Abstract
Objectives
Establishing more effective treatment of pancreatic cancer requires an understanding of the molecular events leading to the onset and progression of this disease. The biology of tumorigenesis may be better understood if cell type–specific genes in the pancreas are more recognized. This recognition may be as important as discovering a disease-responsible gene. Identification of a ductal epithelium–specific gene can contribute not only to our knowledge of pancreatic tumorigenesis, tumor marker discovery, and effective drug targeting but also is crucial for making a reliable animal model.
Methods
We used the x-Profiler engine online to compare the SAGE (Serial Analysis of Gene Expression) libraries derived from 2 short-term cultures of normal human ductal epithelial cells from the pancreas against 34 other SAGE libraries generated from other normal human tissues to identify the best candidate gene specific for the ductal epithelium of the pancreas.
Results
We identified 3 genes, ribosomal protein L38 (RPL38), uridine phosphorylase (UPP1), and FOS-like antigen-1 (FOSL1), predominantly expressed in the pancreatic ductal epithelium. The expression patterns of these 3 genes were confirmed by virtual Northern analysis, semi-quantitative RT-PCR, and in situ hybridization.
Conclusion
Although the expressions of these 3 genes are not completely restricted to the ductal epithelium of the pancreas, we showed that they have more specific expression patterns than CK19 and MUC1. We also demonstrated that all 3 genes are highly expressed in a panel of pancreatic cancer cell lines and can potentially be useful in tumor targeting or as tumor markers.
Keywords: SAGE, pancreas-specific, RPL38, FOSL1, UPP1
Pancreatic cancer is associated with poor prognosis and short-term survival and is often diagnosed after the cancer has metastasized. An effective treatment of the advanced disease is still unavailable.1,2 To unravel the molecular basis of this disease and to identify the disease-causing genes, it is essential to define a subset of genes that are specifically expressed in each cell type of the pancreas. This will not only help us to understand the tissue development and disease biology, but it also is useful for targeting and treating the disease. Tissue-specific genes are valuable for making reliable animal models in that successful creation of transgenic or conditional knockout mice largely depends on the tissue specificity of a promoter.
Although most human pancreatic neoplasms have a ductal morphology,3 none of the currently available promoter sequence drives expression in a pancreatic ductal epithelium–specific manner. The commonly used promoters favored by the pancreatic cancer research community fall into 3 categories: First, the promoter is pancreas specific but not restricted to the ductal epithelium in the pancreas. The elastase and p48 promoters are prime examples of this first category. Elastase 1 (ELA1) expression is predominantly found in the acinar cells of the pancreas.4 ELA1 promoter induces expression in the acinar and/or islet cells in additional to the ductal epithelium in the pancreas. P48 is part of hetero-oligomeric transcription factor PTF1 that directs the expression of genes in the exocrine pancreas and is required for committing cells to exocrine fate.5 P48 is expressed in the pancreatic primordium in the embryo and is restricted to the exocrine lineage in adult pancreas.6 Second, the promoter is restricted to the ductal epithelium in the pancreas but is not specific to the pancreas. Cytokeratin 19 (CK19) and mucin 1 (MUC1) genes are representatives of this second category. Cytokeratins serve as immunocytochemical markers for epithelial cells,7 and cytokeratins 7, 19, and 20 are expressed in neonatal and adult pancreata. In the pancreas, intense expressions of cytokeratins 19 and 20 are localized to proliferative ductal epithelial and associated islet cells.8 However, CK19 is also expressed in the epithelium of other organs.9,10 Third, the promoters drive expression in both the exocrine and endocrine lineages of the neonatal pancreas but are inactive in exocrine cells of the adult pancreas. For example, Pdx1-expressing cells are detected early during embryogenesis and subsequently give rise to differentiated ductal, islet, and acinar cells.11–13 In adult pancreas, Pdx-1 is mostly in mature β cells and at a lower level in mature acinar cells.14,15
A public database, SAGEmap, was created as a component of the Cancer Genome Anatomy Project (CGAP) to provide a central location for depositing, retrieving, and analyzing human gene expression data.16,17 This database uses serial analysis of gene expression to quantify transcript levels in both malignant and normal human tissues. By accessing SAGEmap (http://www.ncbi.nlm.nih.gov/SAGE), the user can compare transcript populations between any of the posted libraries.
In search of a pancreatic ductal epithelium-specific gene or a gene whose promoter is potentially more specific than what the existing promoters such as CK19, MUC1, and ELA1 can offer us, we used the SAGE database to compare a large group of normal tissues with short-term cultures of pancreatic ductal epithelial cultures. Similar use of this database has resulted in the identification of thyroid- and breast-specific genes.18,19
METHODS
Cell Lines and Tissues
The normal pancreatic epithelial cell line HPDE6 was kindly provided by Dr. M. S. Tsao,20 and pancreatic cancer cell lines (Panc1, MiaPaCa2, AsPC-1, BxPC3, Hs766T, Colo357, CaPan-1, CFPAC-1) were obtained from the American Type Culture Collection (ATCC) and maintained according to the ATCC guidelines. The low-passage pancreatic carcinoma cell lines (PL4, PL8, and PL9) were kindly provided by Dr. Elizabeth M. Jaffee.21 All cell lines were grown in a monolayer in Dulbecco’s modified Eagle media (Invitrogen Corp.), supplemented with 10% fetal bovine serum and 1% Pen/Strep (Invitrogen Corp.) and maintained at 37°C in air containing 5% CO2. Normal tissues of the pancreas, spleen, kidney, breast, heart, colon, placenta, liver, lung, and ovary were obtained from the archives of the Division of Surgical Pathology at The Johns Hopkins Hospital. In general, the tissues were derived from grossly normal areas of specimens removed for pathologic process in that organ or in an attached organ.
