Abstract
Gre factors enhance the intrinsic endonucleolytic activity of RNA polymerase to rescue arrested transcription complexes and are thought to confer the high fidelity and processivity of RNA synthesis. The Gre factors insert the extended α-helical coiled-coil domains into the RNA polymerase secondary channel to position two invariant acidic residues at the coiled-coil tip near the active site to stabilize the catalytic metal ion. Gfh1, a GreA homolog from Thermus thermophilus, inhibits rather than activates RNA cleavage. Here we report the structure of the T. thermophilus Gfh1 at 2.4 Å resolution revealing a two-domain architecture closely resembling that of GreA. However, the interdomain orientation is strikingly distinct (~162° rotation) between the two proteins. In contrast to GreA, which has two acidic residues on a well fixed self-stabilized α-turn, the tip of the Gfh1 coiled-coil is flexible and contains four acidic residues. This difference is likely the key to the Gre functional diversity, while Gfh1 inhibits exo- and endonucleolytic cleavage, RNA synthesis, and pyrophosphorolysis, GreA enhances only the endonucleolytic cleavage. We propose that Gfh1 acidic residues stabilize the RNA polymerase active center in a catalytically inactive configuration through Mg2+-mediated interactions. The excess of the acidic residues and inherent flexibility of the coiled-coil tip might allow Gfh1 to adjust its activity to structurally distinct substrates, thereby inhibiting diverse catalytic reactions of RNA polymerase.
Bacterial RNA polymerase (RNAP)3 catalyzes four chemical reactions: nucleotide addition and its reverse reaction, pyrophosphorolysis (1), as well as two transcript cleavage reactions, exo- and endonucleolytic digestion of the nascent RNA (2, 3). All these reactions are thought to occur in the same active site (AS) and conform to the universal two Mg2+-mediated mechanism (4).
The RNAP AS is connected to the enzyme surface by the secondary channel, through which diverse regulatory molecules gain access to the AS and modulate transcription (5–11). Among these, the bacterial factors GreA, GreB, and DksA all employ a long α-helical coiled-coil (CC) domain protruding through the secondary channel to target functionally crucial Mg2+ ions near the RNAP AS with the two invariant acidic residues at the CC tips (8–10, 12). Despite this structural similarity, the functions of these proteins are strikingly distinct; the Gre factors directly remodel the RNAP AS to stimulate the endonucleolytic cleavage of the RNA (8, 10, 12), whereas DksA amplifies the activity of the “magic spot” (ppGpp), the regulator of stringent response in bacteria, but has no direct effect on catalysis (9, 13).
The Thermus thermophilus genome does not encode a ortholog of GreB, instead it contains a gene for a unique GreA paralog, Gfh1 (14). Despite a significant sequence conservation including the two mechanistically important acidic residues (Fig. 1A), this Gre-like factor does not stimulate but instead inhibits the intrinsic and GreA-mediated endonucleolytic RNA cleavage (14) as well as the RNA synthesis (15).
FIGURE 1. The Gfh1 structure and comparison with GreA and DksA.
