradation. Thus, the N-terminal Pro residue of MOS is not a recognition determinant for a Ub ligase, in agreement with the earlier evidence that Pro is a stabilizing residue in the N-end rule (Varshavsky, 1996). We showed that when a stabilizing residue other than Pro, e.g. Gly, is present at the N-terminus of MOS, it results in a much higher net phosphorylation of Ser-2 than occurs in the presence of N-terminal Pro, thereby accounting for rapid stabilization of Gly-Ser-MOS. Yet another outcome of this extensive analysis (Figures 1–4 and 6) was the demonstration that the 20-residue N-terminal region of MOS can function as a portable, phosphorylation-regulated degron if it is linked to an unrelated protein expressed in oocytes (Figures 3 and 6).
The resulting advances in understanding, which accounted for both the earlier data (Nishizawa et al., 1992, 1993) and the findings of the present work, are summarized in Figure 5, which depicts the partitioning of an X-Ser-MOS (X = Pro or Gly) between unphosphoryl ated X-Ser-MOS and its phosphorylated (at Ser-2) counterpart. Our findings (Figures 1–4 and 6) indicated that unphosphorylated X-Ser-MOS is short-lived in stage VI oocytes, irrespective of whether its N-terminal residue is Pro or another stabilizing residue, such as Gly. By contrast, phosphorylated X-Ser(P)-MOS is metabolically stable, again irrespective of whether the N-terminal X residue is Pro or Gly (Figure 5). The wild-type N-terminal Pro of MOS is important, in the context of MOS containing Ser at position 2 (encoded Ser-3), because the presence of Pro downregulates the proteolysis-halting net phosphorylation of Ser-2 in Pro-Ser-MOS. By contrast, the N-terminal Gly residue, in Gly-Ser-MOS, increases net phosphorylation of Ser-2, thereby rapidly stabilizing MOS against degradation. We showed that replacement of Ser-2 with a non-phosphorylatable residue such as Gly abrogates the dependence of MOS degradation on the presence of N-terminal Pro (Figure 6). Thus, it is the relative amounts of phosphorylated (long-lived) versus non-phosphorylated (short-lived) species of X-Ser-MOS that are altered upon the conversion of N-terminal Pro to Gly, because N-terminal Pro suppresses the action of kinases at Ser-2 and/or facilitates the action of phosphatases at the same residue (Figure 5).
(Met)-Pro-Ser-Pro, the four-residue encoded N-terminal sequence of MOS, is conserved among all vertebrates examined (Sagata, 1997). The N-terminal Met of nascent MOS is rapidly removed in vivo by MetAPs (Bradshaw et al., 1998), yielding N-terminal Pro (see Results). A parsimonious interpretation is that N-terminal Pro of MOS was selected, during MOS evolution, as a physiologically optimal N-terminal residue for modulating the extent of net phosphorylation of the adjacent Ser-2 residue, a step that halts MOS degradation in oocytes (Figure 5). The portable, phosphorylation-regulated, apparently oocyte-specific degron of MOS is now a particularly well understood example of a degradation signal downregulated by phosphorylation (Figure 5). A Ub ligase (E3) that recognizes this degron remains to be identified.
Materials and methods
Plasmids
The Xenopus and mouse MOS genomic DNA clones were a gift from Dr G.F.Vande Woude. The PCMV-containing pcDNA3 (InVitrogen) was used to construct the sets of plasmids pcDNA3fDHFRUbR48Xpr and pcDNA3fDHFRUbR48MXpr that encoded, respectively, fDHFR- UbR48–X-[test protein] and fDHFR-UbR48–Met-X-[test protein] (Figure 1A and B), ‘f’ being the flag epitope (Ausubel et al., 2002) and ‘X’ a variable residue at the Ub–protein junction (Varshavsky, 2000). The plasmids were constructed by ligating PCR-produced DNA fragments to HindIII–XbaI-cut pcDNA3. For expression of Xenopus MOS in E.coli, the plasmids pET11cMOSfhk were constructed by ligating NdeI–BamHI-cut pET-11c vector (Stratagene) with PCR fragments that encoded wild-type or mutant Xenopus MOS (MOSfhk) bearing the C-terminal extension GTSDYKDDDDKGGSHHHHHHRRASV (Figure 1G), denoted as fhk, which contained the C-terminal flag tag (f), His6 tag (h) and the kinase A recognition site (k). Some intermediate constructs and all of the final plasmids were verified by sequencing.
