Abstract
The RecJ exonuclease from Escherichia coli degrades single-stranded DNA (ssDNA) in the 5′–3′ direction and participates in homologous recombination and mismatch repair. The experiments described here address RecJ's substrate requirements and reaction products. RecJ complexes on a variety of 5′ single-strand tailed substrates were analyzed by electrophoretic mobility shift in the absence of Mg2+ ion required for substrate degradation. RecJ required single-stranded tails of 7 nt or greater for robust binding; addition of Mg2+ confirmed that substrates with 5′ tails of 6 nt or less were poor substrates for RecJ exonuclease. RecJ is a processive exonuclease, degrading ∼1000 nt after a single binding event to single-strand DNA, and releases mononucleotide products. RecJ is capable of degrading a single-stranded tail up to a double-stranded junction, although products in such reactions were heterogeneous and RecJ showed a limited ability to penetrate the duplex region. RecJ exonuclease was equally potent on 5′ phosphorylated and unphosphorylated ends. Finally, DNA binding and nuclease activity of RecJ was specifically enhanced by the pre-addition of ssDNA-binding protein and we propose that this specific interaction may aid recruitment of RecJ.
INTRODUCTION
RecJ enzyme is an Mg2+-dependent single-stranded DNA (ssDNA) exonuclease that degrades its substrates in the 5′–3′ direction (1). RecJ is the only known ssDNA-specific 5′ exonuclease and has been reported to degrade abasic residues during base excision repair, as a ‘dRPase’ activity (2). Identified for its essential role in recBCD-independent recombination pathways (3), RecJ, in combination with a DNA helicase such as RecQ, may produce 3′ ssDNA tails required to initiate recombination from a double-strand break. Acting alone, RecJ may trim 5′ ends to produce blunt-ended substrates required for other recombination enzymes or to trim 5′ tails after synapsis. RecJ also assists RecBCD-promoted recombination (4–6), presumably by post-synaptic degradation of a displaced 5′ end that competes for pairing with the invading strand. RecJ is also one of four exonucleases that mediate the excision step of methyl-directed mismatch repair after unwinding by UvrD helicase (7,8). RecJ orthologs are found in almost all eubacterial species and in archaea (9–11), suggesting that RecJ function arose early in evolution and plays an important biological function
RecJ has a strong specificity for ssDNA. Double-strand DNA (dsDNA) is neither a substrate nor a competitive inhibitor of ssDNA exonuclease activity (1). Indeed, the crystal structure of a truncated Thermus thermophilus RecJ (ttRecJ) shows that the active center resides in a cleft that is too narrow to accommodate dsDNA (12). We investigate here the number of nucleotides of ssDNA that is required by Eco RecJ for DNA binding and degradation. We examine the processivity of RecJ and the nature of its digestion products, including its ability to degrade dsDNA located in cis to single-stranded regions. In the bacterial cell, ssDNA is likely to be bound by ssDNA-binding protein (SSB). We show that prebinding of ssDNA in vitro by SSB does not inhibit RecJ degradation but, rather, enhances the activity of RecJ protein. Both RecJ and SSB have been shown to enhance RecA strand exchange by sequestering or degrading competing DNA (13,14) and the two may work in concert to accomplish this task.
MATERIALS AND METHODS
Reagents
Synthesized oligonucleotides and dinucleotide d(GA) were obtained from New England Biolabs (MA) or the Molecular Biology Core Facility at the Dana Farber Cancer Institute (MA). T4 polynucleotide kinase, terminal deoxynucleotidyl transferase, λ exonuclease, T4 gp 32 protein, MBP-RecJ(RecJf) and restriction endonucleases were obtained from New England Biolabs and SSB was purchased from Promega. Sources of labeled nucleotides were Perkin Elmer Life Science for [γ-33P]ATP, [α-32P]dATP, [α-32P]dGTP, 5,5-3H dCTP and Amersham Pharmacia Biotech for [α-32P]ddATP. Unlabeled 2′-deoxyadenosine 5′-monophosphate (dAMP), 2′,3′-dideoxyadenosine 5′-monophosphate (ddAMP) and 2′-deoxyguanosine 5′-monphosphate (dGMP) were purchased from Sigma. Rabbit polyclonal antibody to SSB was kindly provided by Roger McMacken.
DNA substrates
Oligonucleotide A was synthesized as a 65mer with the following composition: 5′-GCCCAGGTGG CAGGCCCTAG GGTGGAGGGG AGGCCGCCGG CATGGGGACG CGATGGGCGG AGGCG. Complementary oligonucleotide B (5′-CGCCTCCGCC CATCGCGTCC CCATGCCGGC GGCCTCCCCT CCACC), C (5′-CGCCTCCGCC CATCGCGTCC CCATGCCGGC GGCCTCCCCT CCACCCTAGG GCCTG) and D (5′-CGCCTCCGCC CATCGCGTCC CCATGCCGGC GGCCTCCCCT CCACCCTAGG GCCTGCCACC T) were also synthesized and annealed to oligonucleotide A to form single-stranded 5′ tails of 20, 10 and 4 nt in length, respectively. Annealing to oligonucleotide E (5′-CGCCTCCGCC CATCGCGTCC CCATGCCGGC GGCCTCCCCT CCACCCTAGG GCCTGCCACC TGGGC) created the completely double-stranded substrate. Similarly, oligonucleotides were created and annealed to oligonucleotide A to yield single-stranded 5′ tails of 5, 6, 7, 8 and 9 nt in length.
