Abstract
The bacterial chromosome is organized into multiple independent domains, each capable of constraining the plectonemic negative supercoil energy introduced by DNA gyrase. Different experimental approaches have estimated the number of domains to be between 40 and 150. The site-specific resolution systems of closely related transposons Tn3 and γδ are valuable tools for measuring supercoil diffusion and analysing bacterial chromosome dynamics in vivo. Once made, the wild-type resolvase persists in cells for time periods greater than the cell doubling time. To examine chromosome dynamics over shorter time frames that are more closely tuned to processes like inducible transcription, we constructed a set of resolvases with cellular half-lives ranging from less than 5 min to 30 min. Analysing chromosomes on different time scales shows domain structure to be dynamic. Rather than the 150 domains detected with the Tn3 resolvase, wild-type cells measured over a 10 min time span have more than 400 domains per genome equivalent, and some gyrase mutants exceed 1000.
Introduction
Defining the compact structure of the bacterial chromosomes inside a living cell has been a long-standing goal of molecular genetics. A variety of assays have been developed to probe this structure including visualization by electron microscopy (Delius and Worcel, 1973; Griffith, 1976; Kavenoff and Bowen, 1976; Kavenoff and Ryder, 1976), UV-induced psoralen cross-linking (Pettijohn and Pfenninger, 1980; Sinden et al., 1980; Sinden and Pettijohn, 1981), measuring output from supercoil-sensitive promoters (Miller and Simons, 1993; Pavitt and Higgins, 1993; Spirito et al., 1994), site-specific recombination between distant chromosomal sites (Higgins et al., 1996; Staczek and Higgins, 1998; Higgins, 1999) and sequence-specific DNA localization (Glaser et al., 1997; Webb et al., 1997; Teleman et al., 1998; Niki et al., 2000; Gordon et al., 2004; Viollier et al., 2004). These assays illuminate different aspects of chromosomal behaviour, but understanding the nature of domain structural changes over time has been an especially daunting challenge (Deng et al., 2004).
One method to analyse time-dependent supercoil dynamics involves recombination between res sites spaced at different intervals (Staczek and Higgins, 1998). After the induction of the Tn3 resolvase, cellular DNA can be isolated at different times and the recombinant products can be detected by Southern blot and polymerase chain reaction (PCR) analyses. In stationary-phase cells, a major fraction of the recombination (60%) occurred within 15 min but a significant segment of recombination (40%) happened slowly, i.e. much later than 4 h after induction of the recombination protein and then only after cells were diluted in fresh medium (Staczek and Higgins, 1998). Following recombination by isolating cellular DNA and carrying out either Southern blot or quantitative PCR analyses is tedious and requires a significant investment in both time and reagents, especially if one desires statistically significant data from many chromosomal sites and from bacteria carrying multiple combinations of genetic mutations.
To make site-specific resolution assays time sensitive, we devised a method to alter the life span of the recombinase. A 11-amino-acid sequence was added to the natural C-terminus of the γδRes protein. This sequence corresponds to a degradation ‘tag’ that the Tm or ssrA RNA system appends to proteins when a ribosome stalls at rare codons or when an mRNA lacks a termination codon (Keiler et al., 1996; Hayes et al., 2002a,b). The SsrA tag changed the half-life of resolvase in exponentially growing Escherichia coli or Salmonella enterica serovar typhimurium from well over 40 min to less than 5 min. By modifying amino acid residues in the SsrA tag sequence (Flynn et al., 2001), enzymes were also made with half-lives of 15 and 30 min. Recombination assays performed with this family of time-sensitive enzymes demonstrate a dynamic aspect of chromosome structure. More than 400 domains per genome equivalent are predicted to exist in exponentially growing bacteria rather than the 40 or 150 domains estimated by time-insensitive methods. Moreover, in cells harbouring mutations in gyrase, the number of domains can rise to 1000.