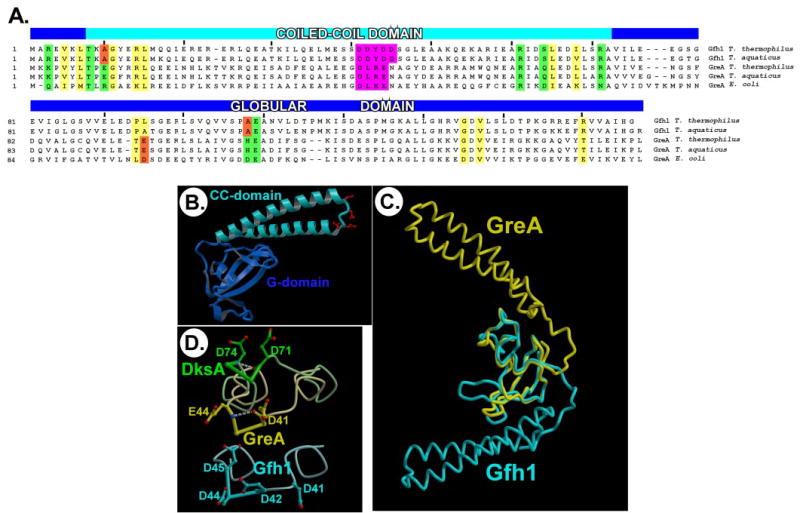
A, alignment of the Gfh1 and GreA sequences. The residues with conserved chemical properties among Gfh1 and GreA sequences that might thus form the Gfh1-like (hydrophobic) and GreA-like (”polar“) interface in both protein families are marked with yellow and green boxes, respectively, whereas the unfavorable substitutions in both Gfh1 and GreA are outlined by red boxes. The potentially functionally crucial segments at the tip of the CC-domains are highlighted with magenta. B, the structure of the Gfh1 protein. The acidic side chains at the CC tip are shown in red. C, the Gfh1 and GreA structures superimposed by the G-domains. D, the CC tips of DksA (green), GreA (yellow), and Gfh1 (cyan). DksA and GreA are superimposed by the CC-domains, and the Gfh1 CC tip is shown in a similar orientation. The hydrogen bonds stabilizing the α-turns in GreA, and DksA are shown as white dashed lines.
To elucidate the molecular mechanism of Gfh1 and the structural features that determine functionality of the CC transcription factors, we have determined the crystal structure of the T. thermophilus Gfh1 at 2.4 Å resolution. The structural and functional analysis suggests that while CC protein regulators appear to utilize similar structural motifs to gain access to the RNAP AS, their dramatically different effects may result from relatively subtle changes in the amino acid composition and conformation of the tip of the CC-domain.
MATERIALS AND METHODS
Crystallization, Structure Determination, and Refinement
The Gfh1 protein was expressed and purified as described (16). The crystals were obtained by a sitting drop vapor diffusion technique followed by macro-seeding (16). The final well crystallization solution contained 3% polyethylene glycol 8000, 33 mm zinc acetate, and 17 mm sodium cacodilate, pH 6.5. The crystals belong to the P43 space group with unit cell dimensions a = b = 59.3, c = 218.9 Å and possess perfect merohedral twinning with the twinning operator {h,-k,-l}. The data for the native protein and three heavy atom derivatives were collected at 100 K on the in house x-ray generator (Rigaku) using a RAXIS-IV++ imaging plate detector and were processed with the HKL2000 program package (17) (supplemental Table 1). The initial phases were obtained by the MLPHARE program and were improved by solvent flattening using the DM program (18). The model was built manually using the O program (19) and refined with the CNS program (20) to a final R-factor of 20.6% (Rfree = 24.5%) at 2.4 Å resolution (supplemental Table 1).
In Vitro Transcription Reactions
Linear DNA template generated by PCR amplification (40 nm), T. thermophilus holo-RNAP (50 nm), and ApU (100 μm; Sigma) were mixed in 45 μl of TGA buffer (20 mm Tris acetate, 20 mm sodium acetate, 2 mm magnesium acetate, 5% glycerol, 14 mm 2-mercaptoethanol, 0.1 mm EDTA, pH 8.0) and incubated for 5 min at 55 °C. ATP, GTP, and UTP (Amersham Biosciences) were added to 2.5 μm, followed by incubation for 5 min at 37 °C and purification through G50 spin columns (Amersham Biosciences) equilibrated in TGA. The obtained A26 transcription elongation complexes (TECs) were incubated with 300 nm [α-32P]CTP (PerkinElmer Life Sciences) for 3 min at 37 °C to form C27 TECs. C27 TECs were incubated at conditions indicated in the legend to Fig. 2. Samples were analyzed as described previously (21).
FIGURE 2. Gfh1 inhibits all catalytic activities of T. thermophilus RNAP.