Oligonucleotide A was 5′ labeled using T4 polynucleotide kinase and [γ-33P]ATP and purified by chromatography with G-50 Sephadex (Boehringer Mannheim), phenol/chloroform extraction, ethanol precipitation and resuspended in TE buffer (pH 8.0) or 10 mM Tris–HCl (pH 8.0). The oligonucleotide was then boiled at 100°C for 5 min with an equal molar amount of complementary oligonucleotide, as appropriate, in a solution containing 20 mM Tris (pH 8.0), 10 mM MgCl2, 50 mM NaCl and 1 mM DTT. The substrates were allowed to cool gradually and anneal overnight at 4°C.
Oligonucleotide A was also labeled at its 3′ terminus with terminal transferase and [α-32P]ddATP and passed over a Sephadex G-50 column. This 3′-labeled substrate was annealed as described above to a 1.5 or equal molar equivalent of oligonucleotide B or other DNAs producing 5, 6, 7, 8 or 9 nt single-stranded tails. To produce 5′ phosphorylated, 3′ end-labeled substrates some of this 3′ labeled oligonucleotide A was also treated with T4 polynucleotide kinase and non-radioactive ATP.
Longer labeled linear DNA substrates consisted of a 2461 bp PCR fragment generated by Taq DNA polymerase amplification (Promega, standard conditions) of plasmid pBR322 template DNA and the following primers: 5′-TAGGCCTGAT AAGCGCAGCG TATCAGGCAA TTTTTATAAT AGATGGCGGA CGCGATGGAT and 5′-ATGATAGCGC CCGGAAGAGA GTCAATTCAG GGTGGTGAAT CTGACGCTCA GTGGAACGAA. Internally labeled PCR fragment DNA was generated by the inclusion of 25 µCi of 3H dCTP or 32P dATP and dGTP in the reaction mixture. Substrates were purified with Qiagen PCR purification kit and were heat-denatured and rapidly cooled on ice immediately before adding to reactions with RecJ. For use in the processivity experiments, internally 3H-labeled PCR DNA was 32P-labeled at each 5′ end with polynucleotide kinase (New England Biolabs) and 10 µCi of [γ-32P]ATP.
RecJ protein expression and purification
Native wild-type RecJ protein was overexpressed from an isopropyl-β-d-thiogalactopyranoside (IPTG) inducible low-copy pWSK29 derived plasmid (9) in a 10 liter Luria–Bertani culture of expression strain STL2329 (recJ284::Tn10 sbcB15 endA DE3 lysogen). Purification of wild-type RecJ protein from induced cultures was performed as described previously (1) with modifications. All column buffers had the following composition: 20 mM Tris (pH 8.0), 10% ethylene glycol, 0.1 mM EDTA and 1 mM DTT with NaCl varying at 0, 50 and 500 mM concentrations. Cultures in log phase were treated to 1 mM IPTG for 4 h at 37°C. Cells were then lysed for crude extract with chicken egg lysozyme as described previously (1). Crude extract was diluted with the above buffer (0 mM NaCl) to a conductivity equivalent to said buffer containing 50 mM NaCl. The extract was applied twice over a 20 ml Cibracon Blue 3GA column (Type 100, Sigma) and eluted by a step gradient of column buffer at 500 mM NaCl. Peak fractions were loaded onto a 1 ml UnoQ column (Bio-Rad) and purified to homogeneity over a 100 ml linear gradient from 50 to 500 mM NaCl, utilizing a Biologic HR Workstation (Bio-Rad). Peak fractions were reapplied to a fresh UnoQ column and eluted similarly to remove trace contaminants. Protein concentration was assayed using the Bradford reagent commercially available from Bio-Rad (15), using BSA as a standard.
Electrophoretic mobility shift assays
Mobility shift assays were performed similarly as described previously (16). RecJ-DNA-binding reactions were typically performed in the absence of Mg2+ catalyst in volumes of 20 µl containing 10 mM Tris (pH 8.0), 50 mM NaCl, 25 mM EDTA, 25% glycerol, 1 mM DTT, 5 nM 5′ radiolabeled DNA substrate, and the varying amounts of purified wild-type RecJ protein or RecJf fusion protein. Binding reactions were incubated on ice for 1 h and then loaded onto non-denaturing acrylamide gels composed of 6% sequencing grade acrylamide (National Diagnostics), 25 mM Tris (pH 8.0), 200 mM glycine, 2.5% glycerol, 0.5 mM DTT, 0.125% ammonium persulfate and 0.1% TEMED. Gels were pre-run in buffer consisting of 25 mM Tris (pH 8.0) and 200 mM glycine for 1 h at 4°C at 75 V. Samples were loaded and subjected to electrophoresis at 4°C at 75 V for 100 min. Gels were then dried onto filter paper (Whatman) for 1 h at 80°C under vacuum and subsequently exposed to X-ray film (Denville Scientific) or analyzed by phosphorimager and Quantity One software (Bio-Rad). For electrophoretic mobility shift experiments with SSB, 2.5 nM 5′-labeled oligonucleotide A substrate was prebound with 15 nM SSB (tetramer) for 20 min before the addition of 40–415 nM RecJ protein to the binding reactions for an additional 40 min. For supershift reactions, 1 µg SSB antibody was added and incubation was continued for 20 min before gel analysis.