Results
Modifying resolvase half-life
One assay to monitor supercoil diffusion in living bacterial chromosomes is based on site-specific recombination catalysed by the Tn3 resolvase (Higgins et al., 1996; Staczek and Higgins, 1998; Deng et al., 2004). Plasmid pJBRescI has the Tn3 tnpR gene expressed from the bacteriophage λ PL promoter under control of the temperature-sensitive cI857 repressor. In this system, resolvase protein expression is induced by a 10-min temperature shift from 30°C to 42°C. res sites are 114 bp DNA sequences that recombine in the presence of either the Tn3 or γδ Res proteins (Krasnow and Cozzarelli, 1983; Benjamin et al., 1996). A burst of resolvase expression will catalyse site-specific recombination between two directly repeated chromosomal res sites if they reside in the same supercoil domain. If resolvase persists in cells at recombination-active concentrations for long time spans, short-lived barriers could be missed. For example, Deng showed that induction of or repression of a strong promoter caused a domain barrier to appear or disappear over a 20 min period (Deng et al., 2004).
To measure the time span over which resolution activity persists after thermoinduction, we developed a plasmid-based assay. The 5.9 kb pRR51 plasmid carries Amp and Tet resistance genes. Two res sites flank the tet gene so that cells become Tet sensitive after the plasmid undergoes resolution. Cells were induced for Res expression by 10 min of growth at 42°C, then returned to 30°C and maintained in exponential phase. At intervals a 1 ml aliquot of the culture was quickly washed in 10% ice-cold glycerol and subjected to electroporation to introduce plasmid pRR51. Cells were spread onto Amp-containing plates to select for cells transformed with plasmid DNA, and then individual transformants were patched onto Tet-containing plates to measure resolution efficiency. Cells had a reproducible resolution pattern. For the first 60 min after the return to 30°C, Tn3 resolution was close to 100% efficient. Then an exponential decay of resolution activity followed with a half-life of about 40 min (Fig. 1, solid triangles). The Tn3 and γδRes proteins are closely related (81% identity and 93% similarity at the amino acid level) and functionally interchangeable (Benjamin et al., 1996). However, the γδ resolvase was consistently more potent, resolving more than 95% of pRR51 DNA for more than 4 h after induction (Fig. 1, closed circles; and data not shown). In previous work, due to an data entry error, we mistakenly thought that a plasmid we obtained encoded the γδ resolvase, but it was the Tn3 resolvase instead (Higgins et al., 1996; Staczek and Higgins, 1998; Scheirer and Higgins, 2001). We discovered the error by sequencing the plasmid and then tested the γδ resolvase, and based on its increased persistence in vivo, chose to modify the C-terminus of this enzyme for chromosome studies.
Fig. 1.
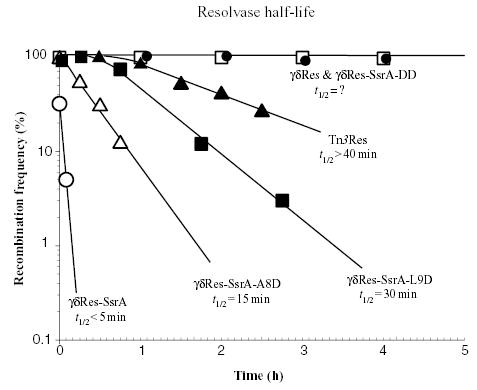
Half-life (t1/2) measurement of modified resolvases. Strains with plasmids expressing different cloned resolvase proteins were grown to mid-log phase (Klett 50) at 30°C, induced at 42°C for 10 min, returned to 30°C and samples were subsequently periodically harvested by centrifugation. After cells were resuspended in 10% ice-cold glycerol, plasmid pRR51 was introduced by electroporation. All experiments were performed in triplicate with at least 300 bacterial colonies examined in each test to determine one standard deviation.
