Abstract
We present an analysis of 203 completed genomes in the Gene3D resource (including 17 eukaryotes), which demonstrates that the number of protein families is continually expanding over time and that singleton-sequences appear to be an intrinsic part of the genomes. A significant proportion of the proteomes can be assigned to fewer than 6000 well-characterized domain families with the remaining domain-like regions belonging to a much larger number of small uncharacterized families that are largely species specific. Our comprehensive domain annotation of 203 genomes enables us to provide more accurate estimates of the number of multi-domain proteins found in the three kingdoms of life than previous calculations. We find that 67% of eukaryotic sequences are multi-domain compared with 56% of sequences in prokaryotes. By measuring the domain coverage of genome sequences, we show that the structural genomics initiatives should aim to provide structures for less than a thousand structurally uncharacterized Pfam families to achieve reasonable structural annotation of the genomes. However, in large families, additional structures should be determined as these would reveal more about the evolution of the family and enable a greater understanding of how function evolves.
INTRODUCTION
A fundamental bridge that enables us to link a protein sequence to its function is knowledge of its structure. It is well known that protein structure tends to be conserved to a greater degree than protein sequence and comparison of protein structure is often able to reveal functional relationships that are hidden at the sequence level (1). The identification of links between structural relatives can be a powerful method to infer function, in many cases it has been shown that a small number of residues within a protein's active site or binding pocket are critical for biological activity and such residues may only appear to be conserved through structural analysis (2,3). Structural biology faces the task of characterizing the shapes and dynamics of the entire protein repertoire of whole genomes in order to facilitate an understanding of biochemical functions and their mechanisms of action within the cell. However, with the ever-growing disparity between the number of known sequences and known structures, the need to structurally and functionally annotate sequence space appears more pressing than ever. Structural genomics projects were instigated to address this issue through the large-scale determination of protein 3D structure (4–8). To solve a structure for each genome sequence would be experimentally, practically and financially prohibitive (9). Rather, many structural genomics initiatives aim to fill in areas of fold space and in doing so, provide structures that will cover surrounding sequence space by acting as a structural template for comparative modelling and fold recognition (1,10,11). Increasing the coverage of structure annotations will reveal new insights between protein sequence, structure and function, which in turn will expedite our understanding of protein function on the molecular level and improve the methods by which we can automatically provide structure-guided functional annotations to new protein structures (12–15).
A variety of structural genomics initiatives are in progress around the world, including the United States, where the Protein Structure Initiative (PSI) funded by the National Institute for General Medical Sciences (NIGMS) under the National Institute of Health (NIH) began its pilot phase in 2000 (16,17). Among its principal aims was the development of bioinformatics-based target selection and monitoring strategies that were able to meet the demands of the large amounts of data required for high-throughput genome-scale structure determination (18,19). Traditional biology has now been solving protein structures for several decades; however, without a ‘global target plan’, solved structures tend to represent the interests of individual researchers, rather than specifically aiming to enrich our knowledge of structure space. Furthermore, a single structure is often solved more than once, bound to different ligands or with a range of amino acid substitutions. While these studies are fundamental to molecular biology, such endeavours would be considered to be redundant under the guise of many structural genomics projects. In order to map protein structure space more efficiently, most structural genomics groups apply a target selection strategy that increases the likelihood that a new structure will exhibit a novel fold or provide a new homologous superfamily in a previously observed fold group. Accordingly, a central step in target selection is the use of comparative sequence analysis to identify and exclude sequences that have a relative of known structure in the Protein Data Bank (PDB) (20). However, there is no guarantee that all the remaining target sequences will be amenable to high-throughput analysis—the high attrition rate of target proteins in high-throughput structural genomics pipelines has been well documented [e.g. (21,22)]. Many target selection protocols have attempted to reduce the number of these difficult proteins by excluding or truncating sequences that are predicted to contain regions of low-complexity, coiled-coils, and transmembrane helices.
Increasingly, target selection is concerned with the organization of genome sequences into protein families (17,23–27). These families can then be prioritized according to a range of properties, such as size, taxonomic distribution and suitability of family representatives for structure determination, directing efforts towards well-considered regions of sequence space. Although these principles of target selection are widely employed in the structural genomics community, a varied array of target selection strategies have also been developed to meet the particular requirements of different initiatives. Such considerations have included the prioritization of representatives from large families or the identification of ORFan sequences for which little is known in terms of their origin and function. Proteins have also been targeted according to their species distribution, which may correlate to their general function, e.g. proteins found in all three super kingdoms of life or those found only in single pathogenic organisms.
Recently, Chandonia and Brenner (27) proposed the Pfam5000 target selection strategy which aims to provide a roadmap for coordinated target selection in the second phase of the PSI. This approach aims to guide the selection of a manageable number of target proteins from a list of the largest 5000 Pfam families, many of which lack a member of known structure. By solving structures, such that the top 5000 largest Pfam families have at least one structural representative, it was shown that at least 1-fold assignment could be achieved for 68 and 61% of all prokaryotic and eukaryotic proteins, respectively.
A diverse range of methods have been applied to the problem of automatically clustering large collections of protein sequences, such as Swiss-Prot/TrEMBL (28), into families. Such methods include ProDom (29), DIVCLUS (30), ProtoNet (31), GeneRAGE (32), SYSTERS (33), ADDA (34) and CHOP (35). These methods first aim to define domain regions in protein sequences which are then clustered based on some measure of relatedness shared between domain sequences. The TribeMCL algorithm, developed by Enright and co-workers (36), aims to assign complete protein sequences into families that correlate closely with overall domain architecture. This family assignment protocol has been used to create the Gene3D database (37,38), which has been used extensively in the target selection pipeline used by the Midwest Center for Structural Genomics (MCSG). Resources such as 3D-GENOMICS (39) and SUPERFAMILY (40) aim to provide domain annotations to genome sequences using sensitive sequence profile methods such as PSI-BLAST (41) and hidden Markov models (HMMs) (42). Similarly, sequences in the Gene3D database are annotated at the domain level, using a HMM library of CATH and Pfam domains, which enables us to provide a wide variety of whole-gene and domain level protein family assignments.
Many families of protein domains, particularly in the case of large families, display divergence in their molecular function (43). It has been proposed that a future direction for target selection in second phase of the protein structure initiative (PSI2) should include fine grain targets, selected from large families of proteins in order to provide a more thorough coverage of functional space as it relates to protein structure. The level of granularity required for the selection of additional targets must consider the number of sequences that can be computationally modelled with ‘useful’ accuracy from each solved structure, based on the medical and biological relevance of individual protein families. It is generally accepted that sequences sharing 30% or more sequence identity are likely to share a similar fold (44) and accordingly, to confidently construct models of reasonable accuracy, at least 30% sequence identity must be shared between the sequence to be modelled and the structural template (11).
Here, we present an analysis of sequences from 203 completed genomes clustered in the Gene3D resource. We analyse the growth in new families together with the distributions of singleton sequences as the number of available completed genomes has increased. Over 90% of the genomes sequences can be provided with CATH, Pfam, transmembrane-helix, coiled-coil, low complexity or N-terminal signal peptide annotations. The mapping of CATH and Pfam domains to the sequences held in Gene3D has enabled us to measure genome coverage provided by the largest CATH and Pfam domain families, where we aim to calculate coverage on the basis of domain sequences, rather than whole gene sequences. We find that many of the remaining unannotated sequence regions appear domain-like in length and belong to a large number of small families that tend to be species specific. The comprehensive domain annotation of a large number of genomes has also enabled us to calculate more accurate estimates of the number of multi-domain proteins in eukaryotes and prokaryotes than previous studies. We also show that while over 70% of domain sequences in 203 completed genomes fall into just 2000 CATH and Pfam domain families, the number of structures that would be required to provide useful homology models for these sequences approaches 90 000, clearly an unachievable demand for structural genomics. With this in mind it will be important to develop and improve methods to identify shifts in function across large superfamilies in order to suggest additional relatives for structure determination.
MATERIALS AND METHODS
Gene3D database
The protein families in the Gene3D database are built using the TribeMCL algorithm developed by Enright and co-workers (36). This method applies the Markov cluster algorithm (45) (http://www.library.uu.nl/digiarchief/dip/diss/1895620/inhoud.htm) that simulates flow in a protein similarity graph and assigns complete protein sequences into families based on the density and strengths of links between them. BLAST sequence comparison is used to identify links between the completed genome sequences using an E-value cutoff of 0.001 (38).