Nuclease assays
DNA substrates containing 5′ single-stranded tails ranging from 5 to 9 nt in length were created with a 5′ radioactive label on oligonucleotide A. Reactions (20 µl) contained 5 nM labeled DNA substrate, 12.5 nM purified RecJ in 50 mM NaCl, 10 mM Tris (pH 8), 1 mM DTT and 10 mM MgCl2. The reactions were incubated for 30 min at 37°C and then loaded upon native acrylamide gels and analyzed as above.
To investigate 5′ phosphate requirements, 5′ OH oligonucleotide A was radioactively labeled at its 3′ end with terminal transferase and [32P]ddATP and a portion of this was subsequently 5′ phosphorylated utilizing non-radioactive ATP and T4 polynucleotide kinase. An aliquot of 0.1 pmol of these 3′-labeled, 5′ OH or 5′ phosphate substrates was treated as described above with increasing amounts of pure RecJ or, after annealing to equimolar complementary oligonucleotide E, to various amounts of λ exonuclease for 30 min at 37°C. Resultant samples were resolved on denaturing polyacrylamide gels as described below. Bio-Rad Quantity One phosphorimager software was utilized to quantify degradation activity. In other nuclease assays, SSB (30 pmol tetramer) or T4 gp32 protein (18 pmol monomer) was allowed to bind to 0.1 pmol 3′-labeled oligonucleotide A or d(ACT)10 homopolymer in the 20 µl reaction buffer for 30 min on ice prior to RecJ addition.
Processivity experiments employed 2461 bp linear ssDNA, labeled internally with 3H and at its 5′ ends with 32P. Competitor DNA was the same heat-denatured PCR fragment, which was unlabeled. RecJ (70 fmol) was preincubated with 15 ng double-labeled substrate DNA on ice in the absence of Mg2+. The reaction was initiated by addition of 10 mM MgCl2 concomitant with addition of 50-fold excess of unlabeled competitor ssDNA to bind any free RecJ. The progress of the reaction from 0.5 to 5 min was monitored by release of TCA-soluble radioactivity. The ratio of internal 3H release to 5′ 32P release reveals the extent to which RecJ degrades the substrate in a single pass. The validity of the cold competition for free RecJ was confirmed in reactions where RecJ was added after the excess unlabeled DNA.
Product analysis by PAGE
Oligonucleotide A was treated with terminal transferase with [α-32P]ddATP to label its 3′ end and annealed to equimolar amounts of complementary oligonucleotide B to yield substrate AB containing a 20 nt 5′ tail. This 5′ tailed substrate (1.7 pmol) was combined with 4 pmol purified RecJ protein in a 10 µl reaction in the presence of 50 mM NaCl, 10 mM Tris (pH 8), 1 mM DTT and 10 mM MgCl2 for 30 min at 37°C. The reaction was terminated by addition of an equal volume of stop solution (95% formamide, 20 mM Tris (pH 8.0), 0.05% bromophenol blue (Sigma), 0.05% xylene cyanol (Sigma) and 3.0 pmol non-radioactive oligonucleotide A. After boiling at 100°C for 5 min, degradation products were resolved on a 15% denaturing acrylamide gel at 65°C containing 1× Tris–borate EDTA buffer (17) and 6 M urea. Gels were dried onto filter paper for 110 min at 80°C and subsequently exposed on X-ray film or phosphorimager screen. Analysis of degradation of a 10-base tailed substrate, produced by labeling and annealing of oligonucleotide A with oligonucleotide C, was performed similarly except with 80 fmol substrate concentration, 170 fmol RecJ and reaction times from 0 to 20 min.
Product analysis by thin layer chromatography (TLC)
RecJ exonuclease reactions were performed on DNA substrates labeled with 32P and DNA degradation products were analyzed for mononucleotides by TLC analysis similarly as described previously (18). Single-strand oligonucleotide A was labeled with 32P on the 5′ guanine or the 3′ adenine as described above for RecJ DNA-binding experiments. Alternatively, a 2.4 kb DNA PCR fragment was internally labeled with 32P on both guanines and adenines as described above for RecJ processivity experiments. The substrates were boiled for 5 min and immediately cooled on ice. Labeled DNA substrates (0.1 pmol) were then treated with 0.8 pmol of RecJ in 20 µl reactions containing 25 mM NaCl, 5 mM Tris–HCl (pH 8.0), 1mM DTT and 5 mM MgCl2. Reactions with the 3′-labeled substrate also included 0.6 pmol of SSB tetramer to stimulate complete digestion of the substrate. All reactions were incubated at 37°C for 40 min and quenched with the addition of excess EDTA. Controls were also run in the absence of RecJ for each experiment. The products were spotted on a polyethyleneimine (PEI) plate that was placed in a covered 500 ml beaker containing a mobile phase of 1 M formic acid and 0.5 M LiCl. The plates were subsequently dried at room temperature and exposed to X-ray film or a phosphoimager screen. dGMP, dAMP and ddAMP were spotted as markers and visualized by an ultraviolet (UV) lamp at 254 nm.