A 36 bp sequence (described in Experimental procedures) was added to the C-terminus of the γδ resolvase in plasmid pγδRes-SsrA. This sequence encodes 11 amino acids that were discovered when the overexpression in E. coli of murine interleukin-6 (IL-6) was found to generate a population of proteins of different sizes all sharing the same C-terminal sequence (Ala–Ala–Asn–Asp–Glu–Asn–Tyr–Ala–Leu–Ala–Ala–COOH) (Tu et al., 1995). The 11 amino acid residues are encoded by a stable RNA (tmRNA) and tagged proteins are efficiently degraded by the ClpXP protease in vivo and in vitro (Keiler et al., 1996; Welty et al., 1997; Gottesman et al., 1998).
When pRR51 was introduced into cells that were harvested immediately after thermoinduction of the γδRes-SsrA protein, 32% of cells transformed with pRR51 plasmid DNA contained recombinant plasmid (Fig. 1, Table 1). Resolution efficiency fell rapidly and after 10 min of growth at 30°C, no cell transformed with pRR51 was recombinant. Thus, the in vivo time-course of resolution can be changed substantially by expressing an SsrA-tagged resolvase.
Table 1.
Recombination activity of the γδRes-SsrA in a wild-type Salmonella typhimurium strain (column 2) and in a strain harbouring a clpP::kan mutation (column 3).
| Time point | LT2/pγδRes-SsrA | LT2, clpP::kan/pγδRes-SsrA | LT2/pγδRes |
|---|---|---|---|
| TP0 (induction) | 32% | 100% | 100% |
| TP1 (1 h) | <1% | 100% | 99% |
| TP2 (2 h) | <1% | 48 ± 6 | 99% |
| TP3 (3 h) | <1% | 35 ± 8 | 98% |
| TP4 (4 h) | <1% | 67 ± 20 | 98% |
The clpP::kan mutation eliminates the ClpP subunit that is part of both the ClpAP and the ClpXP proteosome systems. As a control, recombination data with the wild-type resolvase are presented (column 4).
To confirm the short half-life of γδRes-SsrA, cellular proteins were extracted with SDS at different times after induction and subjected to SDS-polyacrylamide gel electrophoresis. SsrA-tagged proteins were visualized on Western blot transfers using rabbit antibodies to the SsrA tag, generously provided by Tania Baker’s lab. A protein band corresponding to the predicted size of γδRes-SsrA appeared after thermoinduction, then disappeared from cell extracts in less than 5 min after cells were returned to 30°C growth conditions (Fig. 2A). Thus, protein half-life and recombination activity were closely correlated.
Fig. 2.
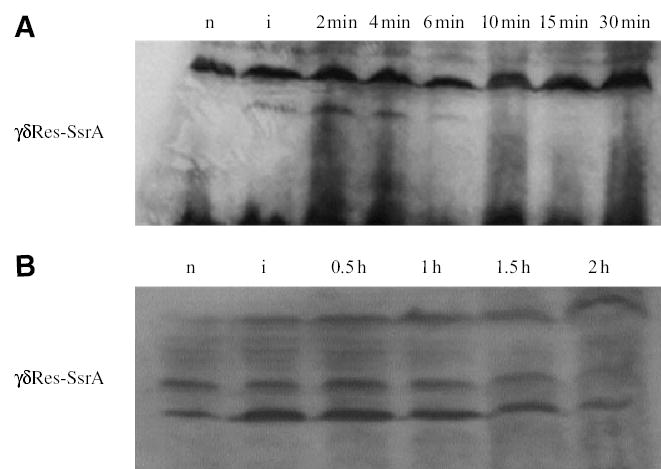
Western blot analysis of resolvase stability. Bacterial cultures (1.5 ml) in mid-log phase (Klett 50) were harvested by centrifugation and lysed in a final volume of 50 μl. Clarified lysate (15 μl) from each preparation was loaded on the gel. Anti-SsrA antibodies identify a band at the predicted size. A non-specific band of higher molecular weight served as the loading control.