Since Gene3D was first developed many more completed genomes have become available while other genomes have been revised. Completed genomes are downloaded from Integr8 (http://www.ebi.ac.uk/integr8/). A new protocol has been developed to update the Gene3D families and provide methods for maintaining the resource. Novel sequences from the updated or newly released genomes are scanned against sequences already held in Gene3D using BLAST. An 80% overlap cutoff is used to select only those matches representing whole chain matches. New sequences are then assigned into the best hit family for each of the new sequences using inCluster (in-house program). If a sequence cannot be assigned to an existing Gene3D family, a new family is created.
Where possible, CATH and Pfam domains are assigned to the genome sequences in Gene3D using Markov models (HMMs). Domain family assignments are made by scanning whole sequences against CATH and Pfam HMM libraries, built using the SAM-T99 protocol developed by Karplus et al. (46) where the fw0.5 script is used as recommended in the SAM-T99 documentation. The domain assignments held in the current release of Gene3D correspond to CATH version 2.6, represented by 4596 models, one for each close sequence family, and Pfam version 18, represented by 7677 models. The model libraries are scanned using hmmscore and domain matches are accepted and processed using the DomainFinder algorithm (47).
Prediction methods
The Memsat program (48) was used to identify transmembrane helices using default thresholds. We used the COILS2 algorithm (49) to identify coiled-coil regions, using a probability cutoff of 0.9 and a window size of 28 residues and the SEG program (50) with default parameters to identify regions of low complexity. The SignalP (51) program was used to predict the presence of signal peptides. SignalP was run with default parameters according to the organism type (gram-positive gram-negative, eukaryotic). SignalP v2.0 runs two prediction methods; a Neural Net (NN) based method and an HMM-based method. A NN prediction was assigned as ‘true’ when both the Y-max score and the S-mean score were above the threshold values. In cases where S-mean is above the threshold, while Y-max is not, it is possible that the protein may contain some sort of membrane anchor (as this can give a high S-mean score if the membrane anchor has a hydrophobic N-terminus). HMM signal peptide predictions were assigned when both the HMM C-max and S-prob are above their threshold values. The NN method tends to assign more accurate cleavage positions for prokaryotic sequences than the HMM method. Correspondingly, in cases where both the NN and HMM method assign a signal peptide, the cleavage position was taken from the NN prediction.
Greedy coverage
A greedy coverage algorithm was run on sequence relatives assigned to CATH, Pfam and NewFam families as follows: Links between family members were assigned, using an implementation of the Needleman and Wunsch global alignment sequence comparison algorithm, in cases where sequence identity and overlap were found to be ≥30% and ≥80%, respectively. The sequence (representing a possible homology modelling template) with the highest number of links is first selected, and removed, along with all those sequences to which it is linked, from further calculations. This step is repeated until no sequences are left in the family.
Calculating homology modelling coverage of CATH superfamilies
The homology modelling coverage of sequence relatives assigned to the top 20 largest CATH domain superfamilies was estimated using a non-identical (N-rep) set of superfamily-specific HMMs taken from CATH version 2.6. The HMMs were constructed using the SAM-T99 software as described above. Here, we calculated homology modelling coverage at the domain level, since the modular nature of many domains means they are often found in different contexts within larger multi-domain structures. As a consequence, we may slightly underestimate our coverage, as the CATH domain database tends to fall behind the PDB in terms of completeness, due to the manual stages that are involved in its curation. Each CATH N-rep HMM was scanned against sequence relatives assigned to its corresponding superfamily and ‘modellable’ sequence regions were assigned where an overlap ≥80% and sequence identity of ≥30% existed between domain template and match sequence. Though the approach taken to calculate homology modelling coverage did not necessitate the identification of remote homologues, SAM-T99 sequence comparison attempts to address the inherent difficulty in identifying sequence relatives of discontinuous domains (46).
RESULTS
Analysis of protein families and singletons in Gene3D
Entire genomes are now sequenced and released on an increasingly frequent basis and in response, resources such as Gene3D must be updated on a regular basis in order to compete with the expansion in sequence data. At the time of writing the Gene3D database currently holds 203 completed genomes; 170 bacteria, 16 archaea and 17 eukaryotes, which are represented by 633 546 non-identical sequences. Whole gene clustering of these sequences formed 51 778 Gene3D families containing two or more sequence members while 158 798 sequences could not be assigned to a Gene3D family. These sequences are described as Gene3D-singletons, i.e. whole gene sequences for which no related sequence can be detected by the TribeMCL algorithm used to construct the Gene3D families.
As the number of clustered genomes increases in the Gene3D database, we are able to assess the rate of discovery of new Gene3D families over time. A previous analysis by Kunin and co-workers (53) demonstrated a constant rate in the discovery of new protein families as the genomes of 80 eukaryotic and prokaryotic organisms were released. This observed trend was contrary to some expectations that the rate of discovery of new protein families would slow as sequence space became better sampled with the increasing number of completed genomes (24,53).
Here, we repeat this analysis for genomes clustered into protein families in Gene3D. We analysed each genome in Gene3D in order of their date of release (as listed on the GOLD website http://www.genomesonline.org) to calculate the rate of increase in new families as assigned by the Gene3D protocol. Where more than one strain of a genome was published, we considered only one strain, reducing the number of genomes analysed from 203 to 169. Despite the fact that over 100 additional genomes were included compared with the analysis by Kunin and co-workers, it appears that, as yet, the increase in the discovery of new families is still relatively constant.
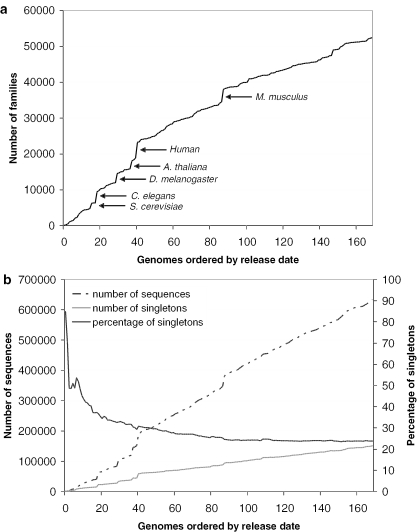
Figure 1a shows the number of Gene3D families after addition of each newly released genome. A steady increase in Gene3D families can be observed as each of the 169 genomes is added to the database, (correlation coefficient with respect to the order of released genomes is R2 > 0.95). The addition of the eukaryotic genomes (the largest of which are indicated in Figure 1a) results in sizeable increases in the number of new Gene3D families; however, analysis of the prokaryotic genomes alone reveals that the rate of discovery of new families in bacteria and archaea is as high (R2 > 0.99) concurring with the observation by Kunin et al. (53) that the sequence diversity across the prokaryotes is as pronounced as it is in the few eukaryotic genomes that have so far been sequenced.
Figure 1.
(a) The accumulation of Gene3D protein families over time, represented by the order in which newly completed genomes have been released. Some of the largest eukaryotic genomes are labelled, each of which contributes a large number of new Gene3D families. (b) With the release of each new genome, the number of singleton sequences is increasing in the Gene3D database, while the overall percentage of sequences assigned as singletons is gradually decreasing.
The continued growth in protein families suggests that sequence space is more diverse than might have initially been thought, with new and distinct protein families appearing in new genomes and taxonomic groups. The additional observation that many genome sequences cannot be assigned to either an existing or new protein families is also suggestive of this diversity. Though it has been argued that some singletons may correspond to highly divergent sequences that we are unable to assign to known families (53), increasing evidence has indicated that many singletons represent sequences unique to each organism representing single member families (52,54–55). Even in cases where structure genomics efforts are largely directed towards the coverage of large families, the structural characterization of singleton sequences will still remain of significant interest, as it is likely that many singleton targets will correspond to real proteins with novel functions or distant homologues of existing families that can only be identified through structure comparison (56).
Earlier work by Siew and Fischer (57) analysed the changes in the distribution of singletons over time as the sequences from 60 completed genomes were released. In a similar analysis, we have calculated the total number of sequences assigned as singletons in Gene3D as each newly released genome has been clustered into the database. The corresponding percentage of singletons, as a fraction of the overall number of sequences in the growing database, has also been calculated. If a sequence that was previously described as a singleton was assigned to a new Gene3D family on addition of a new genome, its status was changed to non-singleton.