Pulse–chase analysis in Xenopus oocytes
pcDNA3fDHFRUbR48Xpr and pcDNA3fDHFRUbR48MXpr were cut with BglII, transcribed in vitro using T7 RNA polymerase (mMESSAGE-mMACHINE kit; Ambion) and capped at 5′ ends to increase translation efficiency. Stage VI Xenopus oocytes were microinjected, using standard procedures (Spector et al., 1997), with ∼4 ng of capped RNA, and thereafter incubated for 1.5 h at 19–21°C in modified Barth solution (MBS) (Gurdon and Wickens, 1983) containing 5 µg/ml progesterone and 1 mCi/ml [35S]methionine (Amersham) (Nishizawa et al., 1992). The oocytes were then transferred to MBS in the presence of 5 µg/ml progesterone and 0.1 mg/ml cycloheximide. At different times afterwards, the sets of six oocytes were lysed together in ice-cold buffer A, at 50 µl/oocyte (buffer A: 1% Triton X-100, 0.15 M NaCl, 5 mM EDTA, 0.2 mg/ml PMSF, 20 mM Tris–HCl pH 7.5). The lysates were centrifuged in a microcentrifuge for 15 min, and the supernatants were precipitated with antibody to Xenopus MOS (Santa Cruz) and/or with anti-flag antibody (Sigma). The antibodies were diluted in ice-cold buffer A and added to a final volume of 0.8 ml, followed by gentle rocking for 30 min at 4°C. Protein G–agarose suspension (25 µl; Pierce) was added to the samples, followed by gentle rocking for 1.5 h at 4°C and 12 s centrifugation in a microcentrifuge at ∼1000 r.p.m. Pellets were washed three times with 0.8 ml of buffer A at 4°C, resuspended in SDS sample buffer, heated at 95°C for 5 min, and subjected to 12% SDS–PAGE and autoradiography, followed by quantitation using a PhosphorImager (Molecular Dynamics).
Assays with oocyte extract, CSF-arrested egg extract and activated egg extract
The extracts from CSF-arrested eggs and activated eggs were prepared as described previously (Murray, 1991). For the preparation of oocyte extract, adult female frogs were injected with 50 IU of pregnant mare serum gonadotropin (PMSG) 5 days before the experiment, and with 25 IU of PMSG 2 days later. Ovaries from frogs treated thus were dissected into clumps and incubated in MBS containing 0.5% collagenase (Sigma) at room temperature for 2–5 h, until oocytes floated free. Oocytes were then rinsed four times with MBS at 20°C and sorted, selecting for stage VI oocytes, which were incubated in MBS containing 5 µg/ml progesterone for 2 h at 20°C, then washed twice with MBS containing protease inhibitors (10 µg/ml each leupeptin, pepstatin and chymostatin), and transferred to 3 ml centrifuge tubes containing MBS plus protease inhibitors and 100 µg/ml cytochalasin B. An excess of MBS was removed, and oocytes were crushed by centrifugation at 10 000 r.p.m. for 10 min at 4°C in a swinging-bucket rotor. The cytoplasmic layer was collected and clarified by centrifugation at 15 000 r.p.m. for 10 min at 4°C in a swinging-bucket rotor. The same protease inhibitors and cytochalasin B were then added from stock solutions, to the same final concentrations, and also 1/20 volume of 20 mM MgCl2, 0.15 M creatine phosphate, 20 mM ATP pH 7.4.
The plasmids pET11cMOSfhk, encoding either wild-type or mutant MOSfhk proteins, were transformed into E.coli BL21 (DE3). Following the addition of isopropyl-1-thio-β-d-galactopyranoside (IPTG) (Ausubel et al., 2002), an induced MOSfhk was purified by Ni-NTA affinity chromatography (Qiagen) under non-denaturing conditions, and rapidly frozen in small aliquots in elution buffer (0.3 M NaCl, 0.25 M imidazole, 50 mM NaH2PO4 pH 8.0).
Purified Ub (Boston Biochem) was added to the extracts to a final concentration of 3 µM. In some experiments, cycloheximide was also added, to a final concentration of 0.1 mg/ml. In proteolytic assays, wild-type and mutant MOSfhk proteins were added to the extract to a final concentration of ∼25 µg/ml, and the reaction mixtures (25 µl) were incubated at 23°C. Samples of 2 µl were withdrawn for each time point and subjected to 12% SDS–PAGE, followed by immunoblotting with anti-flag antibody.
Pulse–chase analysis with cell lines
Mouse NIH 3T3 cells were grown, transfected, and assayed by pulse–chase as described previously (Kwon et al., 2001), except that cycloheximide was not added to the chase medium. Xenopus fibroblast XTC cells (Pudney et al., 1973) were grown in 70% Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and antibiotics in the presence of 7% CO2 at 26°C. Cells were lysed in ice-cold buffer A, and the lysates were immunoprecipitated with antibody to either mMOS (Santa Cruz) or Xenopus MOS (Santa Cruz) and/or anti-flag antibody, followed by 12% SDS–PAGE and autoradiography.