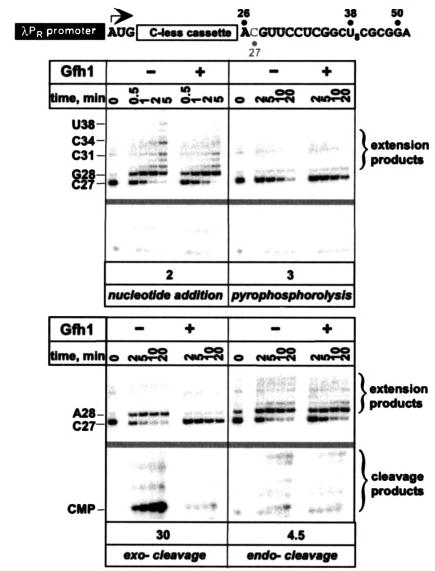
Top, transcript generated from the λPR promoter on pIA253; transcriptional start site is indicated by an arrow and is followed by a 26-nucleotide C-less cassette. Bottom, halted unlabeled A26 TECs were purified through G50 columns (Amersham Biosciences) and incubated with [α-32P]CTP to form 3′-end-labeled C27 TECs. C27 complexes were incubated at 37 °C with 200 nm UTP and GTP (nucleotide addition), 10 μm PPi (pyrophosphorolysis), 500 μm ATP, and 8 mm MnCl2 (exo-cleavage) or at 55 °C (endo-cleavage). Gfh1 was added to 500 nm where indicated. Aliquots were withdrawn at times indicated above each panel, quenched with 5 m urea, 10 mm EDTA, and separated on a 13% denaturing urea-acryl-amide (19:1) gel. The portion of the gel lacking radioactive signals (between the C27 and the cleavage products) was deleted to conserve space (gray bar). Gfh1 inhibitory effect (-fold, shown below each panel) on each catalytic reaction was determined by comparing the rate of reaction in the absence to that in the presence of Gfh1. For nucleotide addition, accumulation of C34 RNA species was quantified. PPi cleavage was measured by the disappearance of the labeled C27 RNA (shorter RNA fragments are not labeled). Endo- and exo- digestion was monitored by the accumulation of the corresponding cleavage products (CMP in case of the exo-nucleolytic cleavage). A representative gel is shown; the experiment was repeated at least three times for each reaction.
RESULTS
The Gfh1 Structure
The Gfh1 structure (Fig. 1B) comprises two domains, globular (G-) and CC, which closely resemble the corresponding domains of GreA (22) (root mean square deviation are 1.1 and 0.85 Å for the G- and CC-domains, respectively). A high quality of the experimental electron density allowed unambiguous modeling of all 156 Gfh1 residues in each of the four molecules in the asymmetric unit of the crystal. The only exception is a linker loop (residues 41–45) intervening the α-helices at the CC tip (Fig. 1, A and B), which was represented by the relatively weak experimental electron density and exhibited high B-factors in a final model, suggesting that this segment is flexible and therefore might adopt alternative conformations. The four independent Gfh1 monomers are arranged as a pair of nearly identical dimers (supplemental Fig. S1), in which the individual molecules are related by a perfect rotational symmetry, suggesting that the dimers might have some physiological relevance. However, the dimers are stabilized almost exclusively through Zn2+-mediated interactions and can represent a crystallization artifact given the high concentration of zinc acetate (33 mm) used for crystallization. On the other hand, judging by 25 well fixed Zn2+ ions predominantly bound to the acidic side chains, Gfh1 apparently has affinity for the divalent cations, implying that at certain conditions its activity might be modulated by metal ions.
Comparison with GreA and DksA
Despite their high structural similarity, there are two essential differences between the Gfh1 and GreA structures. First, the interdomain orientations are strikingly distinct (~162° rotation), resulting in completely different orientations of the CC-domains and the inter-domain interfaces (Fig. 1C). Interestingly, the Gfh1 interacting surface is more extensive (1238 Å2 via 1030 Å2 in GreA) and involves mostly hydrophobic interactions, whereas the GreA interface is determined exclusively by hydrogen bonding (supplemental Fig. 2). The Gfh1 structural organization is reminiscent of that of DksA, whose G- and CC-domains also form a common hydrophobic core that, similarly to Gfh1, masks a big portion of the CC-domain, thereby likely restricting its conformational flexibility (9). The Gfh1 and GreA conformations are compact and are stabilized by specific interdomain interactions, suggesting that they represent physiologically relevant states of the proteins rather than being artificially induced by crystal packing.