As genomes are added to the database (Figure 1b) there is a gradual increase in the total number of singleton sequences. While there are new matches to old singletons, forming new Gene3D families, their number do not cancel out the addition of new singletons provided by each new genome. However, the percentage of all singletons is gradually declining, having fallen to ∼24% of all sequences in the protein family database. The number of singletons initially found in each genome and subsequently reassigned to new families on addition of further genomes is likely to be related to the taxonomic distance between genomes added to the Gene3D database. It is of note that while there are three times as many genomes in our analysis, our observations describe a continuation in the trends observed by Siew and Fischer (57) using the sequences of 60 completed genomes. The steady growth in singleton sequences, rather than a general reduced presence in the more recently sequenced organisms, suggests that these novel sequences form a fundamental part of the genomes, with recent analysis showing that many of these singletons represent real, functional and sometimes essential proteins (55).
Assignment of CATH and Pfam domain annotations
In addition to the division of sequences into gene families, Gene3D aims to determine the domain architecture (domain content and their order in sequence), of each member sequence. In the current version of Gene3D, we search a library of HMMs relating to 1572 CATH version 2.6 (58) and 7677 Pfam version 18 (59) families against the sequences of the completed genomes using the SAM protocol (46). These well established domain databases provide a comprehensive set of domain families whose members are classified using automated protocols together with a considerable amount of manual validation.
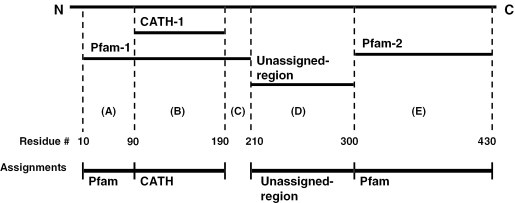
It is necessary to take a hierarchical approach to domain assignment in cases where CATH and Pfam domain annotations are found to overlap. Such conflicts were resolved by calculating a hybrid of CATH and Pfam assignments. CATH domain matches are given priority over Pfam domain matches since domains in the CATH database are identified from both sequence and structure, which is generally considered to be a more reliable approach for protein domain delineation than their identification from sequence. The general approach is illustrated in Figure 2, and described in more detail in the corresponding legend. We found that over 80% of the CATH domain families overlap a Pfam domain assignment by over 90% of their length (just under 10% having no Pfam counterpart). A total of 26% of Pfam domain families overlapped 90% or more residues in a corresponding CATH assignment, while 60% of Pfam domain families showed little or no overlap with a CATH domain mapping.
Figure 2.
Assignment of CATH, Pfam and unassigned-regions to Gene3D sequences. A hierarchical scheme is used, where CATH domains are first assigned, followed by non-overlapping Pfam domain assignments. In this example, a single CATH domain and two Pfam domains have matched a single sequence. CATH-1 and Pfam-1 overlap, with Pfam-1 extending beyond the CATH domain by 80 residues towards the N-terminus and 20 residues towards the C-terminus. According to the hierarchy, the CATH assignment (region B) is given priority and therefore assigned. Pfam-1 is then considered. When assigning domains we apply a conservative length cut-off of ≥50 residues. Consequently residues 10–89 corresponding to a fragment of Pfam-1 (region A), are assigned as a Pfam-1-subdomain whereas residues 191–201 (region C) are not. There is no such clash for Pfam-2 (region E) and this assignment is accepted. Residues 211–300 (region D) cannot be assigned to a CATH or Pfam family, yet form a sequence fragment of ≥50 consecutive residues. We refer to such sequences as ‘unassigned-regions’.
Annotation coverage of genome sequences
In addition to the assignment of CATH and Pfam domains, a series of prediction-based sequence annotations were also made to each sequence; transmembrane helices are predicted using the MEMSAT algorithm (48), coiled-coils using COILS2 (50), regions of low complexity by the SEG program (50) and N-terminal signal peptides by SignalP (51). The annotation coverage of the non-redundant set of sequences from the 203 genomes in Gene3D is tabulated in Table 1. Coverage is calculated on both a whole-sequence and residue basis with the contribution of each feature type given cumulatively and non-cumulatively. Cumulative totals are calculated on a hierarchical basis as ordered in column 1.
Table 1.
Annotation coverage of 663 546 sequences from 203 completed genomes in Gene3D
| Annotation | Coverage per sequence | Coverage per residue | ||||||
|---|---|---|---|---|---|---|---|---|
| Non-cumulative | Cumulative | Non-cumulative | Cumulative | |||||
| No. of sequences | Percentage of sequences | No. of sequences | Percentage of sequences | No. of residues | Percentage of residues | No. of residues | Percentage of residues | |
| CATH | 317 055 | 47.8 | 317 055 | 47.8 | 78 877 234 | 33.3 | 78 877 234 | 33.3 |
| Pfam | 462 820 | 69.7 | 170 297 | 25.7 | 109 103 870 | 46.1 | 52 509 661 | 22.2 |
| Transmembrane helicesa | 175 096 | 26.4 | 34 119 | 5.1 | 16 474 777 | 7.0 | 5 385 960 | 2.3 |
| Coiled-coil | 38 038 | 5.7 | 10 207 | 1.5 | 2 451 346 | 1.0 | 1 796 539 | 0.8 |
| Regions of low complexity | 404 232 | 60.9 | 60 684 | 9.1 | 17 043 439 | 7.2 | 9 406 682 | 4.0 |
| N-terminal signal peptide | 72 261 | 10.9 | 5894 | 0.9 | 2 053 702 | 0.9 | 998 697 | 0.4 |
| Total | — | — | 598 256 | 90.2 | — | — | 148 974 773 | 62.9 |
Coverage is calculated as a percentage of sequences and percentage of residues. Values are also shown on a non-cumulative basis, showing the relative contribution of each annotation type, and cumulatively, calculated on a hierarchical basis, as ordered in column 1. See Materials and Methods for details of the prediction methods used.
aTransmembrane helices were only considered in cases where two or more were assigned to a sequence.
The contribution of a particular feature, as a function of sequences, gives a higher value than the per residue counterpart. For example, while over 26% of sequences from all genomes were predicted to contain two or more transmembrane helices, this corresponds to 7% of residues in the 203 completed genomes. Over 90% of all sequences, corresponding to nearly 63% of residues, can be assigned one or more feature types. CATH domain annotations can be provided for 47.8% of sequences (33.3% of residues) with just under 70% of sequences (46.1% of residues) having one or more matches to a Pfam domain family. The higher level of domain annotation by Pfam is expected as the CATH domain database classifies only sequences of known structure. Assigning CATH domains first, and subsequently counting only non-overlapping domain-like Pfam annotations, provides 76.5% of sequences (55.5% of residues) with a CATH and/or Pfam domain assignment.
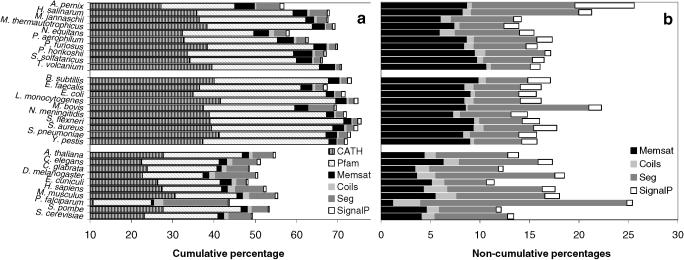
Figure 3a and b shows annotation coverage for 30 randomly selected genomes in the Gene3D database, 10 from each kingdom of life. In this case, values are expressed on a per residue basis, as this may be considered to give a more meaningful measure of coverage.
Figure 3.
Residue annotations of 30 representative completed genomes; 10 archaeal (top), 10 bacterial (middle) and 10 eukaryotic (bottom). (a) Cumulative residue coverage. (b) Non-cumulative residue coverage. The contribution of each type of annotation is shown, to account for the fact that a given residue may be annotated by more than one prediction method. Accordingly, the totals may not reflect the percentage of annotated residues.
The reliability of membrane-helix prediction methods permits us to make some general observations on the distribution of multi-spanning membrane proteins. As reported in Table 1, 26.4% of the proteins encoded by the 203 completed genomes are predicted to contain transmembrane helices, in agreement with previous estimates (60–62). Comparable distributions of membrane proteins were also observed across the prokaryotic and eukaryotic genomes with respective averages of 26.5 and 24.7%. A series of studies have previously demonstrated a roughly linear relationship between the number open reading frames (ORFs) in each genome and the number of helical membrane proteins (60,62). A similar effect was observed in our analysis—it is notable however that although a similar percentage of ORFs appear to encode membrane proteins across the single and multi-cellular genomes, the overall fraction of residues assigned to membrane-helices (as a percentage of all residues in a genome) appears lower for the eukaryotic genomes, see Figure 3. This might in part be an indication that many membrane-spanning proteins in the eukaryotes have an additional series of globular domains.