RESULTS
DNA specificity
Previous studies had shown that exonuclease activity of Escherichia coli RecJ was at least 10 000-fold greater on ssDNA than dsDNA. RecJ was also able to degrade 5′ ssDNA tails produced by digestion of dsDNA with exonuclease III (1).
To define the substrate requirements of RecJ more precisely, we determined the ability of RecJ to bind ssDNA of defined lengths. In the absence of Mg2+, RecJ binding could be detected by mobility shift on native gels without substrate loss due to degradation. As expected, no binding was detected for the double-stranded substrate (Figure 1A). A 4 nt single-stranded 5′ tail was also a poor substrate, with no mobility shift detected (Figure 1B). However, substrates with 10 and 20 nt tails did demonstrate binding with RecJ protein (Figure 1C and D). A shift in mobility could be detected at 5 nM RecJ, with the intensity of the slow mobility species increasing as RecJ concentration was increased. RecJ bound to the 10 nt tailed substrate with an apparent Kd of 9.5 nM. The apparent Kd of RecJ with the 20 nt tail substrate was slightly weaker at 33 nM, and we attribute this decrease to greater potential for secondary structure formation in the longer single strand that would preclude RecJ binding.
Figure 1.
RecJ binds 5′ single-stranded tails. Mobility shift assays were performed with purified wild-type RecJ protein complexed to 33P-labeled DNA substrates. Complexes were resolved on a native acrylamide gel. Assays were performed with 5 nM of a 65 bp double-stranded substrate (A) or 5′ single-stranded tailed substrates with 4 (B), 10 (C) and 20 (D) unpaired 5′ nucleotides. RecJ concentration varies from 5 to 200 nM.
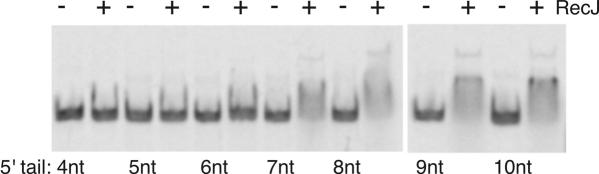
To determine the limit length of the single-stranded tail required for RecJ binding, 5′ single-stranded tail lengths of 5, 6, 7, 8 and 9 nt were examined. In the absence of Mg2+, wild-type RecJ protein bound robustly to tails of 7 nt and longer, but yielded no binding to 5 nt tails and very little to 6 nt tails, even at high RecJ protein concentrations (Figure 2). In a separate experiment, addition of 10 mM MgCl2 in parallel reactions revealed that exonuclease substrate specificity reflected this binding specificity. RecJ had little nuclease or binding activity on the 5′ radioactively labeled end of ssDNA tails of 5 nt or less and only partial cleavage of 6 nt tailed substrates to which it binds weakly (Figure 3). Complete cleavage of the labeled 5′ end was observed on tailed substrates of seven or more nucleotides.
Figure 2.
RecJ binds a 7 nt tail. Mobility shift assays were performed upon DNA substrates with 5′ single-stranded tails ranging from 4 to 10 nt in length. Reactions were performed at 100 nM RecJ protein and 5 nM substrate DNA. Control reactions (−) contained no protein.
Figure 3.
RecJ cleaves the terminal nucleotides on 5′ tails. DNA substrates with 5′ single-stranded tails of 5, 6, 7, 8 and 9 nt in length were subjected to conditions where RecJ binding only could be observed [(B), no Mg2+] and conditions where nuclease activity could be detected [(C), 10 mM MgCl2] by loss of radioactive label at the 5′ end of the single-stranded tail. The 5 nM RecJ protein was combined with 5 nM substrate in binding reactions whereas 12.5 nM protein was combined with 5 nM substrate in cleavage reactions. Control reactions (A) contained no protein.
Whether more than one RecJ molecule binds to an oligonucleotide substrate was investigated by mixing experiments using RecJ proteins of different molecular weights. We tested binding of purified native wild-type RecJ (∼60 kDa in molecular weight) and a RecJ fusion to maltose-binding protein (RecJf, New England Biolabs; 100 kDa) to oligonucleotide A or a 10 nt 5′ single-stranded tailed substrate. Both RecJ species are equally active as nucleases (data not shown) and both bind to oligonucleotide A (A) or a 10 nt 5′ tail (B) substrate (Figure 4) in the absence of Mg2+, with the larger protein producing a greater electrophoretic mobility shift. Upon mixing the two proteins with substrate, no intermediate shifted species was detected, suggesting only one RecJ molecule binds per substrate molecule. This suggests that RecJ has specific recognition for a 5′ single-strand end, for which there is only one in each of these substrates.
Figure 4.
RecJ binds monomerically to DNA. Electrophoretic mobility shift assays with 5 nM oligonucleotide A treated with 100 nM RecJ alone, RecJf alone or with a mixture of the two (A). An identical experiment, except using the 10 nt 5′ tailed substrate (B). Two distinct shifts are noted, owing to the ∼40 kDa difference in molecular weight between RecJ and RecJf.
5′ End phosphorylation
To determine any preference of RecJ for phosphorylation status of the 5′ ends, we prepared 3′ radioactively labeled oligonucleotide A with either a 5′ OH or with a 5′ phosphate terminus. Both substrates were treated with increasing amounts of RecJ and were degraded equally well (Figure 5A). To confirm the phosphorylation status of these substrates, both oligonucleotide A preparations were annealed to a complementary DNA strand and subjected to digestion with λ exonuclease, a 5′–3′ dsDNA exonuclease with a known requirement for 5′ phosphorylated ends (19). As expected, the phosphorylated substrate but not the unphosphorylated substrate was sensitive to λ Exo degradation (Figure 5B). We conclude that a 5′ phosphate is not required for binding or hydrolysis of ssDNA substrates.