SAGE Database
The online SAGE database included 141 human SAGE libraries at the time of analysis. These libraries consisted of 2,319,815 total tags, 460,638 unique tags, and 87,493 UniGene clusters. The database is on the Web site of the National Center for Biotechnology Information and is open to public use. SAGE was chosen to create a comprehensive quantitative expression database for the CGAP because this assay provides absolute transcript numbers in a digital format and can be easily adapted to provide statistical comparisons of data from multiple laboratories. The basis of SAGE is to count expressed transcripts by sequencing a 10-bp ‘‘tag’’ of a gene, which is normally sufficient for gene identification. Custom tools were created for this Web site to analyze the expression of a single gene in multiple tissues or to compare the full expression pattern between tissues.22,23
Multiplex RT-PCR
RNA was isolated from HPDE6 and pancreatic cancer cell lines using Trizol (Invitrogen Corp.) according to the manufacturer’s instructions. cDNAs were derived using dNTP mix (5 mmol/L each of dNTP), oligo (dT)16, and 2.5 U of SuperScript II RNase H reverse transcriptase (Invitrogen Corp.) at 37°C for 1 hour. For semi-quantitative PCR, the linear range of the PCR product was determined for each target gene by varying PCR cycles. The PCR cycles of RPL38, FOSL1, and UPP1 in linear range were determined to be 18, 24 and 26, respectively. G3PDH was used as the internal control in all the multiplex RT-PCR. Human Multiple Tissue cDNA panels I and II (BD-Clontech) were used for the comparison of the gene expression in different tissues. cDNAs were amplified by PCR using specific primer pairs and 2.5 U of AmpliTaq DNA polymerase in a 50-μL reaction mix (cycles of denaturation at 95°C for 2 minutes, annealing at 60°C for 30 seconds, and extension at 72°C for 60 seconds). All the primer sequences used in the PCR and in situ were listed in Table 1.
TABLE 1.
Primer Sequences Used for PCR and In Situ Hybridization
| UPP1-F | ACTGCCCAGGTAGAGACTATC |
| UPP1-R | CTGCACCAGCTTCTTGTTAAG |
| FOSL1F | AAGCATCAACACCACCATGAGTG |
| FOSL1R | GGAGATACAAGGTACAG |
| RPL38-F | GCCATGCCTCGGAAAATTG |
| RPL38-R | GTGTTCTGGATTCATATTCAAG |
| T7UPP1-F | CTAATACGACTCACTATAGGG ACTGCCCAGGTAGAGACTATC |
| T7UPP1-R | CTAATACGACTCACTATAGGG CTGCACCAGCTTCTTGTTAAG |
| T7FOSL1-F | CTAATACGACTCACTATAGGG AAGCATCAACACCACCATGAGTG |
| T7FOSL1-R | CTAATACGACTCACTATAGGG GGAGATACAAGGTACAG |
| T7RPL38-F | CTAATACGACTCACTATAGGG GCCATGCCTCGGAAAATTG |
| T7RPL38-R | CTAATACGACTCACTATAGGGCAGTTTCCAATCAGTGTGTCTG |
Preparation of Riboprobes for Nonradioactive In Situ Hybridization
PCR was used to generate 400- to 500-bp sized DNA templates for antisense or sense riboprobes by incorporating the T7 promoter (5′CTAATACGACTCACTATAGGG 3′) into the 5′ end of the antisense or sense primer. Antisense and sense riboprobes were prepared by using a DIG RNA labeling kit (Boehringer Mannheim) according to the manufacturer’s instructions. Probes were stored in TE buffer and used within 2 months.
In Situ Hybridization Procedure
All steps before and during hybridization were conducted under RNase-free conditions. Sections were deparaffinized with xylene (5 minutes × 2 times) and rehydrated through descending ethanol concentrations (100%, 90%, 80%, 70%, and 50%; 5 minutes each). The sections were treated with proteinase K, 15 μg/mL in TBS, pH 7.4 at 37°C for 30 minutes. The slides were incubated with hybridization solution (DAKO, cat. no. S3304), which contained approximately 0.5-μg/mL DIG-labeled probes. On each slide, 300-μL hybridization solutions were applied. Slides were then incubated in a humidified chamber at 40°–50°C for 16 hours. After hybridization, slides were subsequently washed with 2 × SSC at 45°C (5 minutes × 2 times) and treated with 250RNase A/T1 cocktail (Ambion) diluted 1/35 in 2 × SSC 37°C for 30 minutes. Slides were then washed with 50% (vol/vol) formamide/2 × SSC at 42°C (20 minutes × 2 times) and 0.08 × SSC. The slides were then incubated with 5% (wt/vol) blocking reagent (Boehringer Mannheim) in buffer 1 (TBS-T) at room temperature for 30 minutes. The slides were incubated in a humidified chamber overnight at 4°C with sheep anti-DIG antibody conjugated with horseradish peroxidase (Boehringer Mannheim), diluted to 1:100 in buffer 1 containing 1% blocking reagent. Slides were washed with TBS (5 minutes) at RT. After washing with TBS, slides were incubated with biotin and then with streptavidin and developed with DAB (DAKO Gen point kit). Slides were counterstained with hematoxylin, dehydrated with alcohol, cleared with xylene, and covered with a coverslip before examination.
RESULTS
Identification of Novel Pancreatic Ductal Epithelium-Specific Genes From SAGEmap
We used the x-Profiler engine online to compare the SAGE libraries derived from 2 short-term cultures of normal human ductal epithelial cells from the pancreas (group A) against 34 other SAGE libraries generated from other normal human tissues to identify the best candidate gene specific for the ductal epithelium of the pancreas. The 34 SAGE libraries included bulk tissues of heart, liver, brain, breast, kidney, peritoneum, prostate, lung, spinal cord, and muscle; cells lines derived from embryos, skin, prostate, blood, and vascular tissue; and epithelia of mammary gland, colon, ovary, and stomach (group B) (Table 2). Except in 2 incidences, the tag numbers of these libraries were within the 10,000—100,000 range (Table 2). The x-Profiler normalized the tag numbers to count per million when comparing two or more libraries with discordant tag numbers.16 222,441 unique tags were generated from groups A and B combined. We computed the online x-Profiler program for tags that were represented 10-fold greater or more in group A than in group B.
TABLE 2.