Another notable structural difference that distinguishes Gfh1 from both GreA and DksA is the local conformation of the linker at the CC tip. In GreA/DksA this region contains a DXX(E/D) motif with the two invariant acidic residues crucial for function (8–10, 12). In both GreA and DksA this segment adopts a well fixed α-turn conformation, which is self-stabilized through the internal main chain interactions and brings the acidic side chains proximal to each other (Fig. 1D). Given the potential functional importance of the acidic residues in other proteins, this motif can be considered ”extended“ in Gfh1 (DDYDD) (Fig. 1A). While similar to GreA in orientation, the Gfh1 linker is apparently missing the self-stabilized α-turn, making it more flexible and allowing three out of four Asp side chains to approach each other in an acidic cluster whose formation would be effectively prohibited in the presence of the α-turn (Fig. 1D).
Gfh1 Inhibits All Catalytic Activities of RNAP
The reactions catalyzed by RNAP utilize different substrates and thus likely have different requirements for the ”optimal“ configuration of the AS. Indeed, alterations in the AS geometry brought by the substitutions of individual Asp residues inhibit different polymerization and cleavage reactions to different extent (23). Similarly, protein factors, like GreA and DksA, may remodel the AS to preferentially impact one but not other activities, thus exerting a specific regulatory effect. In contrast, Gfh1 was shown to inhibit two distinct catalytic reactions (14). To test whether Gfh1 would have a similar effect on all types of catalytic reactions in a single TEC, we prepared C27 TECs that were uniquely labeled at their 3′-terminal C residue (Fig. 2).
C27 RNA was extended upon addition of GTP and UTP; at the low NTP concentrations used here, transcription arrests at U38. C27 RNA was also degraded in the presence of PPi releasing CTP (which co-migrated with the excess [α-32P]CTP; data not shown). At 55 °C, both the endonucleolytic activity that leads to the release of short 3′-labeled RNA fragments (cleavage products) and the ability to scavenge trace nucleotides to extend C27 RNA (extension products) became pronounced. Similarly to published studies (14, 15), we found that Gfh1 moderately (from 2- to 4.5-fold effects) inhibited all these three reactions (Fig. 2).
Upon addition of Mn2+ and non-cognate ATP (3), a fraction of C27 RNA was extended to A28 as a result of misincorporation (ATP preparation used is not contaminated with GTP; data not shown) and the remaining RNA was cleaved, releasing 3′-terminal CMP. The exonucleolytic cleavage was essentially blocked by Gfh1 (CMP synthesis was inhibited 30-fold; Fig. 2). Thus, although Gfh1 inhibits all RNAP reactions, the exonucleolytic cleavage is particularly sensitive to inhibition. This selectivity may be relevant to the physiological role of Gfh1, which remains obscure.
DISCUSSION
The sequence and structural homology between Gfh1 and GreA and the observation that Gfh1 affects all intrinsic (factor-independent) catalytic activities of RNAP (Fig. 2) suggests that Gfh1 utilizes the Gre/DksA-like structural mechanism of action: protruding the CC-domain through the secondary channel toward the RNAP catalytic center, where the CC-tip acidic residues remodel the AS (perhaps through Mg2+-mediated interactions with the catalytic residues), thereby modulating the RNAP activities.