Figure 5.
RecJ recognizes phosphorylated and non-phosphorylated 5′ ends. The 3′ 32P-labeled oligonucleotide A was either phosphorylated with T4 polynucleotide kinase on the 5′ end or left unphosphorylated. (A) In a 20 µl reaction, 0.1 pmol of each substrate was added to various amounts of purified RecJ or, after annealing with equimolar complementary oligonucleotide B, λ exonuclease. (B) Phosphorimager analysis of three independent nuclease assays with RecJ and 5′ phosphorylated substrate (circles) and 5′ OH substrate (triangles).
Processivity
The extent of RecJ processivity was measured on a denatured 2461 base linear DNA fragment, which had been labeled internally with 3H deoxynucleotides and on its 5′ end with 32P. RecJ was allowed to bind to the labeled substrate prior to initiation of the reaction with addition of Mg2+, and at the same time, 50-fold excess unlabeled competitor DNA. Any RecJ bound to the labeled substrate would therefore initiate degradation 5′–3′, releasing first the 32P-label and subsequently 3H-labeled nucleotides as it traversed the substrate. Upon its first dissociation from the labeled substrate, RecJ would be titrated by the excess unlabeled competitor DNA and unable to contribute significantly to additional release of radioactivity. (RecJ was also added after the unlabeled excess DNA in a control reaction to confirm the competition for free RecJ.) Because the rate of RecJ digestion is quite rapid, at 1000 nt per minute (1), we monitored the release of radioactivity from 0.5 to 5 min after Mg2+ addition. The ratio of the acid-soluble fraction for the 3H internal label to the 32P end-label leveled to a value of 0.41 at 1–2 min into the reaction. This indicates that for every 5′ end attacked, 41% of the 2461 nt substrate is digested, revealing the extent of processivity to be ∼1000 nt.
Degradation products
We also investigated the ability of RecJ to penetrate a region of duplex DNA after initiating degradation on an ssDNA-tailed substrate. Because RecJ is a processive exonuclease it was of interest whether RecJ could degrade duplex DNA after degradation had been initiated on an ssDNA end in cis. The substrate had a 20 nt 5′ tail and a 45 bp duplex region, with a 3′ 32P-label on the longer strand. After digestion with RecJ, there was a heterogeneous set of products. However, a predominant product appeared to comigrate with an ssDNA marker of 45 nt, indicating degradation to the double-stranded junction (Figure 6A). Therefore, RecJ sometimes terminated digestion upon encountering duplex DNA. However, we noted other products shorter than 45 nt, indicating RecJ digestion into the duplex region of the substrate. These may result from transient denaturation (‘fraying’) of the duplex substrate at the ends, exposing single-strand for RecJ digestion. Minor products longer than 45 nt were also evident, one of which was ∼52 nt. This is 7 nt from the junction, the size tail at which RecJ began to lose affinity in the previous binding experiments. In a similar experiment with a 10 nt ssDNA-tailed substrate, where we monitored the reaction progress over time, we also saw a major product arise early in the reaction, which persisted at longer digestion times (Figure 6B), at which time almost no full-length substrate remained. The size of this product is consistent with degradation close to the point at which duplex would be encountered on the substrate. However, this product accounts for only a minority of the starting substrate, indicating that the reaction is heterogeneous in terms of the final product and that many substrates may be degraded more completely.
Figure 6.
RecJ degrades DNA to a junction. (A) DNA substrate (1.7 pmol) containing a 5′ single-stranded tail of 20 nt in length and a radiolabel on the 3′ end was degraded with 4 pmol purified native RecJ protein (+RecJ). The products were then resolved on a denaturing acrylamide gel alongside a control with no degradation (−RecJ). Single-stranded size markers of 65, 61, 55 and 45 nt in length were resolved in adjacent lanes. (B) DNA substrate (80 fmol) containing a single-stranded tail of 10 nt and a radiolabel on the 3′ end was degraded with 170 fmol purified native RecJ protein for the indicated times (0–20 min) and resolved as before. Adjacent size markers were 65, 55, 45 and 30 nt.
To investigate the products formed during RecJ digestion of ssDNA, 5′ or 3′ radioactively labeled oligonucleotide A was treated with RecJ in the presence of Mg2+ ion and the products resolved by TLC (Figure 7A and C). With the 3′-labeled substrate, SSB was added to promote degradation to the 3′ end and prevent secondary structure formation (see below). RecJ digestion of internally 32P-labeled 2461 nt ssDNA was also analyzed (Figure 7B). The labeled reaction products comigrated with non-radiolabeled dGMP (Figure 7A and B) or dAMP (Figure 7B and C) that were spotted at the same origin on the TLC plate and visualized by UV shadowing with a 254 nm wavelength lamp. Therefore, RecJ appears to release mononucleotides throughout its digestion of ssDNA, even as a free 3′ end is encountered on the terminal end.
Figure 7.