Pulse–chase analysis of Xenopus MOS in reticulocyte lysate
The TNT T7 Quick Coupled Transcription/Translation System (Promega) is a rabbit reticulocyte lysate premixed with most of the reaction components for carrying out transcription/translation in the lysate, including all amino acids except Met. A pcDNA3fDHFRUbR48MXpr plasmid (1 µg) encoding a fDHFR-UbR48–MX-[test protein] fusion and 20 µCi of [35S]methionine (Amersham) were added to this system, and the sample (40 µl) was incubated at 30°C for 30 min. Both cycloheximide and unlabeled Met were then added, to final concentrations of 0.1 mg/ml and 0.7 mM, and the sample was incubated at 30°C for 2 h. Samples of 5 µl were withdrawn at various times, and analyzed by 12% SDS–PAGE and autoradiography.
Acknowledgments
Acknowledgements
We thank G.F.Vande Woude for the gifts of Xenopus and mouse MOS genomic DNA, and for anti-MOS antibodies that were used at early stages of this study. We are grateful to H.Lester for allowing the use of his laboratory’s facility for microinjection of oocytes, and thank Y.Tong for his advice on microinjection. We also thank the current and former members of the Varshavsky laboratory, especially Y.Xie, H.Wang, G.Turner, Y.T.Kwon and F.Du, for helpful discussions and suggestions. This study was supported by grants from the National Institutes of Health to A.V. (GM31530 and DK39520) and W.G.D. (GM43974). W.G.D. is an Investigator in the Howard Hughes Medical Institute.
References
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Smith,J.A., Seidman,J.G. and Struhl,K. (eds) (2002) Current Protocols in Molecular Biology. Wiley-Interscience, New York, NY.
- Bachmair A. and Varshavsky,A. (1989) The degradation signal in a short-lived protein. Cell, 56, 1019–1032. [DOI] [PubMed] [Google Scholar]
- Bachmair A., Finley,D. and Varshavsky,A. (1986) In vivo half-life of a protein is a function of its amino-terminal residue. Science, 234, 179–186. [DOI] [PubMed] [Google Scholar]
- Bradshaw R.A., Brickey,W.W. and Walker,K.W. (1998) N-terminal processing: the methionine aminopeptidase and Nα-acetyl transferase families. Trends Biochem. Sci., 23, 263–267. [DOI] [PubMed] [Google Scholar]
- Castro A., Peter,M., Magnaghi-Jaulin,L., Vigneron,S., Galas,S., Lorca,T. and Labbe,J.-C. (2001) Cyclin B/cdc2 induces c-Mos stability by direct phosphorylation in Xenopus oocytes. Mol. Biol. Cell, 12, 2660–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth A., Ridge,J.A., King,L.A., MacNicol,M.C. and MacNicol,A.M. (2002) A novel regulatory element determines the timing of Mos mRNA translation during Xenopus oocyte maturation. EMBO J., 21, 2798–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov I.V. and Varshavsky,A. (2000) RGS4 is arginylated and degraded by the N-end rule pathway in vitro. J. Biol. Chem., 275, 22931–22941. [DOI] [PubMed] [Google Scholar]
- deGroot R.J., Rümenapf,T., Kuhn,R.J. and Strauss,J.H. (1991) Sindbis virus RNA polymerase is degraded by the N-end rule pathway. Proc. Natl Acad. Sci. USA, 88, 8967–8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesbery N.S. and Vande Woude,G.F. (2002) An arresting activity. Nature, 416, 804–805. [DOI] [PubMed] [Google Scholar]
- Dupré A., Jessus,C., Ozon,R. and Haccard,O. (2002) Mos is not required for the initiation of meiotic maturation in Xenopus oocytes. EMBO J., 21, 4026–4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell J.E. (1999) Xenopus oocyte maturation: new lessons from a good egg. BioEssays, 21, 833–842. [DOI] [PubMed] [Google Scholar]
- Ferrell J.E. (2002) Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr. Opin. Chem. Biol., 6, 140–148. [DOI] [PubMed] [Google Scholar]
- Freeman R.S., Meyer,A.N., Li,J. and Donoghue,D.J. (1992) Phosphorylation of conserved serine residues does not regulate the ability of mos protein to induce oocyte maturation or function as cytostatic factor. J. Cell Biol., 116, 725–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonda D.K., Bachmair,A., Wünning,I., Tobias,J.W., Lane,W.S. and Varshavsky,A. (1989) Universality and structure of the N-end rule. J. Biol. Chem., 264, 16700–16712. [PubMed] [Google Scholar]