List of SAGE Libraries Used to Identify Pancreatic Ductal Epithelium-Specific Candidate Genes
| Library Name | Source | No. of Unique Tags | Total No. of Tags | No. of Sequences |
|---|---|---|---|---|
| Group A | ||||
| 1. H126 | Pancreas epithelium ductal normal cell line short-term culture | 12479 | 32512 | 940 |
| 2. HX | Pancreas epithelium ductal normal cell line short-term culture | 12485 | 32226 | 925 |
| Group B | ||||
| 3. 293-CTRL | Normal kidney cell line embryo | 18676 | 43527 | 1945 |
| 4. BB542_whitematter | Normal brain | 32026 | 94876 | 3867 |
| 5. Br_N | Normal mammary gland epithelium ductal antibody purified | 16297 | 37642 | 1754 |
| 6. Chen_Normal_Pr | Normal prostate | 20776 | 66483 | 2884 |
| 7. Duke_40N | Normal mammary gland epithelium ductal | 3618 | 7165 | 372 |
| 8. Duke_48N | Normal mammary gland epithelium ductal | 5725 | 12142 | 770 |
| 9. Duke_BB542_normal_cerebellum | Normal cerebellum | 24932 | 58826 | 2403 |
| 10. Duke_HMVEC | Normal vascular endothelium cell line short-term culture | 20819 | 52579 | 1967 |
| 11. Duke_Kidney | Normal kidney | 16626 | 41857 | 1995 |
| 12. Duke_leukocyte | Normal white blood cells | 15245 | 48523 | 2049 |
| 13. Duke_precrisis_fibroblasts | Normal cell line intestine fibroblast | 4741 | 8851 | 890 |
| 14. Duke_thalamus | Normal bulk thalamus | 16852 | 48548 | 3170 |
| 15. HMEC_B41 | Normal mammary gland epithelium | 955 | 1430 | 142 |
| 16. HOSE_4 | Normal short-term culture ovary epithelium | 16317 | 48552 | 2009 |
| 17. IOSE29_11 | Normal cell line ovary epithelium | 18143 | 48586 | 1685 |
| 18. NC1 | Normal colon epithelium | 17913 | 50179 | 2640 |
| 19. NC2 | Normal colon epithelium | 16569 | 49593 | 3265 |
| 20. NHA_5th | Normal brain | 21199 | 52261 | 2177 |
| 21. PERITO-13 | Normal peritoneum | 23146 | 54096 | 1587 |
| 22. PR317_normal_prostate | Normal microdissected prostate | 18876 | 59553 | 1520 |
| 23. TSU | Normal cell line prostate | 6238 | 11377 | 607 |
| 24. Mammary_epithelium | Normal antibody purified mammary gland epithelium ductal | 18576 | 49281 | 2136 |
| 25. Normal_cerebellum | Normal cerebellum bulk | 24729 | 51280 | 2099 |
| 26. Normal_lung | Normal lung | 24962 | 89143 | 2404 |
| 27. Normal_pool_6th | Normal brain | 25399 | 63208 | 2780 |
| 28. Normal_prostate | Normal prostate | 6719 | 13302 | 1271 |
| 29. Breast_myoepithelial | Normal myoepithelial antibody purified mammary gland | 26994 | 58444 | 1792 |
| 30. Normal_gastric_body_epithelial | Normal myoepthelial mammary gland | 9137 | 25302 | 1691 |
| 31. Normal_liver | Normal liver | 15722 | 66861 | 2113 |
| 32. Normal_pediatic_cortex_H1571 | Normal cortex | 24356 | 77968 | 2072 |
| 33. Normal Human Muscle_old | Muscle | 12836 | 53853 | NONE |
| 34. Normal Human Muscle_young | Muscle | 11382 | 53875 | NONE |
| 35. AKUnaffected_Epidermis_skin | Skin | 5861 | 11563 | NONE |
| 36. Normal spinal cord | Normal spinal cord | 20058 | 55422 | 1544 |
We identified more than 20 tags with reliable gene assignments to represent more than 10 times greater in number of transcripts in group A than in group B. Six of these candidate genes, regulator of G-protein signaling 4 (RGS4), ribosomal protein L38 (RPL38), uridine phosphorylase (UPP1), FOS-like antigen-1 (FOSL1), DEK oncogene (DEK), and zinc finger protein 36, C3H type-like 1 (ZFP36L1), were selected for further analysis based on their tag number and virtual Northern blot supplied by the National Institutes of Health (NIH) (Table 3). The tag numbers and virtual Northern blot showed that RPL38 had the highest number of transcripts in the 2 pancreatic ductal epithelial cultures among the 6 genes. The tag numbers of RGS4, DEK, and FOSL1 were lower compared with RPL38 but more specific to the pancreatic ductal epithelium (Table 3). Since none of the 6 genes appeared to be specific to the ductal epithelium of the pancreas at the first glance, to help us better assess the potential usefulness of these 6 genes, the tag numbers and virtual Northern blots of MUC1 and CK19 were extracted from the SAGE database for comparison. The results showed that the tag numbers of the 6 candidate genes were significantly higher in the 2 pancreatic ductal epithelial cultures than MUC1 and CK19 by at least 9-fold (DEK) to over 300-fold (RPL38) (Table 3). While the highest expression level of each of our 6 candidate genes resided in the ductal epithelium of the pancreas (group A), both MUC1 and CK19 were be found more highly expressed in other tissue types (group B) (Table 3). The expression patterns of the MUC1 and CK19 were equally if not more unspecific to the pancreas compared with our candidate genes, with significantly much lower tag numbers in the 2 pancreatic ductal epithelial cultures (Table 3).
TABLE 3.