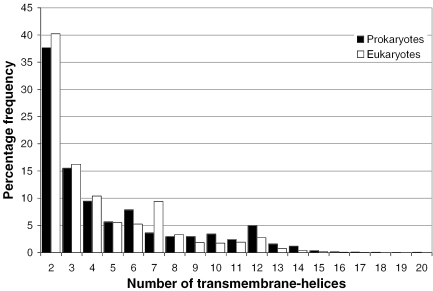
Figure 4 shows the distribution of membrane spanning helices across proteins encoded by prokaryotic or eukaryotic genomes. As described in Materials and Methods, proteins predicted to contain a single membrane helix are treated as a separate class in our target selection pipeline, as these may well be membrane-anchored globular domains. While the number of membrane proteins falls off rapidly with an increasing number of spanning helices, a number of exceptions are apparent; eukaryotic proteins have a greater representation of 7-transmembrane-helices (G-protein coupled receptors), prokaryotes have a greater fraction of 6, 10 and 12-transmembrane-helical proteins (permease and transporter-like proteins). Similar biases have been reported before, despite the use of different transmembrane prediction algorithms and smaller genome datasets (25,60–63).
Figure 4.
The distribution of the number of membrane spanning helices (n) in prokaryotic and eukaryotic proteins. The percentage frequency of whole-gene sequences predicted to contain n membrane helices is shown on the y-axis. Eukaryotes appear to have a greater number of 7-transmembrane-helices (7-TM-helices), while prokaryotes tend to require a higher fraction of 6, 10 and 12-TM-helical proteins.
Coiled-coils were predicted in 5.7% of the sequences across all genomes. Eukaryotic proteins tend to have, on average, a larger fraction of proteins with coiled-coil regions, with 10.6% of eukaryotic sequences predicted to contain coiled-coils compared with 3.8% of the prokaryotic sequences. The vast majority of coiled-coil regions were over 28 residues in length. Fewer than 11% of sequences were assigned an N-terminal signal peptide by the SignalP algorithm, though a number of genomes contained a significantly higher number of secreted proteins, including Aeropyrum pernix. Altogether, 69% of sequences (458 228 of 663 546) were predicted to contain one or more regions corresponding to transmembrane helices, coiled-coils, regions of low complexity or N-terminal signal peptide predictions.
Clustering and filtering the unassigned-regions
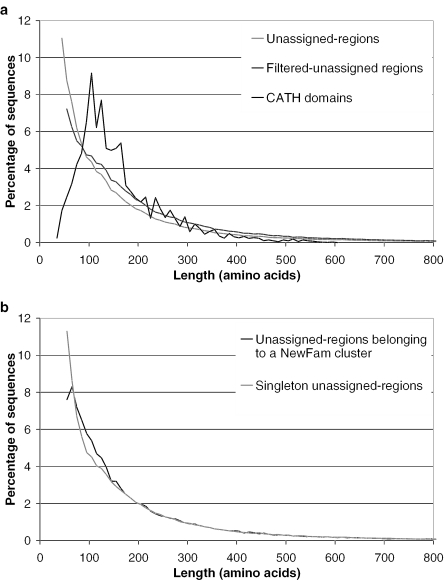
As described above, the mapping of CATH and Pfam domains to the genome sequences leaves a subset of sequences for which a domain family assignment cannot be provided, even by sensitive sequence comparison algorithms, such as HMMs. This subset is populated by whole sequences lacking CATH or Pfam domain assignments, or partial sequences that remain after CATH and/or Pfam domain assignments have been calculated. The length distribution of these unassigned-regions is shown in Figure 5a, (grey line) and can be compared with the length distribution of domains assigned in the CATH domain database (black line). A significant proportion of unassigned-region sequences appear domain-like in their length when compared with the length distribution of domains assigned in the CATH database, though it is also notable that nearly 7% of the unassigned sequence regions are greater than 450 residues in length (roughly three times the CATH domain length average) and are likely to represent many of the whole sequences to which no CATH or Pfam domain assignments could be made.
Figure 5.
(a) The length distribution of unassigned regions in Gene3D sequences compared with domains in the CATH domain database. (b) Distribution of lengths for sequences belonging to NewFam clusters compared to NewFam singleton sequences.
We filtered the unassigned regions by including only those sequences that contained 50 or more consecutive residues free from any transmembrane helix, coiled-coil, low complexity or signal peptide annotation, a similar approach has previously been described by Liu and Rost (25). Such sequences (referred to as filtered-unassigned-regions) might be expected to have an increased likelihood of forming a globular state on folding and as such are more desirable for structure determination. This filtering reduced the number of shorter unassigned-regions, Figure 5a dark-grey line, with an overall reduction of the unassigned sequences from 468 860 to 371 059, with nearly 30% of residues in the unassigned-regions assigned to one of the feature types. The mean sequence length for domains in CATH is just above 160 residues, while the unassigned-regions and filtered unassigned-regions have mean lengths of ∼200 and 220 residues, respectively. A large proportion of the unassigned regions have a similar length distribution to single domain proteins, but without further analysis (such as more rigorous domain boundary identification), we cannot be certain that all unassigned regions are single domain. Nevertheless, for simplicity we approximate each unassigned region as a domain. As such, hereinafter we describe all CATH, Pfam and unassigned sequence assignments as domain sequences.
Once identified, all 468 860 unassigned-regions were clustered using the TribeMCL algorithm (see methods) to generate 56 854 families, described as NewFam clusters, containing two or more members (corresponding to 204 414 sequences) with 256 446 domain sequences remaining unclustered, described as NewFam singletons. The largest NewFam cluster contained 366 sequences. The length distribution of the NewFam members and singletons is shown in Figure 5b, where it is apparent that many of the shortest unassigned-regions remained unclustered by the TribeMCL algorithm.
CATH, Pfam and NewFam coverage of the genome sequences
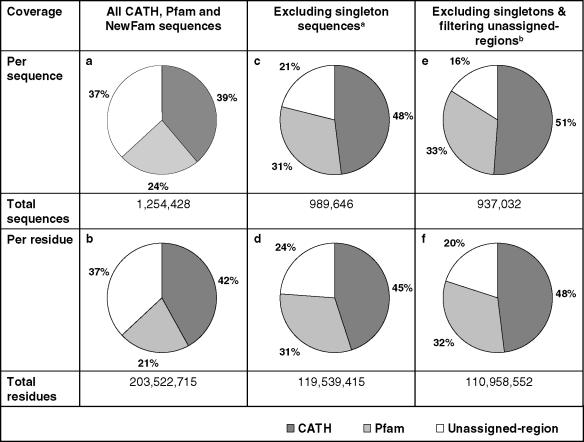
Generating NewFam clusters enabled the calculation of CATH, Pfam and NewFam coverage of sequences from the 203 completed genomes, summarized in Table 2. Coverage is calculated as the percentage of all domain sequences, rather than whole sequences. Table 2a shows that 39 and 24% of sequence fragments are assigned to CATH and Pfam domains, respectively, with per residue coverage illustrated in Table 2b. Table 2c gives the CATH and Pfam domain coverage as a percentage of non-singleton sequence fragments; that is CATH, Pfam and NewFam singletons are excluded from this calculation. The targeting of such domain sequences that cannot be assigned to a family is given a low priority in family based target selection strategies. As expected, an increase in coverage of both CATH and Pfam domains is seen, with 48 and 31% of sequence fragments being assigned to their respective CATH and Pfam families. Finally, Table 2e and f shows the per sequence and residue assignment with the exclusion of singleton sequences and using filtered-unassigned-regions. An increase in the percentage in CATH and Pfam coverage is seen, in accordance with the decrease in the total number of domain-like sequences, with 84% of the domain sequences covered by CATH and non-overlapping Pfam domain families.
Table 2.
Coverage of CATH and Pfam domain assignments
aSingletons are defined as those sequences belonging to single member CATH, Pfam or NewFam families.
bThe unassigned-regions were filtered by including only those sequences containing 50 or more consecutive residues devoid of any transmembrane-helix, coiled-coil or low complexity annotations.