A low resolution (15 Å) electron density (12) demonstrates that the RNAP secondary channel accommodates the GreB CC-domain but does not allow one to distinguish between the alternative G-domain orientations: the GreA and Gfh1 G-domains would likely fit equally well to the same electron density if the proteins are superimposed by their CC-domains (supplemental Fig. S3). The published models for RNAP-Gre factor interactions agree on the overall topology of the complex but differ in their details (8, 10, 12), and substitutions in RNAP that directly affect the Gre factors binding have not been reported. The sequence analysis and structural modeling show that with the exception of a few unfavorable amino acid substitutions (Fig. 1A), which likely make the observed conformations preferential for the particular protein in its free form, the residues forming the interdomain interfaces in Gfh1 and GreA are conserved among both protein families. At the same time the interdomain contacts in both proteins are not very extensive and could be disrupted under certain conditions to allow a ”flip“ between the two observed orientations. In principle, either of the two conformations might be induced for each protein in the course of its recruitment to RNAP. These considerations suggest two alternative modes of Gfh1/GreA binding to RNAP. First, the proteins may maintain their free form conformations. Assuming that in both cases the G-domains contain the major binding determinants (8, 24), while the CC-domains would be positioned essentially similar given the restrictions imposed by the size and shape of the secondary channel on their orientations, this scenario implies distinct RNAP binding modes for Gfh1 and GreA (they may utilize different structural elements to bind to the same site on RNAP or bind to different sites on RNAP); consistently, the G-domains surfaces exposed for the potential interactions with RNAP differ both in shape and sequence. Alternatively, both proteins may adopt one of the observed conformations induced upon binding to RNAP and in this case would likely bind to the same site on RNAP in a similar fashion; this also seems possible in light of their sequence and apparent structural similarity. Interestingly, changes in the relative positions of the two GreB domains upon binding to RNAP and TECs were detected by hydroxyl radical footprinting analysis (24).
Structural comparison of the Gfh1, GreA, and DksA proteins allows us to propose that their dramatically different effects on transcription in vitro (Fig. 2 and Refs. 8, 9, and 14) are likely determined by the sequence and conformation of the short linker between the α-helices at the tip of the CC (Fig. 1D). In GreA and DksA, the well fixed nearly identical α-turns strictly fix the orientations of the two functional acidic side chains, allowing them to coordinate a single Mg2+ ion but at the same time restricting their mobility to focus their activity on a single particular target. The distinct orientations of the α-turns in GreA and DksA (Fig. 1D) likely reflect the differences in the positions of their substrates thus avoiding functional overlap. We propose that, although the Gre/DksA-like stabilization of the Mg2+ ions in the vicinity of the RNAP AS is also a key element of the Gfh1 function, the flexibility of the Gfh1 CC linker missing the stable α-turn and the excess of the acidic residues in this region might allow Gfh1 to coordinate the Mg2+ ions occupying somewhat distinct positions (likely a characteristic of different reactions catalyzed by RNAP) without significant alterations of the CC orientation. This hypothesis is consistent with the broad functional specificity of Gfh1, which inhibits all RNAP catalytic reactions (Fig. 2). Indeed, the three-residue acidic cluster observed in the Gfh1 structure may readily stabilize one Mg2+ ion providing three coordination bonds (in contrast to only two in GreA/DksA) (Fig. 3A). Under certain conditions, however, one of these three Asp residues may flip its side chain to approach the adjacent fourth Asp, this would allow for coordination of the Mg2+ ion located in a substantially distinct position (Fig. 3B). In fact, the apparent flexibility of the Gfh1 CC linker might allow for additional conformations and subsequent spatial regrouping of the four Asp side chains, resulting in a number of the alternative binding modes of the Mg2+ ions.
FIGURE 3. Proposed mechanisms of the Gfh1 action.
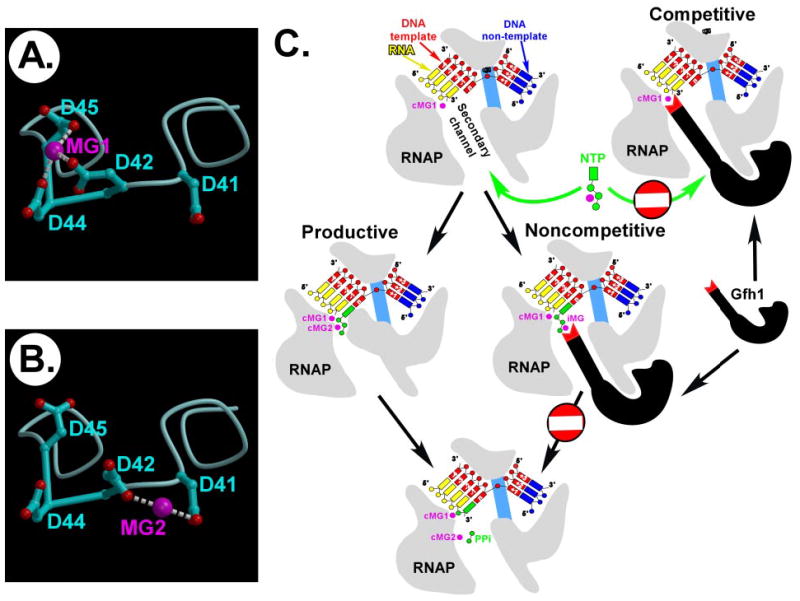
A and B, the flexibility and the number of the acidic residues at the CC tip of Gfh1 allow for the alternative binding (MG1 or MG2) of the Mg2+ ions. C, two hypothetical pathways (competitive and noncompetitive) by which the Gfh1 protein may inhibit nucleotide addition. cMg1 and cMg2 are the two catalytic metals, whereas iMg is a putative inhibitory Mg2+ ion.