The Expression Profiles of the Six Pancreatic Ductal Epithelium-specific Candidate Genes Resulted From the Analysis of the SAGE Libraries and Their Comparisons to MUC1 and CK19
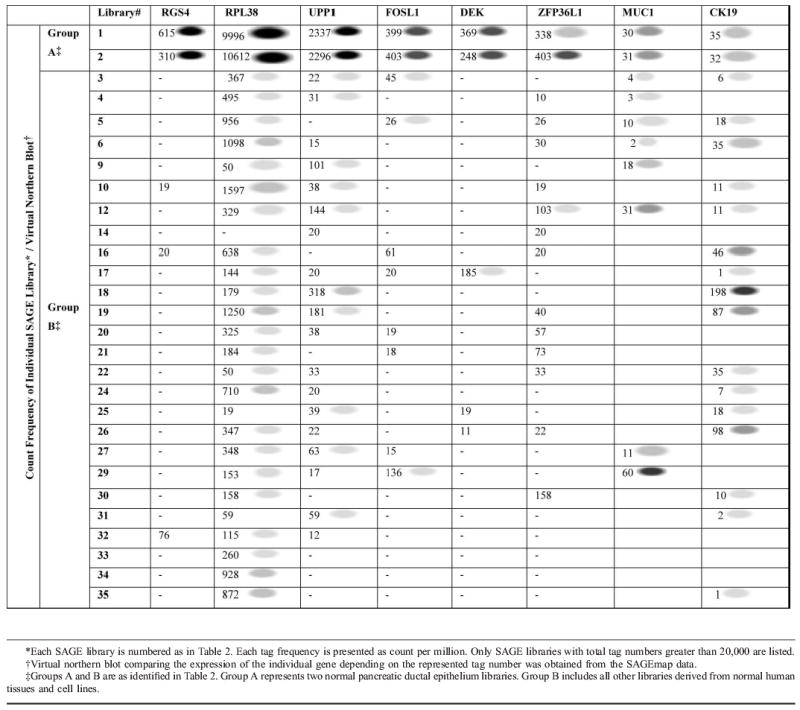
RPL38 Is Abundantly Expressed in the Ductal Epithelium of the Pancreas
Each of the 6 candidate genes was then further verified by semi-quantitative RT-PCR on tissue cDNA panels to confirm their predicted expression. The multiplex RT-PCR analyses were performed on Human Multiple Tissue cDNA panels I and II. An immortalized human ductal epithelium cell line, HPDE6,20 and a pancreatic adenocarcinoma cell line, Panc-1, were used as positive controls.
We used the exponential log phase of the PCR cycle to compare the difference of the expression of various tissues. Similar to the tag number analysis, PCR results showed that the expressions of RPL38, UPP1, and FOSL1 were far more abundant in the ductal epithelium of the pancreas compared with other tissues. There was no or less expression found in the nonpancreatic tissue samples (Fig. 1). Consistent with the tag number analysis, the expression of RPL38 (18 PCR cycles) was higher than those of UPP1 and FOSL1 in the pancreas (26 and 24 PCR cycles, respectively). The expression of RGS4, DEK, and ZFP36L1 was not higher in the ductal epithelium of the pancreas than in other tissue types (Fig. 1). Semi-quantitative RT-PCR analyses were also performed using CK19- or MUC1-specific primers for comparison. The expression profiles of the RPL38, UPP1, and FOSL1 in tissues appeared to be more restricted to pancreatic ductal epithelium than those of CK19 and MUC1 (Fig. 1). Again, consistent with the tag number analysis, the expression levels of CK19 and MUC1 (30 PCR cycles each) were significantly lower in the ductal epithelium of the pancreas than those observed for RPL38, FOSL1, and UPP1.
FIGURE 1.
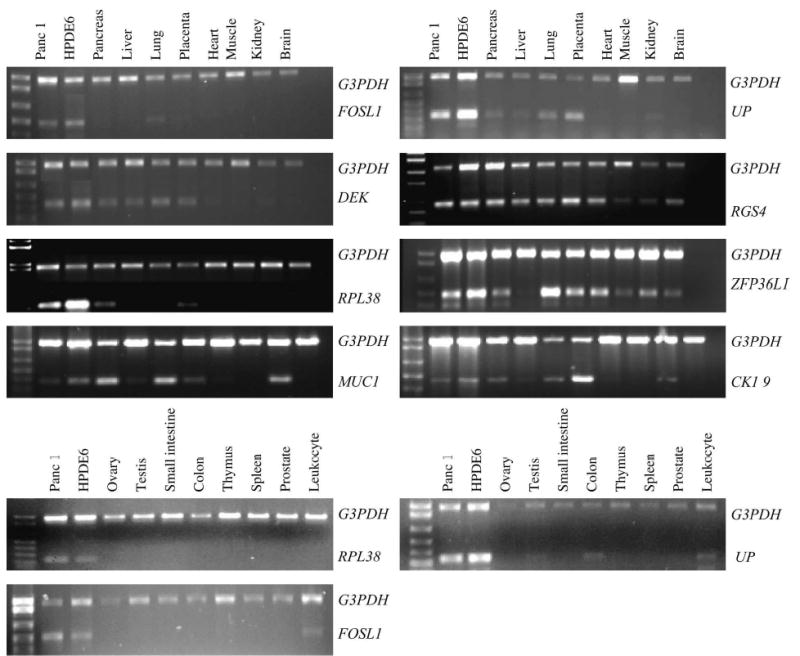
The Multiplex RT-PCR analysis of the candidate genes in the Human Multiple Tissue cDNA panels. Panc-1 pancreatic cancer cell line and HPDE6 immortalized normal human pancreatic ductal epithelial cells served as positive controls for each PCR reaction. Multiplex RT-PCRs were performed, with G3PDH primers serving as an internal control. A previously determined log phase PCR cycle for individual gene expression was performed for each gene.
In Situ Analysis of RPL38, UPP1, and FOSL1 Expression
We used in situ hybridization, which would allow us to identify cell type–specific expression in each tissue, to further verify the RT-PCR results. In situ hybridization with a digoxigenin-labeled probe specific to RPL38, UPP1, or FOSL1 was performed on a panel of paraffin-embedded human normal tissues including pancreas, spleen, kidney, breast, heart, colon, placenta, liver, lung, and ovary. We found that RPL38 was highly expressed in the ductal epithelium of the pancreas, with less expression in the acinar and islet cells. High expression of RPL38 was also detected in the stroma of the ovary. Low expression was observed in the epithelial lining of the colon and breast (Fig. 2, Table 4). No significant expression was detected in the other tissues and cell types. UPP1 expression was restricted to the pancreas with some expression in the ovary. The other tissues showed no or very little expression with the in situ gene-specific probe to UPP1 (Table 4). FOSL1 expression was mainly detected in the pancreas and epithelium of the colon and lung. However, the RNA messages detected by the gene-specific probes to FOSL1 and UPP1 were low in abundance compared with RPL38, as might have been predicted by the SAGE and RT-PCR analyses (Table 3, Fig. 1). Previous studies showed that MUC1 and CK19 are less preferably expressed in the pancreatic ductal epithelium than RPL38, UPP1, and FOSL1 (Table 4).10,24–31 In addition to those tissues listed in Table 4, CK19 is also highly expressed in the bladder, skin, stomach, uterus, and some glands.10,29,30 MUC1 is also abundantly expressed in the uterus, prostate, and stomach.24–26 However, it is worth noting that CK19 is the only candidate gene not expressed by acinar cells in the pancreas.