Frequency of multi-domain proteins in the three kingdoms of life
Using the extensive domain annotations described above, we have attempted to estimate the number of number of multi-domain proteins in the three kingdoms of life. Previous studies (60,64) suggested that ∼80% of proteins in the eukaryotes and 65% of proteins in prokaryotes contain more than one domain. Work by Harrison et al. (65) estimated that almost 66% of eukaryotic genome sequences form multi-domain proteins. However, these measurements were extrapolated from a few genomes based only on the subset of proteins for which domain assignments could be provided.
In our analysis, we first identified sequences fully covered by a CATH or Pfam domain assignment, which we described as single-domain, and those sequences assigned a combination of two or more CATH, PFam and NewFam domains, which we described as multi-domain. For this calculation, we also attempted to further subdivide the remaining sequences for which no CATH or Pfam assignment could be made, as some of these proteins may contain more than one domain. The mean length and corresponding standard deviation of the single-domain proteins found in each individual genome was calculated to provide a genome-specific length threshold (mean length plus 1 standard deviation) that was used to subdivide, where possible, the whole-gene unannotated sequences. Though a relatively simple approach, this method was considered to achieve a better overall estimate of the number of multi-domain sequences, as protein domains have been shown to follow relatively narrow length distributions (66,67). A significant majority of whole-gene unassigned-regions were assigned as single-domain proteins.
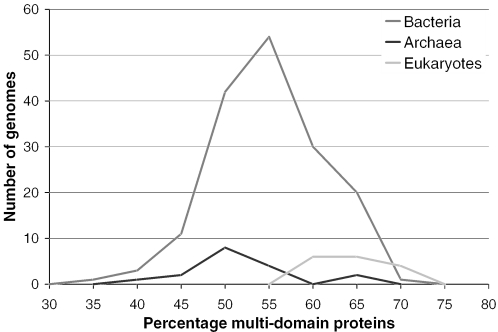
We estimate that 67% of eukaryotic proteins are multi-domain with 54 and 57% of archaeal and bacterial sequences forming multi-domain proteins. Interestingly, we also found that the percentage of multi-domain proteins within individual eukaryotic genomes is relatively unvaried compared to bacteria, see Figure 6. In fact, our estimates show that the number of multi-domain proteins in bacterial genomes vary widely, ranging from ∼32% of sequences to almost 75% (as high as some eukaryotes). It also appears likely that the distribution of multi-domain proteins in archaeal genomes would follow a similar distribution to that of the bacteria if a similar number of archaeal genomes were available for analysis, (i.e. hundreds rather than tens). Though a similar discrepancy in sample size also exists between bacterial and eukaryotic genomes, the differences in average percentages of multi-domain proteins between eukaryotes and prokaryotes appear significant. Recently, work by Bjorklund et al. (68) calculated that 65% of proteins in eukaryotes and 40% of proteins in prokaryotes are multi-domain. However, their lower numbers are likely to be attributable to the lower minimum domain length cutoff they use (100 residues) and also the use of 21 genomes in their analysis compared to the 203 used in this study. Their results are very similar—though possibly slightly lower—when they use, in our view, a more reasonable sequence length cutoff of 50 residues, as applied in this analysis.
Figure 6.
Percentage of multi-domain protein sequences in each of 203 completed genomes from the three kingdoms of life.
Identification of domain families for structure determination
It is quite clear that structural genomics cannot support the experimental determination of all proteins, and so it is hoped that by solving a few thousand well chosen structures we can go someway towards filling in the remaining structure space by computational homology modelling (1). Structural genomics must therefore sample a broad range of sequence families in order to optimize the number of sequences that can be modelled. Here, we aim to deduce the number of experimental structures that are required to provide structural annotation to the genome sequences. Coverage is calculated on a domain sequence and residue basis.
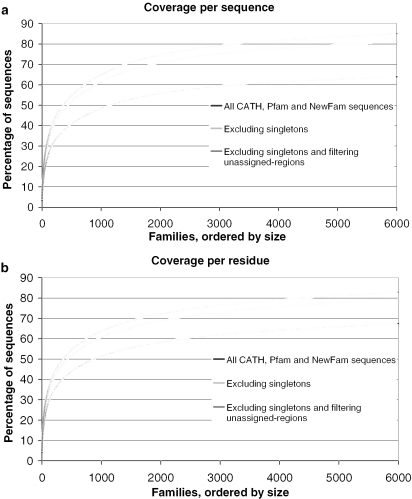
The coverage of space, as represented by the 203 completed genomes, in CATH, Pfam and NewFam domain families is shown in Figure 7a (per domain sequence) and Figure 7B (per residue), where domain families are ordered according to size on the x-axis, with the largest first. The exclusion of singletons from the coverage calculations suggests that if the top 2000 largest domain families include at least one member of known structure (with 1328 already doing so), we can cover ∼70% of domain family members (including unassigned-regions belonging to NewFam clusters). If only prokaryotic targets are used, the coverage of the correspondingly largest 2000 families only falls to 67% of the domain sequences. Alternatively, ensuring that the 2000 largest domain families found in the Human genome have at least one member of known structure would provide coverage for 57% of all domain family members in the Human genome.
Figure 7.
Coverage of protein space, represented by 203 complete genomes. CATH, Pfam and NewFam families are ordered by size (x-axis). Coverage is shown on a sequence (a) and residue (b) basis.
Increasing the number of families to 5000 gives a comparatively lower increase of coverage to 80% of all the non-singleton domain sequences assigned in the 203 genomes, Figure 7a. A rapid gain in sequence coverage is initially achieved by targeting the largest families first, an effect that tails off as smaller families are considered. Coverage is also shown for all sequences, including singletons (black line), representing a rather more pessimistic view, with coverage of a little over 55% for the 2000 largest families. Figure 7b shows coverage on a per residue basis, with similar values to the coverage given on a sequence level.
Calculation of coverage on the basis of one structure per domain family provides a lower bound in terms of effort required by the structural genomics initiatives. The domain families represent a ‘coarse-grained’ division of sequence space into broad sequence families containing all relatives sharing a common ancestor. Targeting large families in Gene3D provides greater structural coverage of all proteins but a large proportion of the structural models generated will only be moderately accurate for distant homologues in the family. It does not account for the fact that many domain families will contain considerable structural and functional variation that cannot be resolved by the determination of a single structure. In view of this, it is necessary to calculate the number of protein structures that are required to provide homology models, of reasonable accuracy, for the remainder of sequence space. Homology modelling uses experimentally determined structures (templates) to predict the 3D structure of another protein (target) with a related amino acid sequence. Protein structure can reveal the chemical principles underlying protein function and this structure-derived functional output can be transferred to related sequences. The level to which such annotations can be achieved in homology modelling is generally considered to be dictated by the level of sequence identity that is shared between template and modelled sequence. We assume that at 30% sequence identity or above, reasonably accurate models can be produced to enable structural variations within a given family to be correlated with putative functional variation.
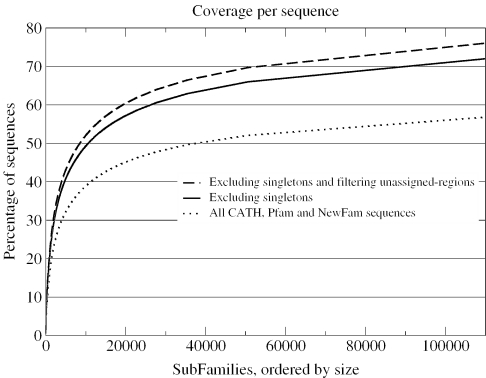
The members of each CATH, Pfam and NewFam family were sub-clustered using the greedy coverage algorithm described by (69). Greedy coverage first identifies the sequence with the largest number of relatives above a given threshold, in this case 30% sequence identity. These sequences are assigned to the first cluster and removed, while the process is repeated iteratively on the remaining sequences until no sequences remain. Such clustering is suitable for the needs of structural genomics, where the prioritization of targets that provide the highest number of homology models as possible is a significant goal of target selection. Figure 8 shows genome coverage on the basis of the subfamilies identified within the domain families. Note that singletons are defined as those sequences belonging to single-member CATH, Pfam and NewFam families rather than single-member subfamily clusters. Excluding such singletons, the number of structures required to provide accurate models (≥30% sequence identity) for 70% of the CATH, Pfam and NewFam sequences is ∼90 000. Clearly, this is significant increase in the number of targets in comparison to the figures presented in Figure 6. Similar analysis, albeit using differing approaches, including those by Vitkup et al. (24), Liu and Rost (62) and Chandonia and Brenner (27) have also demonstrated large-scale requirements for the significant coverage of sequence space at similar modelling densities. Achieving these numbers of structural determinations is clearly not a realistic goal for any structural genomics effort.