RecJ degrades DNA to mononucleotides. An aliquot of 0.1 pmol of oligonucleotides (A) labeled with 32P on the 5′ guanine, (B) labeled internally with 32P on guanines and adenines or (C) labeled with 32P on the 3′ adenine were treated in the presence or absence of 0.8 pmol of RecJ for 40 min. Products of the reactions were then spotted on PEI plates and resolved with TLC. For reactions with the 3′-labeled substrate, 0.6 pmol of SSB tetramer was added to stimulate complete digestion. Positions of dGMP, dAMP and ddAMP controls are indicated.
SSB enhancement of RecJ binding and degradation
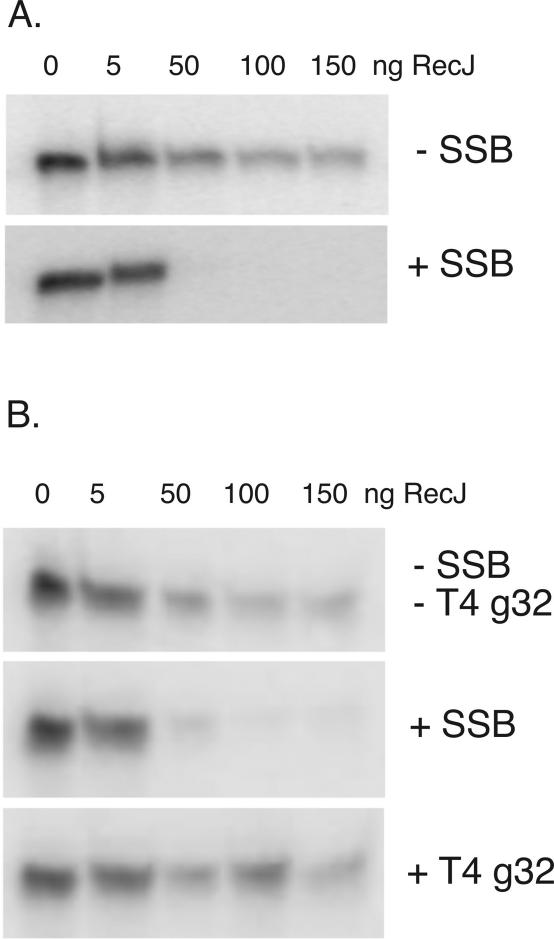
In reactions with 3′-labeled oligonucleotide A, the 3′ end was poorly digested by RecJ, with a fraction of substrate resistant to degradation. The inefficiency of digestion of single-strand oligonucleotides was somewhat sequence-specific (E.S. Han, V.A. Sutera, Jr. and S.T. Lovett, unpublished data) and may be due to the transient formation of secondary structure that interferes with RecJ binding to GC-rich oligonucleotides such as oligonucleotide A. Indeed, addition of SSB to these reactions, which would remove such secondary structures, stimulated digestion of these substrates (Figure 8A). To minimize the contribution of secondary structure, we also assayed digestion of a homopolymeric substrate, (ACT)10. With this substrate, we observed similar enhancement of RecJ digestion by SSB (Figure 8B). Moreover, addition of T4 gp 32 protein, likewise an SSB protein, did not facilitate RecJ digestion. (Control electrophoretic mobility shift experiments showed that under these conditions, the oligonucleotide was fully bound by gp32; data not shown). This may reflect a specific interaction between RecJ and SSB or a mode of binding distinct between the two single-strand binding proteins.
Figure 8.
SSB enhances RecJ degradation. An aliquot of 0.1 pmol of oligonucleotide (A) with radioactive 3′ label was treated with increasing amounts of pure RecJ in the absence, (−SSB), and presence, (+SSB, 3 pmol tetramer), prebound SSB protein and analyzed on denaturing acrylamide gel (A). Likewise, 0.1 pmol of a 30 nt homopolymer 5′d(ACT)10 was treated with RecJ with and without prebound SSB3 pmol tetramer) or T4 Gene 32 protein (600 ng) (B).
We performed RecJ mobility shift binding experiments with oligonucleotide A substrate, with and without prebound SSB. In the absence of SSB, RecJ binds this substrate extremely poorly (Figure 9A) with no detectable electrophoretic mobility shift. SSB binds this olignucleotide more tightly (Figure 9A). When the ssDNA was prebound with SSB, addition of RecJ produced a more slowly-migrating species, absent when RecJ was heat-denatured prior to addition (Figure 9, rightmost lane), consistent with a ternary complex of SSB, RecJ and DNA. Addition of antibody directed to SSB produced a super-shifted species (Figure 9B), confirmed the presence of SSB in this complex.
Figure 9.
(A) SSB enhances RecJ binding to ssDNA. An aliquot of 0.05 pmol of oligonucleotide A was incubated with no protein, RecJ alone (8.3 pmol) or preincubated with SSB (0.3 pmol, tetramer) and then treated with increasing amounts of RecJ (0.8–8.3 pmol, monomer) or heat-inactivated RecJ (8.3 pmol) alone. Samples were split and either analyzed for a mobility shift (A) or for supershift by SSB antibody (B).
DISCUSSION
To bind stably and to initiate DNA degradation, E.coli RecJ exonuclease requires a 5′ single-strand of 7 nt or longer. Upon encountering dsDNA, RecJ digestion tends to terminate. The products of such a reaction are not precise: although RecJ often degrades to the position of the junction, it may stop several nucleotides before or after beginning of duplex DNA. RecJ appears to be a processive exonuclease, digesting ∼1000 nt before it dissociates from its substrate. RecJ releases mononucleotide products during degradation of ssDNA.