FIGURE 2.

The expression of RPL38 in normal human tissues by in situ hybridization. The paraffin sections of human tissues were hybridized with a digoxigenin-labeled probe corresponding to RPL38 cDNA sequences. High expression of RLP38 was detected by the antisense probe in the ductal epithelium of the pancreas (A). Low expression was also observed in the acinar cells of the pancreas (B), the epithelial ducts in the breast (D), and at the epithelium of the colon (E). No expression was detected in the kidney (F). Arrows point to epithelial cells that expressed RLP38 mRNA. Each serial tissue section was also incubated with the sense probe as the negative control. C, A representative of the sense control. No hybridization was detected in the sense control of the pancreas (C).
TABLE 4.
Expression of RPL38, FOSL1, and UPP1 in Normal Human Tissues Using In Situ Hybridization and Their Comparisons with MUC1 and CK19
| Tissues | RPL38 | FOSL1 | UPP1 | MUC1* | CK19* |
|---|---|---|---|---|---|
| Pancreas | |||||
| Acini | +++ | + | ++ | +/− | − |
| Islet cells | ++ | − | + | − | +/− |
| Duct cells | ++++ | +/− | ++ | + | +++ |
| Spleen | +/− | + | |||
| White pulp | − | − | − | ||
| Red pulp | − | − | − | ||
| Kidney | ++ | ||||
| Glomeruli (epithelial) | +/− | − | +/− | +++ | |
| Tubules (epithelial) | +/− | − | − | +++ | |
| Interstitium (among blood vessels) | − | − | − | − | |
| Adrenal (cortex) | − | − | − | ||
| Adrenal (medulla) | − | − | − | − | |
| Breast | N/A | ||||
| Duct | + | − | − | ++ | |
| Lobules | + | +/− | − | ++ | |
| Heart | − | +/− | − | +/− | N/A |
| Colon | |||||
| Epithelium | ++ | ++ | + | +/− | ++ |
| Placenta | NA | ||||
| Trophoplast | − | − | − | + | |
| Villi | − | − | − | − | |
| Liver | − | ||||
| Hepatocytes | − | + | − | − | |
| Bile duct | +++ | + | − | ++ | |
| Lung | |||||
| Respiratory epithelium | ++ | − | − | ++ | ++ |
| Pneumocytes | − | − | − | − | − |
| Interstitium | − | + | − | − | − |
| Ovary | ++ | + | |||
| Stroma | +++ | − | ++ | ||
| Blood vessel | − | − | + | ||
| Epithelium | +/− | − | ++ | − | |
RPL38, UPP1, and FOSL1 Are Highly Expressed in Pancreatic Cancer Cell Lines
A previous publication has shown that Rpl38 is one of the ribosomal proteins that were elevated in cancer cell lines.32 Similarly, FOSL1, which belongs to the nontransforming family of the c-FOS group of oncogenes, was elevated in tumor cell lines.33 Although, UPP1’s role in uridine synthesis had been well described, recent evidence suggested that UPP1 is directly regulated by p53 and is increased in surgical tumor tissues.34,35 These data led us to investigate the possible roles of these genes in pancreatic cancer in addition to their abundant expression in the nontransformed ductal epithelium of the pancreas. We studied the expression of RPL38, FOSL1, and UPP1 in the 11 pancreatic cancer cell lines, using multiplex RT-PCR. cDNA from normal pancreatic tissue was used as a control. We found all 3 genes were equally or more abundantly expressed in many of the 11 pancreatic cancer cells than in the normal control (Fig. 3).
FIGURE 3.
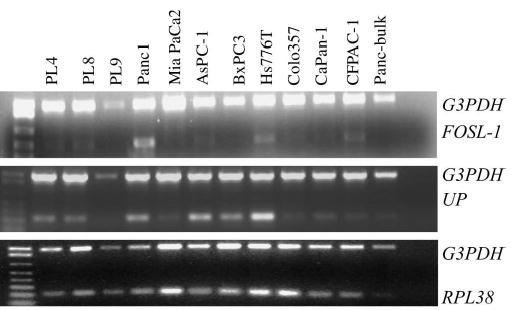
The Multiplex RT-PCR analysis of expression of RPL38, FOSL1, and UPP1 in pancreatic cancer cell lines. Bulk pancreas cDNA obtained from a normal pancreas was used as nontransformed control. RPL38, FOSL1, and UPP1 were abundantly expressed in multiple pancreatic cancer cell lines.
DISCUSSION
Differences in only 2% of the 15,000 distinct transcripts of an individual cell are sufficient to differentiate cells of closely related origins.18 Recognizing the distinguishing subset of the genes that is specific to each cell type is valuable for understanding disease origin and identifying potential disease markers. Such markers are especially important for pancreatic cancer, a disease that currently lacks useful assays for early detection.
A tissue-specific promoter is critical in applications such as drug delivery and gene therapy. Reliable animal models, whether transgenic or conditional knockout, are also largely dependent on the specificity of the promoter used for gene expression. Transgenic mice utilizing CK19 or ELA1 to drive oncogene expression have resulted in the development of phenotypes similar to pancreatic metaplasia and acinar carcinoma but not ductal adenocarcinoma,9,10,36,37 while recent utilization of p48 and PDX-1 promoters have resulted in successful mouse models for pancreatic ductal adenocarcinomas.38,39 The successful application of p48 and PDX-1 promoters in mouse modeling validates the notion that mouse models of human pancreatic ductal adenocarcinoma are feasible given the right combination of promoters and targeted gene.