Figure 8.
Fine-grained coverage of protein space. We applied a greedy coverage algorithm to calculate the number of structures required to model all members of CATH, Pfam and NewFam families based on a cutoff ≥30% sequence identity. This measure suggests many more structural determinations will be required to enable accurate modelling of a large percentage of protein sequences compared with the numbers suggested by a coarse-grained coverage approach.
A more rational methodology is required that targets additional family members in a manner that will provide broader structural insights in the most functionally diverse domain families. Comparative genome analysis has shown that a large proportion of these families are very large, having expanded significantly during evolution through extensive gene duplication within a genome (70–72). Increasing our knowledge on the diverse functional roles across subfamilies will have a greater medical and biological value than random fine-grained target selection.
Already, many of the largest domain superfamilies in nature are classified in CATH and SCOP (73), since they have at least one relative of known structure. However, for many of these large superfamilies, this structural data can only be extrapolated to a small percentage of the remaining sequences in the superfamily through homology modelling. This can be highlighted by calculating the homology modelling coverage of the twenty most frequently occurring CATH domain superfamilies in the 203 completed genomes, illustrated in Figure 9. Homology modelling coverage was measured at the domain level, using a subset of non-identical superfamily-specific domains from CATH to act as structural templates, using an overlap of 80% and a minimum sequence identity of 30% to be shared between the template and target structure (see Materials and Methods). This may provide a slight underestimate in the fraction of sequences for which a model can be provided, since the CATH database is partially reliant on manual curation, and is not entirely up to date with the PDB.
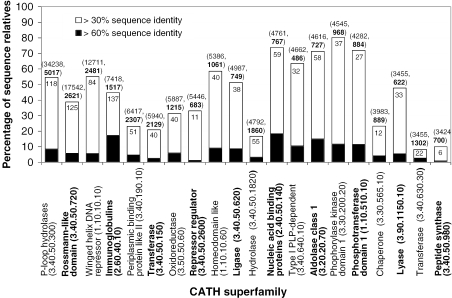
Figure 9.
Homology modelling coverage calculated for sequence relatives in the twenty largest CATH domain families found in 203 completed genomes in Gene3D. Each bar of the histogram represents the percentage of superfamily sequence relatives that can be modelled at relatively high accuracy (≥30% sequence identity shared between target and template). The black regions represent the subset of these sequences that can be modelled at ≥60% sequence identity. The value shown within each histogram bar indicates the number of known unique-domain structures (sharing <30% sequence identity) within each superfamily. The number of sequences relatives assigned to each superfamily, and the number of sequence families they cluster into (at 30% sequence identity) is shown in brackets above each bar. Though these superfamilies contain many relatives of known structure, we are still unable to accurately model many of the sequence relatives, suggesting additional experimentally determined structures are still required to increase our structural understanding of these highly recurrent domain families.
The level of modelling coverage varies across the superfamilies: while as many as 80% of sequence relatives in the Phosphorylase kinase domain 1 superfamily (3.30.200.20) can be modelled based on a sequence identity of 30% or more, structural data are extrapolated to a much smaller percentage of the sequence members in many other of the largest CATH domain families. For example, just 16% of the sequence relatives in the equivalently sized Hydrolase superfamily (3.40.50.1820) can be structurally annotated. This effect is more pronounced when calculating modelling coverage of these large superfamilies above 60% sequence identity. Such a level of sequence identity is required to give higher quality models that are essential for reliable drug design and ligand docking.
A closer examination shows that the level of modelling coverage within each superfamily is tends to be underpinned by the number of unique known structures and sequence diversity (here measured by the number of 30% sequence identity families, Figure 9, values in bold). In general, those superfamilies with a similar number of unique solved structures and a similar level of sequence diversity share comparable levels of modelling coverage. In contrast, a higher sequence diversity in a given superfamily can lead to a lower modelling coverage despite a similar number of known structures. For example, the Hydrolases (3.40.50.1820) and the Aldolase Class-1 superfamily (3.20.20.70) differ greatly in modelling coverage (18 and 69%, respectively) while containing a similar number of unique solved structures (55 and 58). In turn, a lower modelling coverage can occur with a lower number of unique solved structures despite similar levels of sequence diversity, e.g. Peptide synthase (3.40.50.980) and Aldolase Class-1 (3.20.20.70).
Though many CATH or SCOP superfamilies contain relatives of known structure, it is clear that we are still unable to accurately model many of the sequence relatives, suggesting additional experimentally determined structures are still required to increase our structural understanding of these highly recurrent domain families. Therefore, to provide good structural models for a significant proportion of all proteome sequences, it will be essential to target additional sequence subfamilies with no structural relatives in the large superfamilies. Determining structures of sequence-distant members of these highly recurrent superfamilies will in turn have a significant impact on the understanding of their structural and functional evolution and is essential for the continued development of tools that enable us to predict function from structure.
DISCUSSION
In the past few years, we have witnessed a steady increase in the number of structural genomics projects, and though many differ in their aims and approaches, all are united by a common goal to extend the repertoire of structure space. Recent analysis of a number of initiatives shows an encouraging start to these endeavours, while also highlighting the requirement for the continued development and application of rigorous target selection strategies to enable the efficient coverage of protein space (18,74).
In this paper, we have described some of the approaches that have been applied within the MCSG consortium, including the use of the Gene3D database to enable a family-based approach to our selection of target sequences. The latest version of Gene3D provides sequence similarity derived protein family clusters for 203 completed genomes, which in turn have been annotated by structural and sequence domain family assignments. These large-scale CATH and Pfam domain assignments have allowed us to extrapolate the extent to which sequence space can be effectively covered by solving new structures, and how suitable targets might be derived from the huge array of uncharacterized sequences that are available to us.
We have shown that at a coarse-level of granularity, a structural representative for each of the 2000 largest CATH and Pfam families will give a coverage of over 70% of the domain-like sequences found in 203 completed genomes. This level of annotation would require the derivation of roughly 1000 structures, corresponding to Pfam families lacking a member of known structure. It is intended that many of the protein structures solved by the structural genomics initiatives will be used as fold templates to build models for additional proteins related by sequence similarity with structural comparison then facilitating the assignment of function from structures with a characterized function. Even though the continued improvement in homology modelling and fold recognition methods will permit structural annotations to be made at increasingly low levels of sequence similarity, the characterization of one structure per domain family will ultimately leave large areas of sequence space beyond the limits of our modelling pipelines. Consequently, we calculated the number of structures that would be required to provide models for a fine-grained coverage of sequence space, as represented by the 203 genomes in Gene3D. By applying a greedy coverage algorithm to each CATH, Pfam and NewFam family in Gene3D, to simulate a modelling density cutoff of 30% sequence identity, we found considerably more structures (over 90 000) would be required to provide reasonable homology models for 70% of domain-like sequences. With this in mind it is clear that the diversity of structures that can be provided for the reliable modelling of sequence space must be brought into context with the requirements of biological and biomedical sciences to enable fold space to be targeted in an effective way.
The data from this analysis, together with related work by Vitkup (24) and Chandonia and Brenner (27), suggest that the derivation of a target list that includes representatives from the largest domain families can serve as a convenient platform for target selection between different structural genomic projects. This coarse-grained target selection should then work along side a fine-grained approach allowing additional targets to be selected from families with particular biomedical or biochemical importance or families that cover diverse areas of sequence and function space. The derivation of new or additional representative structures for sequence families should result in iterative updates of such target lists to concentrating on the most relevant areas of sequence space.
We are currently investigating the use of simple methods such as sequence clustering, comparison of domain architecture, and functional annotation [using GO (www.geneontology.org), COG (75) and KEGG (76)] to enable the identification of sequences likely to represent structurally uncharacterized function space. For example, the 2000 largest CATH and Pfam domain superfamilies contain 4790 COG functional groups that have no close structural homologue (>30% sequence identity). The more specific annotations and groupings tend to belong within a limited number of broader categories, though there is not a neat hierarchical nesting of all clusters and some overlap between levels is seen. Scoring schemes may therefore be required to prioritize targets according to their membership of different categories of diversity within a given family. This would provide a more directed approach to pick out additional high-value targets within large sequence families, as opposed to a more random approach.