Using the nucleotide addition reaction as an example, we suggest two hypothetical pathways, namely competitive and noncompetitive, by which Gfh1 may inhibit RNAP catalysis (Fig. 3C). In the competitive mode, Gfh1 may directly stabilize both catalytic Mg2+ ions that are required for substrate binding and catalysis with its four Asp residues. This would enhance the Gfh1 affinity to RNAP on one hand and sterically exclude substrate binding in the AS on the other. In the noncompetitive pathway, Gfh1 may fix an additional, non-catalytic Mg2+ ion in the site adjacent to, but not overlapping with, the AS. While this would not block substrate binding, competition of this inhibitory Mg2+ ion with the catalytic metal for binding of the substrate phosphates might result in a formation of a stable substrate-bound, but catalytically inactive, transcription complex. A similar mechanism of inhibition was recently proposed for the RNAP inhibitor tagetitoxin, which ”donates“ an additional Mg2+ ion bound in the vicinity of the AS (11). Given a broad Gfh1 specificity, it may utilize either or both of these pathways to modulate distinct RNAP catalytic reactions. Undoubtedly, the proposed mechanisms remain hypothetical and would require further biochemical and structural verification.
Gfh1 represents a third example of the rapidly growing family of the CC transcription factors operating through the RNAP secondary channel. All of them share a long CC-domain to deliver their active structural elements to the common target, the RNAP AS, but confer strikingly distinct effects on transcription. Remarkably, this functional diversity is achieved through relatively subtle variations in the conformation and sequence of a short linker segment located at the tip of the CC-domain that presumably targets only a few metal ions in the RNAP AS. The CC factors are thus reminiscent of a set of surgical tools, many of which are nearly identical in size and shape and differ only by their tips designed for highly specialized, non-overlapping applications. As transcription is a highly regulated process, we may await discovery of other CC ”tools“ ”operating“ on the AS of RNAP. Moreover, the CC-domain can be replaced with another extended protein segment (7), further expanding the repertoire of the delivery tools.
Finally, it is worth noting that the studies of the mechanisms of the CC transcription factors are still in their infancy. Although the previous analyses and this work have shed significant light on the structural organization and interactions with RNAP, allowing us and other groups (8–10, 12) to suggest several plausible molecular mechanisms of these factors, a number of important questions concerning their binding sites on RNAP, detailed mechanisms of the AS modulation, and their precise regulatory effects remain unanswered. A combination of biochemical and structural analyses of the CC proteins bound to their respective targets (transcription complexes) is required to address these questions.
Supplementary Material
Footnotes
This work was supported in part by National Institutes of Health Grants GM74252 and GM74840 (to D. G. V.) and GM67153 (to I. A).
The on-line version of this article (available at http://www.jbc.org) contains supplemental data, Table 1, and Figs. S1–S3.
The abbreviations used are: RNAP, RNA polymerase; AS, active site; CC, coiled-coil; G-domain, globular domain; TEC, transcription elongation complex.
The atomic coordinates and structure factors (code 2EUL) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Note Added in Proof
While this paper was under review the article describing the homologous structure of the T. aquaticus Gfh1 protein was communicated to us (25). Both the results and implications of this work are in good agreement with our data.