Here we identified 3 genes that are predominantly expressed in the ductal epithelium of the pancreas using the CGAP SAGE database. Although their expression patterns are not completely restricted to the ductal epithelium of the pancreas, the combination of their abundance in the pancreas and their restricted transcription pattern makes them attractive genes for future studies. RPL38, the most abundant of the 3, has at least 6.2-fold higher expression in the pancreatic ductal epithelial cultures than other tissue types by tag analysis (9996 counts in H126, 1597 counts in Duke_HMVEC=6.2), while MUC1 and CK19 are expressed higher elsewhere than in the pancreatic ductal epithelium (Table 3). Furthermore, the expression level of RPL38 in the pancreatic ductal epithelium is also more than 300-fold greater than that of both MUC1 and CK19 (Table 3). These observations were supported by both semi-quantitative RT-PCR analysis and in situ hybridization results (Fig. 1, Table 4). The expression of FOSL1 and UPP1 are 12-and 72-fold higher than both MUC1 and CK19 in the pancreatic ductal epithelium (Table 3). As such, these 3 genes, particularly RPL38, should prove to be useful for diagnosis, drug delivery, and gene targeting.
RPL38 was cloned as one of the ribosomal proteins overexpressed in a colon cancer cell line, HF-29.32 Although it is ubiquitously expressed in some of the intestinal and pancreatic cancer cell lines, its expression pattern in normal tissues has not been broadly studied.32 Several reports show that contrary to the common belief, certain ribosomal proteins are expressed in a tissue-specific manner. By data-mining 6 unbiased human tissue cDNA libraries available on UniGene, Bortoluzzi et al40 found that among 89 ribosomal protein genes expressed in these tissues, 13 of them appeared to be differentially expressed among the tissues. These data show that regulation of the transcriptional activity of ribosomal protein genes in differential human tissues seems to be regulated with less concordance than previously supposed. Differential expression of ribosomal proteins has also been shown at the protein level.41 In studying the expression profiles of 12 ribosomal proteins in the colon by immunohistochemistry, 10 ribosomal proteins were found to be highly expressed in the normal mucosal epithelial, while 2 were not. One of the 2 exceptions, ribosomal protein L7, displayed a specific expression in the secretory granules of the enterochromaffin cells in normal colorectal mucosa.41 L39-2 is another ribosomal protein reported to have testis-specific expression.42 Our study shows that RPL38 is predominantly expressed in the normal ductal epithelium of the pancreas and each of the pancreatic cancer cell lines examined. Together, these data show that differential expression of ribosomal proteins is not uncommon and suggest that ribosomal proteins may perform extraribosomal functions that might require tissue-specific expression. The possible role of the Rpl38 protein in pancreatic development and neoplastic transformation remains to be investigated. However, the highly abundant expression and specificity of RPL38 imply potential roles in both the development and transformation of the ductal epithelium of the pancreas.
Fosl1, formerly known as c-Fos–related protein-1, is a member of the Fos protein family. Members of this group are differentially activated in response to external stimuli and possess different structural features. Importantly, while c-Fos and FosB contain multiple transactivation modules in their N-and C-terminal domains, transactivation domains are absent in the nontransforming Fos protein Fosl1. Although Fosl1 has not been found to trigger oncogenic transformation in vitro, it is expressed abundantly in ras- and src-transformed murine and chicken fibroblasts, in neoplastic thyroid cells, and in highly malignant mouse adenocarcinoma cells.33 Our studies also show that FOSL1 is expressed in multiple pancreatic cancer cell lines at levels that are significantly higher than in the normal bulk pancreas. Although FOSL1 has an established role in bone development,43 specific functions for FOSL1 protein in the maintenance and progression of the transformed state have not yet been identified.
Uridine phosphorylase is responsible for the reversible phosphorylation of uridine to uracil. Uridine, a pyrimidine nucleoside essential for the synthesis of RNA and biomembranes, is a crucial element in the regulation of normal physiologic processes as well as pathologic states.34,35 Uridine plays a role at the clinical level in modulating the cytotoxic effects of fluoropyrimidines in both normal and neoplastic tissues. The concentration of uridine in plasma and tissues is tightly regulated by UPP1.34 The characterization of the promoter region of UPP1 indicated a direct regulation of its expression by the p53 tumor suppressor. The evaluation of human surgical specimens showed elevated UPP1 activity in tumor tissue compared with paired normal tissue, similar to our results.35 Further studies are needed to explore the significance of elevation of UPP1 in tumor tissues.
Although the expressions of UPP1 and FOSL1 are more restricted to the pancreas, their expression levels in the ductal epithelium of the pancreas are much lower than that of RPL38, as indicated by their SAGE tag counts and our semiquantitative RT-PCR. If their expression levels can be increased by manipulating their promoter without changing the tissue specificity, they may be potential candidate genes specific for the pancreas.
Although many genes showed promising SAGE profiles, their tissue-specific expression was not verified by the RT-PCR analyses. There are several reasons why a promising SAGE profile may not be duplicated in RT-PCR analyses. SAGE sequences are usually only single-pass sequenced and therefore have 1% error rate on average, which can either increase or decrease the correct tag count of an already established tag or establish and count a tag that does not really exist. Such sequencing errors are not compensated for by the x-Profiler. Considering that some genes have more than one tag, more than one gene can share the same tags, and only 0.1% of the human genome has well-characterized mRNA/cDNA sequences; these factors can all affect the accuracy of tag-to-UniGene assignments. Another source of discrepancy can come from 3′ ESTs terminating elsewhere than the terminus of the single longest consensus sequence. This will generate incomplete transcripts whose truncated sequences will be wrongly assigned.16 Verification is therefore a critical step in our approach.
In this paper, we identified 3 genes whose expression is predominantly restricted to the pancreatic ductal epithelium. RPL38 is expressed at particularly high levels in the normal ductal epithelium of the pancreas. Although its expression is not completely restricted to the ductal epithelium of the pancreas, RPL38 may possess a more desirable expression pattern than the currently available promoters such as CK19 and MUC1. The remarkably high expression level of RPL38 in the pancreas begs for further investigations. It is possible that a more tissue-specific element can be isolated from the promoter region of RPL38 in future studies. The expression patterns of the other 2 genes, FOSL1 and UPP1, are more specific to the pancreas, although their expression levels are lower compared with that of the RPL38. We also showed that these genes are highly expressed in a panel of pancreatic cancer cell lines. We believe that these genes may prove to be valuable in applications that require a strong pancreas-specific promoter. More importantly, their expression patterns in the normal ductal epithelial of the pancreas and in pancreatic cancer cell lines indicate that these genes may be important in the normal development and tumor formation of pancreatic tissue. Their potential uses as tumor and imaging markers also remain to be further investigated.