While structural genomics has often been considered to be synonymous with the pursuit of new folds, it is clear that the characterization of structures distantly related to existing domain superfamilies or forming novel superfamilies is of equal importance, because it enables us to gain new viewpoints on the evolution of protein structure and function. In turn, the need for a more comprehensive coverage of particular sequence families from a modelling point of view will be of increasing importance. By aiming to understand the structure and function of each gene product, we can also begin to unravel the huge variety of protein interactions and assemblies that occur within the cellular environment. It is clear that future structure genomics efforts must target proteins that fulfil many of these requirements.
Acknowledgments
The authors thank Steve Brenner and colleagues for discussions on these topics. The authors also wish to thank Tony Lewis, Mark Dibley and Timothy Dallman for valuable comments and discussion. This work was funded by the NIGMS of the NIH through the Midwest Center for Structural Genomics, the Wellcome Trust and the EU funded BioSapiens project.
Conflict of interest statement. None declared.
REFERENCES
- 1.Chance M.R., Bresnick A.R., Burley S.K., Jiang J.S., Lima C.D., Sali A., Almo S.C., Bonanno J.B., Buglino J.A., Boulton S., et al. Structural genomics: a pipeline for providing structures for the biologist. Protein Sci. 2002;11:723–738. doi: 10.1110/ps.4570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Russell R.B. Detection of protein three-dimensional side-chain patterns: new examples of convergent evolution. J. Mol. Biol. 1998;279:1211–1227. doi: 10.1006/jmbi.1998.1844. [DOI] [PubMed] [Google Scholar]
- 3.Kasuya A., Thornton J.M. Three-dimensional structure analysis of PROSITE patterns. J. Mol. Biol. 1999;286:1673–1691. doi: 10.1006/jmbi.1999.2581. [DOI] [PubMed] [Google Scholar]
- 4.Kim S.H. Shining a light on structural genomics. Nature Struct. Biol. 1998;5:643–645. doi: 10.1038/1334. [DOI] [PubMed] [Google Scholar]
- 5.Burley S.K., Almo S.C., Bonanno J.B., Capel M., Chance M.R., Gaasterland T., Lin D., Sali A., Studier F.W., Swaminathan S. Structural genomics: beyond the human genome project. Nature Genet. 1999;23:151–157. doi: 10.1038/13783. [DOI] [PubMed] [Google Scholar]
- 6.Burley S.K. An overview of structural genomics. Nature Struct. Biol. 2000;7:932–934. doi: 10.1038/80697. [DOI] [PubMed] [Google Scholar]
- 7.Brenner S.E. A tour of structural genomics. Nature Rev. Genet. 2001;2:801–809. doi: 10.1038/35093574. [DOI] [PubMed] [Google Scholar]
- 8.Stevens R.C., Yokoyama S., Wilson I.A. Global efforts in structural genomics. Science. 2001;294:89–92. doi: 10.1126/science.1066011. [DOI] [PubMed] [Google Scholar]
- 9.Sali A. 100000 protein structures for the biologist. Nature Struct. Biol. 1998;5:1029–1032. doi: 10.1038/4136. [DOI] [PubMed] [Google Scholar]
- 10.Sanchez R., Pieper U., Melo F., Eswar N., Marti-Renom M.A., Madhusudhan M.S., Mirkovic N., Sali A. Protein structure modelling for structural genomics. Nature Struct. Biol. 2000;7:986–990. doi: 10.1038/80776. [DOI] [PubMed] [Google Scholar]
- 11.Baker D., Sali A. Protein structure prediction and structural genomics. Science. 2001;294:93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 12.Moult J., Melamud E. From fold to function. Curr. Opin. Struct. Biol. 2000;10:384–389. doi: 10.1016/s0959-440x(00)00101-9. [DOI] [PubMed] [Google Scholar]
- 13.Skolnick J., Fetrow J.S., Kolinski A. Structural genomics and its importance for gene function analysis. Nat. Biotechnol. 2000;18:283–287. doi: 10.1038/73723. [DOI] [PubMed] [Google Scholar]
- 14.Teichmann S.A., Murzin A.G., Chothia C. Determination of protein function, evolution and interactions by structural genomics. Curr. Opin. Struct. Biol. 2001;11:354–363. doi: 10.1016/s0959-440x(00)00215-3. [DOI] [PubMed] [Google Scholar]
- 15.Laskowski R.A., Watson J.D., Thornton J.M. From protein structure to biochemical function? J. Struct. Funct. Genomics. 2003;4:167–177. doi: 10.1023/a:1026127927612. [DOI] [PubMed] [Google Scholar]
- 16.Norvell J.C., Machalek A.Z. Structural genomics programs at the US National Institute of General Medical Sciences. Nature Struct. Biol. 2000;7:931. doi: 10.1038/80694. [DOI] [PubMed] [Google Scholar]
- 17.Terwilliger T.C. Structural genomics in North America. Nature Struct. Biol. 2000;7:935–939. doi: 10.1038/80700. [DOI] [PubMed] [Google Scholar]
- 18.O'Toole N., Grabowski M., Otwinowski Z., Minor W., Cygler M. The structural genomics experimental pipeline: insights from global target lists. Proteins. 2004;56:201–210. doi: 10.1002/prot.20060. [DOI] [PubMed] [Google Scholar]
- 19.Bray J.E., Marsden R.L., Rison S.C., Savchenko A., Edwards A.M., Thornton J.M., Orengo C.A. A practical and robust sequence search strategy for structural genomics target selection. Bioinformatics. 2004;20:2288–2295. doi: 10.1093/bioinformatics/bth240. [DOI] [PubMed] [Google Scholar]
- 20.Westbrook J., Feng Z., Chen L., Yang H., Berman H.M. The Protein Data Bank and structural genomics. Nucleic Acids Res. 2003;31:489–491. doi: 10.1093/nar/gkg068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goh C.S., Lan N., Douglas S.M., Wu B., Echols N., Smith A., Milburn D., Montelione G.T., Zhao H., Gerstein M. Mining the structural genomics pipeline: identification of protein properties that affect high-throughput experimental analysis. J. Mol. Biol. 2004;336:115–130. doi: 10.1016/j.jmb.2003.11.053. [DOI] [PubMed] [Google Scholar]
- 22.Canaves J.M., Page R., Wilson I.A., Stevens R.C. Protein biophysical properties that correlate with crystallization success in Thermotoga maritima: maximum clustering strategy for structural genomics. J. Mol. Biol. 2004;344:977–991. doi: 10.1016/j.jmb.2004.09.076. [DOI] [PubMed] [Google Scholar]
- 23.Brenner S.E. Target selection for structural genomics. Nature Struct. Biol. 2000;7:967–969. doi: 10.1038/80747. [DOI] [PubMed] [Google Scholar]
- 24.Vitkup D., Melamud E., Moult J., Sander C. Completeness in structural genomics. Nature Struct. Biol. 2001;8:559–566. doi: 10.1038/88640. [DOI] [PubMed] [Google Scholar]
- 25.Liu J., Rost B. Target space for structural genomics revisited. Bioinformatics. 2002;18:922–933. doi: 10.1093/bioinformatics/18.7.922. [DOI] [PubMed] [Google Scholar]
- 26.Frishman D. Knowledge based selection of targets for structural genomics. Protein Eng. 2002;15:160–183. doi: 10.1093/protein/15.3.169. [DOI] [PubMed] [Google Scholar]
- 27.Chandonia J.M., Brenner S.E. Implications of structural genomics target selection strategies: Pfam5000, whole genome, and random approaches. Proteins. 2005;58:166–179. doi: 10.1002/prot.20298. [DOI] [PubMed] [Google Scholar]
- 28.Boeckmann B., Bairoch A., Apweiler R., Blatter M.C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O'Donovan C., Phan I., et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bru C., Courcelle E., Carrere S., Beausse Y., Dalmar S., Kahn D. The ProDom database of protein domain families: more emphasis on 3D. Nucleic Acids Res. 2005;33:D212–D215. doi: 10.1093/nar/gki034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park J., Teichmann S.A. DIVCLUS: an automatic method in the GEANFAMMER package that finds homologous domains in single- and multi-domain proteins. Bioinformatics. 1998;14:144–150. doi: 10.1093/bioinformatics/14.2.144. [DOI] [PubMed] [Google Scholar]
- 31.Kaplan N., Sasson O., Inbar U., Friedlich M., Fromer M., Fleischer H., Portugaly E., Linial N., Linial M. ProtoNet 4.0: a hierarchical classification of one million protein sequences. Nucleic Acids Res. 2005;33:D216–D218. doi: 10.1093/nar/gki007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Enright A.J., Ouzounis C.A. GeneRAGE: a robust algorithm for sequence clustering and domain detection. Bioinformatics. 2000;16:451–457. doi: 10.1093/bioinformatics/16.5.451. [DOI] [PubMed] [Google Scholar]