The crystal structure of RecJ suggests that its strong specificity for ssDNA is determined by steric factors. The structure of the C-terminally truncated RecJ ortholog in T.thermophilus reveals a DNA-binding cleft that is too narrow to contain dsDNA (12). In this respect, RecJ resembles E.coli Exonuclease I (20), a processive 3′–5′ exonuclease on ssDNA. Like RecJ, its structure presents a narrow cleft suitable for ssDNA but not duplex DNA. Noted for the cleft of the truncated Tth RecJ was a positively charged binding surface that could accommodate 4 nt of ssDNA. Our results with the full-length E.coli RecJ suggest more extensive contacts with substrate: at least 7 nt are required both for optimal binding and for hydrolysis. This extended surface of binding may explain the processivity of the enzyme, allowing it to remain bound to substrate after each hydrolytic reaction.
Once presented with an ssDNA substrate of suitable length, RecJ appears to degrade its substrates completely to mononucleotides. Likewise, RecJ can degrade ssDNA tails to or past the junction of dsDNA, substrates it cannot stably bind to initiate degradation. This is likely explained by the fact that the initial binding and cleavage reaction differs from subsequent reactions during the processive cycle. Upon encountering 5′ ssDNA of sufficient length, RecJ may convert from a loosely bound to a tightly bound conformation that can mediate potent and processive nuclease function. Tth RecJ contains two connected globular domains with a narrow binding groove, bridged by a single ∼12-turn helix (12), which may act as a hinge for association of the two flanking globular domains, dependent on the presence of ssDNA in the cleft.
RecJ binds as a monomer to ssDNA substrates—we found no evidence for higher-order bound complexes. This was of interest because both E.coli and Thermus RecJ form homo-oligomeric species in solution. However, only monomeric RecJ is active as a nuclease [S. T. Lovett, unpublished data and (21)] and oligomerization may serve to inactivate the protein in vivo. We cannot rule out the possibility that RecJ interacts with other protein partners, although no strong complex has been yet isolated.
Interactions with SSB protein, either direct or indirect, are likely to be important for RecJ function. In vitro, successive cleavage events to liberate the 3′ terminus of a 65 nt ssDNA substrate were clearly enhanced by SSB. Because of RecJ's specificity for ssDNA, formation of secondary structure should be inhibitory to the nuclease activity of RecJ. SSB may, in part, stimulate the nuclease activity of RecJ by facilitating its processivity by removal of such secondary structures. Interestingly, although bacteriophage T4 gene 32 protein can bind likewise to ssDNA and remove secondary structure, it did not stimulate RecJ. This suggests that specific contacts between RecJ and SSB, or a distinct structure formed by SSB may enhance the activity of RecJ. E.coli exonuclease I, an exonuclease similar to RecJ but of the opposite polarity, also forms specific interactions with SSB as detected by gel filtration and yeast two-hybrid interactions (22,23). Unlike the case for ExoI, we found no evidence for SSB-RecJ interaction in solution in the absence of ssDNA (E. S. Han and S. T. Lovett, unpublished data) although both RecJ and ExoI have been shown recently to copurify with affinity-tagged SSB (24). In gel shift experiments, RecJ binding was enhanced to ssDNA when it was SSB-coated, producing a novel species suggestive of a co-complex of SSB and RecJ. During processive ssDNA degradation, RecJ must displace the tightly bound SSB to move ssDNA into its cleft, a process that could be facilitated by specific interactions between the two proteins. The C-terminal domain of RecJ, which is not essential for the nuclease activity of Tth RecJ and leads to the aggregation of RecJ (21), is a good candidate for the site of SSB interaction.
In vivo, the interplay of that bind ssDNA proteins may determine whether ssDNA is fated for RecJ-mediated degradation or for RecA-mediated strand exchange. Binding of SSB would enhance RecJ degradation; binding of RecA would prevent such degradation and promote recombinational interactions. Both SSB and RecJ have been shown to enhance in vitro RecA strand exchange by sequestering or degrading the ssDNA displaced during the process, which may compete for pairing with the invading strand (13,14).