Acknowledgments
The authors thank Drs. Richard Baer, Tom Hei, and George Juang for their help in the preparation of the manuscript.
Footnotes
Supported by a Lustgarten Foundation grant, The Michael Rolfe Foundation, NIH grant R03 CA93202, the NCI Temin Award CA95434, and NIH grant GI-SPORE CA62924.
References
- 1.Kern SE, Hruban RH, Hidalgo M, et al. An introduction to pancreatic adenocarcinoma genetics, pathology and therapy. . Cancer Biol Ther. 2002;1:607–613. doi: 10.4161/cbt.307. [DOI] [PubMed] [Google Scholar]
- 2.Jaffee EM, Hruban RH, Canto M, et al. Focus on pancreas cancer. . Cancer Cell. 2002;2:25–28. doi: 10.1016/s1535-6108(02)00093-4. [DOI] [PubMed] [Google Scholar]
- 3.Hruban RH, Adsay NV, Albores-Saavedra J, et al. Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. . Am J Surg Pathol. 2001;25:579–586. doi: 10.1097/00000478-200105000-00003. [DOI] [PubMed] [Google Scholar]
- 4.MacDonald RJ, Swift GH. Analysis of transcriptional regulatory regions in vivo. . Int J Dev Biol. 1998;42:983–994. [PubMed] [Google Scholar]
- 5.Krapp A, Knofler M, Ledermann B, et al. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. . Genes Dev. 1998;12:3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krapp A, Knofler M, Frutiger S, et al. K. WP: The p48 DNA-binding subunit of transcription factor PTF1 is a new exocrine pancreas-specific basic helix-loop-helix protein. . EMBO J. 1996;15:4317–4329. [PMC free article] [PubMed] [Google Scholar]
- 7.Rafiee P, Ho SB, Bresalier RS, et al. Characterization of the cytokeratins of human colonic, pancreatic, and gastric adenocarcinoma cell lines. . Pancreas. 1992;7:123–131. doi: 10.1097/00006676-199203000-00001. [DOI] [PubMed] [Google Scholar]
- 8.Bouwens L, Wang RN, De Blay E, et al. Cytokeratins as markers of ductal cell differentiation and islet neogenesis in the neonatal rat pancreas. . Diabetes. 1994;43:1279–1283. doi: 10.2337/diab.43.11.1279. [DOI] [PubMed] [Google Scholar]
- 9.Brembeck FH, Schreiber FS, Deramaudt TB, et al. The mutant K-ras oncogene causes pancreatic periductal lymphocytic infiltration and gastric mucous neck cell hyperplasia in transgenic mice. . Cancer Res. 2003;63:2005–2009. [PubMed] [Google Scholar]
- 10.Grippo PJ, Sandgren EP. Highly invasive transitional cell carcinoma of the bladder in a simian virus 40 T-antigen transgenic mouse model. . Am J Pathol. 2000;157:805–813. doi: 10.1016/S0002-9440(10)64594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright CV, Schnegelsberg P, De Robertis EM. XlHbox 8: a novel Xenopus homeo protein restricted to a narrow band of endoderm. . Development. 1989;105:787–794. doi: 10.1242/dev.105.4.787. [DOI] [PubMed] [Google Scholar]
- 12.Jonsson J, Carlsson L, Edlund T, et al. Insulin-promoter-factor 1 is required for pancreas development in mice. . Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 13.Song SY, Gannon M, Washington MK, et al. Expansion of Pdx1-expressing pancreatic epithelium and islet neogenesis in transgenic mice overexpressing transforming growth factor alpha. . Gastroenterology. 1999;117:1416–1426. doi: 10.1016/s0016-5085(99)70292-1. [DOI] [PubMed] [Google Scholar]
- 14.Guz Y, Montminy MR, Stein R, et al. Expression of murine STF-1, a putative insulin gene transcription factor, in beta cells of pancreas, duodenal epithelium and pancreatic exocrine and endocrine progenitors during ontogeny. . Development. 1995;121:11–18. doi: 10.1242/dev.121.1.11. [DOI] [PubMed] [Google Scholar]
- 15.Peshavaria M, Gamer L, Henderson E, et al. XIHbox 8, an endoderm-specific Xenopus homeodomain protein, is closely related to a mammalian insulin gene transcription factor. . Mol Endocrinol. 1994;8:806–816. doi: 10.1210/mend.8.6.7935494. [DOI] [PubMed] [Google Scholar]
- 16.Lash AE, Tolstoshev CM, Wagner L, et al. SAGEmap: a public gene expression resource. . Genome Res. 2000;10:1051–1060. doi: 10.1101/gr.10.7.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riggins GJ, Strausberg RL. Genome and genetic resources from the Cancer Genome Anatomy Project. . Hum Mol Genet. 2001;10:663–667. doi: 10.1093/hmg/10.7.663. [DOI] [PubMed] [Google Scholar]
- 18.Moreno JC, Pauws E, van Kampen AH, et al. Cloning of tissue-specific genes using serial analysis of gene expression and a novel computational substraction approach. . Genomics. 2001;75:70–76. doi: 10.1006/geno.2001.6586. [DOI] [PubMed] [Google Scholar]
- 19.Miksicek RJ, Myal Y, Watson PH, et al. Identification of a novel breast-and salivary gland-specific, mucin-like gene strongly expressed in normal and tumor human mammary epithelium. . Cancer Res. 2002;62:2736–2740. [PubMed] [Google Scholar]
- 20.Ouyang H, Mou L, Luk C, et al. Immortal human pancreatic duct epithelial cell lines with near normal genotype and phenotype. . Am J Pathol. 2000;157:1623–1631. doi: 10.1016/S0002-9440(10)64800-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaffee EM, Schutte M, Gossett J, et al. Development and characterization of a cytokine-secreting pancreatic adenocarcinoma vaccine from primary tumors for use in clinical trials. . Cancer J Sci Am. 1998;4:194–203. [PubMed] [Google Scholar]