- Gurdon J.B. and Wickens,M.P. (1983) The use of Xenopus oocytes for the expression of cloned genes. Methods Enzymol., 101, 370–386. [DOI] [PubMed] [Google Scholar]
- Hochegger H., Klotzbücher,A., Kirk,J., Howell,M., Guellec,K., Fletcher,K., Duncan,T., Sohail,M. and Hunt,T. (2001) New B-type cyclin synthesis is required between meiosis I and II during Xenopus oocyte maturation. Development, 128, 3795–3807. [DOI] [PubMed] [Google Scholar]
- Kwon Y.T., Xia,Z., Davydov,I.V., Lecker,S.H. and Varshavsky,A. (2001) Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3a) of the N-end rule pathway. Mol. Cell. Biol., 21, 8007–8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y.T., Kashina,A.S., Davydov,I.V., Hu,R.-G., An,J.Y., Seo,J.W., Du,F. and Varshavsky,A. (2002) An essential role of N-terminal arginylation in cardiovascular development. Science, 297, 96–99. [DOI] [PubMed] [Google Scholar]
- Mendez R., Hake,L.E., Andresson,T., Littlepage,L.E., Ruderman,J.V. and Richter,J.D. (2000) Prosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature, 404, 302–307. [DOI] [PubMed] [Google Scholar]
- Murray A.W. (1991) Cell cycle extracts. Methods Cell Biol., 36, 581–605. [PubMed] [Google Scholar]
- Nebreda A.R. and Ferby,I. (2000) Regulation of the meiotic cell cycle in oocytes. Curr. Opin. Cell Biol., 12, 666–675. [DOI] [PubMed] [Google Scholar]
- Nishizawa M., Okazaki,K., Furuno,N., Watanabe,N. and Sagata,N. (1992) The second-codon rule and autophosphorylation govern the stability and activity of Mos during the meiotic cell cycle in Xenopus oocytes. EMBO J., 11, 2433–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa M., Furuno,N., Okazaki,K., Tanaka,H., Ogawa,Y. and Sagata,N. (1993) Degradation of Mos by the N-terminal proline (Pro-2)-dependent ubiquitin pathway on fertilization of Xenopus eggs: possible significance of natural selection for Pro-2 in Mos. EMBO J., 12, 4021–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham C.D., Vuyyuru,V.B., Yang,Y., Bai,W. and Singh,B. (1999) Evidence for an important role of serine 16 and its phosphorylation in the stabilization of c-Mos. Oncogene, 18, 4287–4294. [DOI] [PubMed] [Google Scholar]
- Pudney M., Varma,M.G.R. and Leake,C.J. (1973) Establishment of a cell line XTC-2 from the South American clawed toad Xenopus laevis. Experientia, 29, 466–467. [DOI] [PubMed] [Google Scholar]
- Rao H., Uhlmann,F., Nasmyth,K. and Varshavsky,A. (2001) Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature, 410, 955–960. [DOI] [PubMed] [Google Scholar]
- Reimann J.D.R. and Jackson,P.K. (2002) Emi1 is required for cytostatic factor arrest in vertebrate eggs. Nature, 416, 850–854. [DOI] [PubMed] [Google Scholar]
- Sagata N. (1997) What does Mos do in oocytes and somatic cells? BioEssays, 19, 13–21. [DOI] [PubMed] [Google Scholar]
- Sagata N., Oskarsson,M., Copeland,T., Brumbaugh,J. and Vande Woude,G.F. (1988) Function of c-mos proto-oncogene product in meiotic maturation of Xenopus oocytes. Nature, 335, 519–525. [DOI] [PubMed] [Google Scholar]
- Spector D.L., Goldman,R.D. and Leinwald,L.A. (1997) Cells: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Suzuki T. and Varshavsky,A. (1999) Degradation signals in the lysine–asparagine sequence space. EMBO J., 18, 6017–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner G.C. and Varshavsky,A. (2000) Detecting and measuring cotranslational protein degradation in vivo. Science, 289, 2117–2120. [DOI] [PubMed] [Google Scholar]
- Turner G.C., Du,F. and Varshavsky,A. (2000) Peptides accelerate their uptake by activating a ubiquitin-dependent proteolytic pathway. Nature, 405, 579–583. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. (1996) The N-end rule: functions, mysteries, uses. Proc. Natl Acad. Sci. USA, 93, 12142–12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. (2000) Ubiquitin fusion technique and its descendants. Methods Enzymol., 327, 578–593. [DOI] [PubMed] [Google Scholar]