- 33.Meinel T., Krause A., Luz H., Vingron M., Staub E. The SYSTERS Protein Family Database in 2005. Nucleic Acids Res. 2005;33:D226–D229. doi: 10.1093/nar/gki030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heger A., Wilton C.A., Sivakumar A., Holm L. A domain database with global coverage of the protein universe. Nucleic Acids Res. 2005;33:D188–D191. doi: 10.1093/nar/gki096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J., Rost B. CHOP proteins into structural domain-like fragments. Proteins. 2004;55:678–688. doi: 10.1002/prot.20095. [DOI] [PubMed] [Google Scholar]
- 36.Enright A.J., Van Dongen S., Ouzounis C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buchan D.W., Shepherd A.J., Lee D., Pearl F.M., Rison S.C., Thornton J.M., Orengo C.A. Gene3D: structural assignment for whole genes and genomes using the CATH domain structure database. Genome Res. 2002;12:503–514. doi: 10.1101/gr.213802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee D., Grant A., Marsden R.L., Orengo C. Identification and distribution of protein families in 120 completed genomes using Gene3D. Proteins. 2005;59:603–615. doi: 10.1002/prot.20409. [DOI] [PubMed] [Google Scholar]
- 39.Fleming K., Muller A., MacCallum R.M., Sternberg M.J. 3D GENOMICS: a database to compare structural and functional annotations of proteins between sequenced genomes. Nucleic Acids Res. 2004;32:D245–D250. doi: 10.1093/nar/gkh064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madera M., Vogel C., Kummerfeld S.K., Chothia C., Gough J. The SUPERFAMILY database in 2004: additions and improvements. Nucleic Acids Res. 2004;32:D235–D239. doi: 10.1093/nar/gkh117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 43.Todd A.E., Orengo C.A., Thornton J.M. Evolution of function in protein superfamilies, from a structural perspective. J. Mol. Biol. 2001;307:1113–1143. doi: 10.1006/jmbi.2001.4513. [DOI] [PubMed] [Google Scholar]
- 44.Sander C., Schneider R. Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins. 1991;9:56–68. doi: 10.1002/prot.340090107. [DOI] [PubMed] [Google Scholar]
- 45.Van Dongen S. University of Utrecht; 2000. Graph clustering by flow simulation. PhD Thesis. [Google Scholar]
- 46.Karplus K., Barrett C., Hughey R. Hidden Markov models (HMMs) for detecting remote protein homologies. Bioinformatics. 1998;14:846–856. doi: 10.1093/bioinformatics/14.10.846. [DOI] [PubMed] [Google Scholar]
- 47.Pearl F.M.G., Lee D., Bray J.E., Buchan D.W., Shepherd A.J., Orengo C.A. The CATH extended protein-family database: providing structural annotations for genome sequences. Protein Sci. 2002;11:233–244. doi: 10.1110/ps.16802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones D.T., Taylor W.R., Thornton J.M. A mutation data matrix for transmembrane proteins. FEBS Lett. 1994;339:269–75. doi: 10.1016/0014-5793(94)80429-x. [DOI] [PubMed] [Google Scholar]
- 49.Lupas A., Van Dyke M., Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 50.Wootton J.C., Federhen S. Analysis of computationally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- 51.Nielsen H., Engelbrecht J., Brunak S., Von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 52.Coulson A.F.W., Moult J. A unifold, mesofold and superfold model of protein fold use. Proteins. 2002;46:61–71. doi: 10.1002/prot.10011. [DOI] [PubMed] [Google Scholar]
- 53.Kunin V., Cases I., Enright A.J., de Lorenzo V., Ouzounis C.A. Myriads of protein families, and still counting. Genome Biol. 2003;4:401–402. doi: 10.1186/gb-2003-4-2-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fischer D., Eisenberg D. Finding families in genomic ORFans. Bioinformatics. 1999;15:759–762. doi: 10.1093/bioinformatics/15.9.759. [DOI] [PubMed] [Google Scholar]
- 55.Siew N., Fischer D. Structural biology sheds light on the puzzle of genomic ORFans. J. Mol. Biol. 2004;342:369–373. doi: 10.1016/j.jmb.2004.06.073. [DOI] [PubMed] [Google Scholar]
- 56.Fischer D. Rational structural genomics: affirmative action for ORFans and the growth in our structural knowledge. Protein Eng. 1999;12:1029–1030. doi: 10.1093/protein/12.12.1029. [DOI] [PubMed] [Google Scholar]
- 57.Siew N., Fischer D. Analysis of singleton ORFans in fully sequenced microbial genomes. Proteins. 2003;53:241–251. doi: 10.1002/prot.10423. [DOI] [PubMed] [Google Scholar]
- 58.Orengo C.A., Michie A.D., Jones S., Jones D.T., Swindells M.B., Thornton J.M. CATH—a hierarchical classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 59.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wallin E., von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jones D.T. Do transmembrane superfolds exist? FEBS Lett. 1998;423:281–285. doi: 10.1016/s0014-5793(98)00095-7. [DOI] [PubMed] [Google Scholar]
- 62.Liu J., Rost B. Comparing function and structure between entire proteomes. Protein Sci. 2001;10:1970–1979. doi: 10.1110/ps.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gerstein M. A structural consensus of genomes: comparing bacterial, eukaryotic and archaeal genomes in terms of protein structure. J. Mol. Biol. 1997;274:562–576. doi: 10.1006/jmbi.1997.1412. [DOI] [PubMed] [Google Scholar]
- 64.Apic G., Gough J., Teichmann S.A. Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J. Mol. Biol. 2001;310:311–325. doi: 10.1006/jmbi.2001.4776. [DOI] [PubMed] [Google Scholar]
- 65.Harrison P.M., Kumar A., Lang N., Snyder M., Gerstein M. A question of size: the eukaryotic proteome and the problems in defining it. Nucleic Acids Res. 2002;30:1083–1090. doi: 10.1093/nar/30.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wheelan S.J., Marchler-Bauer A., Bryant S.H. Domain size distributions can predict domain boundaries. Bioinformatics. 2000;7:613–618. doi: 10.1093/bioinformatics/16.7.613. [DOI] [PubMed] [Google Scholar]
- 67.Marsden R.L., McGuffin L.J., Jones D.T. Rapid protein domain assignment from amino acid sequence using predicted secondary structure. Protein Sci. 2002;12:2814–2824. doi: 10.1110/ps.0209902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bjorklund A.K., Ekman D., Light S., Frey-Skott J., Elofsson A. Domain rearrangements in protein evolution. J. Mol. Biol. 2005;353:911–923. doi: 10.1016/j.jmb.2005.08.067. [DOI] [PubMed] [Google Scholar]
- 69.Hobohm U., Sander C., Scharf M., Schneider R. Selection of representative protein datasets. Protein Sci. 1992;1:409–417. doi: 10.1002/pro.5560010313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koonin E.V., Wolf Y.I., Karev G.P. The structure of the protein universe and genome evolution. Nature. 2002;420:218–223. doi: 10.1038/nature01256. [DOI] [PubMed] [Google Scholar]
- 71.Chothia C., Gough J., Vogel C., Teichmann S.A. Evolution of the protein repertoire. Science. 2003;300:1701–1703. doi: 10.1126/science.1085371. [DOI] [PubMed] [Google Scholar]
- 72.Ranea J.A., Buchan D.W., Thornton J.M., Orengo C.A. Evolution of protein superfamilies and bacterial genome size. J. Mol. Biol. 2004;336:871–887. doi: 10.1016/j.jmb.2003.12.044. [DOI] [PubMed] [Google Scholar]
- 73.Andreeva A., Howorth D., Brenner S.E., Hubbard T.J., Chothia C., Murzin A.G. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Todd A.E., Marsden R.L., Thornton J.M., Orengo C.A. Progress of structural genomics initiatives: an analysis of solved target structures. J. Mol. Biol. 2005;348:1235–1260. doi: 10.1016/j.jmb.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 75.Tatusov R.L., Natale D.A., Garkavtsev I.V., Tatusova T.A., Shankavaram U.T., Rao B.S., Kiryutin B., Galperin M.Y., Fedorova N.D., Koonin E.V. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. The KEGG resources for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]