- 22.Velculescu VE, Zhang L, Vogelstein B, et al. Serial analysis of gene expression. . Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 23.Zhang L, Zhou W, Velculescu VE, et al. Gene expression profiles in normal and cancer cells. . Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 24.Ho SB, Niehans GA, Lyftogt C, et al. Heterogeneity of mucin gene expression in normal and neoplastic tissues. . Cancer Res. 1993;53:641–651. [PubMed] [Google Scholar]
- 25.Xing PX, Lees C, Lodding J, et al. Mouse mucin 1 (MUC1) defined by monoclonal antibodies. . Int J Cancer. 1998;76:875–883. doi: 10.1002/(sici)1097-0215(19980610)76:6<875::aid-ijc18>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 26.Flucke U, Steinborn E, Dries V, et al. Immunoreactivity of cytokeratins (CK7, CK20) and mucin peptide core antigens (MUC1, MUC2, MUC5AC) in adenocarcinomas, normal and metaplastic tissues of the distal oesophagus, oesophago-gastric junction and proximal stomach. . Histopathology. 2003;43:127–134. doi: 10.1046/j.1365-2559.2003.01680.x. [DOI] [PubMed] [Google Scholar]
- 27.Buisine MP, Devisme L, Copin MC, et al. Developmental mucin gene expression in the human respiratory tract. . Am J Respir Cell Mol Biol. 1999;20:209–218. doi: 10.1165/ajrcmb.20.2.3259. [DOI] [PubMed] [Google Scholar]
- 28.Brembeck FH, Moffett J, Wang TC, et al. The keratin 19 promoter is potent for cell-specific targeting of genes in transgenic mice. . Gastroenterology. 2001;120:1720–1728. doi: 10.1053/gast.2001.24846. [DOI] [PubMed] [Google Scholar]
- 29.Ichinose Y, Hashido K, Miyamoto H, et al. Molecular cloning and characterization of cDNA encoding mouse cytokeratin no. 19. . Gene. 1989;80:315–323. doi: 10.1016/0378-1119(89)90295-3. [DOI] [PubMed] [Google Scholar]
- 30.Bader BL, Franke WW. Cell type-specific and efficient synthesis of human cytokeratin 19 in transgenic mice. . Differentiation. 1990;45:109–118. doi: 10.1111/j.1432-0436.1990.tb00464.x. [DOI] [PubMed] [Google Scholar]
- 31.Lussier M, Ouellet T, Lampron C, et al. Mouse keratin 19: complete amino acid sequence and gene expression during development. . Gene. 1989;85:435–444. doi: 10.1016/0378-1119(89)90437-x. [DOI] [PubMed] [Google Scholar]
- 32.Espinosa L, Martin M, Nicolas A, et al. Primary sequence of the human, lysine-rich, ribosomal protein RPL38 and detection of an unusual RPL38 processed pseudogene in the promoter region of the type-1 angiotensin II receptor gene. . Biochim Biophys Acta. 1997;1354:58–64. doi: 10.1016/s0167-4781(97)00124-3. [DOI] [PubMed] [Google Scholar]
- 33.Tulchinsky E. Fos family members: regulation, structure and role in oncogenic transformation. . Histol Histopathol. 2000;15:921–928. doi: 10.14670/HH-15.921. [DOI] [PubMed] [Google Scholar]
- 34.Pizzorno G, Cao D, Leffert JJ, et al. Homeostatic control of uridine and the role of uridine phosphorylase: a biological and clinical update. . Biochim Biophys Acta. 2002;1587:133–144. doi: 10.1016/s0925-4439(02)00076-5. [DOI] [PubMed] [Google Scholar]
- 35.Miyashita H, Takebayashi Y, Eliason JF, et al. Uridine phosphorylase is a potential prognostic factor in patients with oral squamous cell carcinoma. . Cancer. 2002;94:2959–2966. doi: 10.1002/cncr.10568. [DOI] [PubMed] [Google Scholar]
- 36.Sandgren EP, Luetteke NC, Palmiter RD, et al. Overexpression of TGF alpha in transgenic mice: induction of epithelial hyperplasia, pancreatic metaplasia, and carcinoma of the breast. . Cell. 1990;61:1121–1135. doi: 10.1016/0092-8674(90)90075-p. [DOI] [PubMed] [Google Scholar]
- 37.Grippo PJ, Nowlin PS, Demeure MJ, et al. Preinvasive pancreatic neoplasia of ductal phenotype induced by acinar cell targeting of mutant Kras in transgenic mice. . Cancer Res. 2003;63:2016–2019. [PubMed] [Google Scholar]
- 38.Hingorani SR, Petricoin EF, Maitra A, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. . Cancer Cell. 2003;6:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 39.Aguirre AJ, Bardeesy N, Sinha M, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. . Genes Dev. 2003;17:3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bortoluzzi S, d’Alessi F, Romualdi C, et al. Differential expression of genes coding for ribosomal proteins in different human tissues. . Bioinformatics. 2001;17:1152–1157. doi: 10.1093/bioinformatics/17.12.1152. [DOI] [PubMed] [Google Scholar]
- 41.Kasai H, Nadano D, Hidaka E, et al. Differential expression of ribosomal proteins in human normal and neoplastic colorectum. . J Histochem Cytochem. 2003;51:567–574. doi: 10.1177/002215540305100502. [DOI] [PubMed] [Google Scholar]
- 42.Nadano D, Notsu T, Matsuda T, et al. A human gene encoding a protein homologous to ribosomal protein L39 is normally expressed in the testis and derepressed in multiple cancer cells. . Biochim Biophys Acta. 2002;1577:430–436. doi: 10.1016/s0167-4781(02)00445-1. [DOI] [PubMed] [Google Scholar]
- 43.Jochum W, David JP, Elliott C, et al. Increased bone formation and osteosclerosis in mice overexpressing the transcription factor Fra-1. . Nat Med. 2000;6:980–984. doi: 10.1038/79676. [DOI] [PubMed] [Google Scholar]