References
- 1.Rozovskaia T, Chenchik A, Bibilashvili R. Mol Biol (Mosk) 1981;15:636–652. [PubMed] [Google Scholar]
- 2.Borukhov S, Sagitov V, Goldfarb A. Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- 3.Sosunov V, Sosunova E, Mustaev A, Bass I, Nikiforov V, Goldfarb A. EMBO J. 2003;22:2234–2244. doi: 10.1093/emboj/cdg193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steitz TA. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 5.Adelman K, Yuzenkova J, La Porta A, Zenkin N, Lee J, Lis JT, Borukhov S, Wang MD, Severinov K. Mol Cell. 2004;14:753–762. doi: 10.1016/j.molcel.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 6.Mukhopadhyay J, Sineva E, Knight J, Levy RM, Ebright RH. Mol Cell. 2004;14:739–751. doi: 10.1016/j.molcel.2004.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kettenberger H, Armache KJ, Cramer P. Cell. 2003;114:347–357. doi: 10.1016/s0092-8674(03)00598-1. [DOI] [PubMed] [Google Scholar]
- 8.Laptenko O, Lee J, Lomakin I, Borukhov S. EMBO J. 2003;22:6322–6334. doi: 10.1093/emboj/cdg610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I, Vassylyev DG. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 10.Sosunova E, Sosunov V, Kozlov M, Nikiforov V, Goldfarb A, Mustaev A. Proc Natl Acad Sci U S A. 2003;100:15469–15474. doi: 10.1073/pnas.2536698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vassylyev DG, Svetlov V, Vassylyeva MN, Perederina A, Igarashi N, Matsugaki N, Wakatsuki S, Artsimovitch I. Nat Struct Mol Biol. 2005;12:1086–1093. doi: 10.1038/nsmb1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Opalka N, Chlenov M, Chacon P, Rice WJ, Wriggers W, Darst SA. Cell. 2003;114:335–345. doi: 10.1016/s0092-8674(03)00600-7. [DOI] [PubMed] [Google Scholar]
- 13.Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. Cell. 2004;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 14.Hogan BP, Hartsch T, Erie DA. J Biol Chem. 2002;277:967–975. doi: 10.1074/jbc.M108737200. [DOI] [PubMed] [Google Scholar]
- 15.Laptenko O, Borukhov S. Methods Enzymol. 2003;371:219–232. doi: 10.1016/S0076-6879(03)71016-7. [DOI] [PubMed] [Google Scholar]
- 16.Perederina, A., Vassylyeva, N. M., Berezin, I. A., Svetlov, V., Artsimovitch, I., and Vassylyev, D. G. (2005) Acta Crystallogr. Sect. F, in press [DOI] [PMC free article] [PubMed]
- 17.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 18.Collaborative Computational Project Number 4. Acta Crystallogr Sect D Biol Crystallogr. 1994;50:760–763. [Google Scholar]
- 19.Jones TA, Zou JY, Cowan SW, Kjeldgaard Acta Crystallogr Sect A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 20.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr Sect D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 21.Artsimovitch I, Patlan V, Sekine S, Vassylyeva MN, Hosaka T, Ochi K, Yokoyama S, Vassylyev DG. Cell. 2004;117:299–310. doi: 10.1016/s0092-8674(04)00401-5. [DOI] [PubMed] [Google Scholar]
- 22.Stebbins CE, Borukhov S, Orlova M, Polyakov A, Goldfarb A, Darst SA. Nature. 1995;373:636–640. doi: 10.1038/373636a0. [DOI] [PubMed] [Google Scholar]
- 23.Sosunov V, Zorov S, Sosunova E, Nikolaev A, Zakeyeva I, Bass I, Goldfarb A, Nikiforov V, Severinov K, Mustaev A. Nucleic Acids Res. 2005;33:4202–4211. doi: 10.1093/nar/gki688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loizos N, Darst SA. J Biol Chem. 1999;274:23378–23386. doi: 10.1074/jbc.274.33.23378. [DOI] [PubMed] [Google Scholar]
- 25.Lamour, V., Hogan, B. P., Erie, D. A., and Darst, S. A. (2005) J. Mol. Biol., in press [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