Acknowledgments
We thank Earl Gillespie and Paul Morrison of the Molecular Biology Core Facility at the Dana Farber Cancer Institute for synthesis of DNA oligonucleotides and Roger McMaken of Johns Hopkins for providing us with SSB antibody. This work was supported by grants from the General Medical Sciences Institute of the National Institutes of Health, RO1 GM43889 to S.T.L. and training grants T32 GM07122 to E.S.H and T32 GM07596-27 to N.S.P. Funding to pay the Open Access publication charges for this article was provided by GM43889.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lovett S.T., Kolodner R.D. Identification and purification of a single-stranded-DNA-specific exonuclease encoded by the recJ gene of Escherichia coli. Proc. Natl Acad. Sci. USA. 1989;86:2627–2631. doi: 10.1073/pnas.86.8.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dianov G., Sedgwick B., Daly G., Olsson M., Lovett S., Lindahl T. Release of 5′-terminal deoxyribose-phosphate residues from incised abasic sites in DNA by the Escherichia coli RecJ protein. Nucleic Acids Res. 1994;22:993–998. doi: 10.1093/nar/22.6.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lovett S.T., Clark A.J. Genetic analysis of the recJ gene of Escherichia coli K-12. J. Bacteriol. 1984;157:190–196. doi: 10.1128/jb.157.1.190-196.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Viswanathan M., Lovett S.T. Single-strand DNA-specific exonucleases in Escherichia coli: roles in repair and mutation avoidance. Genetics. 1998;149:7–16. doi: 10.1093/genetics/149.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Razavy H., Szigety S.K., Rosenberg S.M. Evidence for both 3′ and 5′ single-strand DNA ends in intermediates in chi-stimulated recombination in vivo. Genetics. 1996;142:333–339. doi: 10.1093/genetics/142.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miesel L., Roth J.R. Evidence that SbcB and RecF pathway functions contribute to RecBCD-dependent transductional recombination. J. Bacteriol. 1996;178:3146–3155. doi: 10.1128/jb.178.11.3146-3155.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burdett V., Baitinger C., Viswanathan M., Lovett S.T., Modrich P. In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair. Proc. Natl Acad. Sci. USA. 2001;98:6765–6770. doi: 10.1073/pnas.121183298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viswanathan M., Burdett V., Baitinger C., Modrich P., Lovett S.T. Redundant exonuclease involvement in Escherichia coli methyl-directed mismatch repair. J. Biol. Chem. 2001;276:31053–31058. doi: 10.1074/jbc.M105481200. [DOI] [PubMed] [Google Scholar]
- 9.Sutera V.A., Han E.S., Rajman L.A., Lovett S.T. Mutational analysis of the RecJ exonuclease of Escherichia coli: identification of phosphoesterase motifs. J. Bacteriol. 1999;181:6098–6102. doi: 10.1128/jb.181.19.6098-6102.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aravind L., Koonin E.V. A novel family of predicted phosphoesterases includes Drosophila prune protein and bacterial RecJ exonuclease. Trends Biochem. Sci. 1998;23:17–19. doi: 10.1016/s0968-0004(97)01162-6. [DOI] [PubMed] [Google Scholar]
- 11.Rajman L.A., Lovett S.T. A thermostable single-strand DNase from Methanococcus jannaschii related to the RecJ recombination and repair exonuclease from Escherichia coli. J. Bacteriol. 2000;182:607–612. doi: 10.1128/jb.182.3.607-612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamagata A., Kakuta Y., Masui R., Fukuyama K. The crystal structure of exonuclease RecJ bound to Mn2+ ion suggests how its characteristic motifs are involved in exonuclease activity. Proc. Natl Acad. Sci. USA. 2002;99:5908–5912. doi: 10.1073/pnas.092547099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corrette-Bennett S.E., Lovett S.T. Enhancement of RecA Strand-transfer activity by the RecJ exonuclease of Escherichia coli. J. Biol. Chem. 1995;270:6881–6885. doi: 10.1074/jbc.270.12.6881. [DOI] [PubMed] [Google Scholar]
- 14.Lavery P.E., Kowalczykowski S.C. A postsynaptic role for single-stranded DNA-binding protein in recA protein-promoted DNA strand exchange. J. Biol. Chem. 1992;267:9315–9320. [PubMed] [Google Scholar]
- 15.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 16.Colbert T., Lee S., Schimmack G., Hahn S. Architecture of protein and DNA contacts within the TFIIIB–DNA complex. Mol. Cell. Biol. 1998;18:1682–1691. doi: 10.1128/mcb.18.3.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ausubel F.A., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K. Short Protocols in Molecular Biology. NY: Greene Publishing and Wiley-Interscience; 1989. [Google Scholar]
- 18.Zhu L., Hailigan B.D. Characterization of a 3′-5′ exonuclease associated with VDJP. Biochem. and Biophys. Res. Comm. 1999;259:262–270. doi: 10.1006/bbrc.1999.0774. [DOI] [PubMed] [Google Scholar]
- 19.Subramanian K., Rutvisuttinunt W., Scott W., Myers R.S. The enzymatic basis of processivity in lambda exonuclease. Nucleic Acids Res. 2003;31:1585–1596. doi: 10.1093/nar/gkg266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breyer W., Matthews B. Structure of Escherichia coli exonuclease I suggests how processivity is achieved. Nature Struct. Biol. 2000;7:1125–1128. doi: 10.1038/81978. [DOI] [PubMed] [Google Scholar]
- 21.Yamagata A., Masui R., Kakuta Y., Kuramitsu S., Fukuyama K. Overexpression, purification and characterization of RecJ protein from Thermus thermophilus HB8 and its core domain. Nucleic Acids Res. 2001;29:4617–4624. doi: 10.1093/nar/29.22.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandigursky M., Franklin W.A. Escherichia coli Single-stranded DNA binding protein stimulates the DNA deoxyribophosphodiestrease activity of exonuclease I. Nucleic Acids Res. 1994;22:247–250. doi: 10.1093/nar/22.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandigursky M., Mendez F., Bases R.E., Matsumoto T., Franklin W.A. Protein-protein interactions between the Escherichia coli single-stranded DNA-binding protein and exonuclease I. Radiat. Res. 1996;145:619–623. [PubMed] [Google Scholar]
- 24.Butland G., Peregrin-Alvarez J.M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]