A. γδRes-SsrA is a short-lived resolvase. The protein is detected at induction (lane 2) and 2 and 4 min after induction (lanes 3 and 4 respectively). After the 6 min post-induction point (lane 5) the protein is at or below detection limits (lanes 6, 7 and 8). The lanes are: n, non-induced; i, induction; 2 min, 4 min, 6 min, 10 min, 15 min and 30 min are post-induction time points. Sample preparation is described under Experimental procedures.
B. In a clpP− background (clpP replaced with a kan gene), γδRes-SsrA becomes a very stable protein, detectable for hours after heat induction. The lanes are: n, non-induced; i, induced; 0.5 h, 1 h, 1.5 h and 2 h are post-induction time points. Sample preparation is described under Experimental procedures.
One potential problem with the SsrA tagging strategy is that catalytic properties of resolvase might be altered by a C-terminal modification. This could be caused by interference with DNA binding or by impeding conformational changes that are required for recombination. To determine whether the enzymatic properties of resolvase were altered by the SsrA tag, three experiments were carried out.
First, ClpP is the protease-active subunit of the ClpAP and the ClpXP proteases. ClpXP is the major cellular degradation pathway for proteins carrying the SsrA tag. Eliminating this protease should stabilize the γδRes-SsrA protein. The clpP gene was deleted from the Salmonella chromosome and replaced with a Kan-resistant marker. Strain NH3566 carries the clpP deletion in combination with plasmid pγδRes-SsrA and cells from this strain are viable with growth properties very similar to wild type (WT). Cultures were heat shocked at 42°C for 10 min and subsequently transformed at different intervals with pRR51 to monitor resolution activity. One problem with Salmonella strains lacking ClpXP protease is that they had a constitutive low level of Res protein expression even at 30°C. The presence of the γδRes-SsrA protein could be detected on a Western blot before heat shock (Fig. 2B), and 35% of the plasmid transformed cells were recombinant even in the absence of heat shock induction. Nonetheless, a temperature-associated increase in the γδRes-SsrAg band was seen and the elevated level of this band persisted for 1 h after thermoinduction. Moreover, the resolution efficiency stayed near 100% for 2 h when plasmid DNAs were introduced by electroporation into an induced strain (Table 1, Fig. 2B).
Second, the two terminal alanine residues of the SsrA tag are crucial for recognition by the Clp system (Keiler et al., 1996). Many proteins tagged with a 11-amino-acid sequence containing two terminal aspartic acid residues (DD) have half-lives comparable to untagged proteins. A variant Res protein was made with both alanines changed to aspartates by two point mutations (γδRes-SsrA-DD). The time-course of resolution for the γδRes-SsrA-DD protein was examined in cells with WT levels of ClpXP and ClpAP. Resolution activity persisted in these cells for >4 h after induction (Fig. 1, open squares).
Third, amino acid residues within the SsrA tag affect ClpXP proteolytic activity (Flynn et al., 2001). Two positions known to reduce ClpXP-dependent degradation are positions at leucine 9 and alanine 8. In plasmid pγδRes-SsrA-L9D and in pγδRes-SsrA-A8D the leucine and ala-nine residues, respectively, were changed to aspartate. Strains harbouring both plasmids were thermoinduced and tested for resolution activity using the pRR51 electroporation procedure. The half-life of γδRes-SsrA-A8D was 15 min (Fig. 1, open triangles) and γδRes-SsrA-L9D had a 30 min half-life (Fig. 1, solid squares). Together, these experiments show that in cells with active ClpXP protease, a family of resolvase enzymes can be used to measure the chromosome dynamics for intervals varying from a few minutes to several hours using the same thermoinduction protocol.
A 400-domain chromosome
The number of supercoil domains represents a basic unit of chromosome organization inside living cells. Estimates based on WT resolvase assays would underestimate the domain number if they appear and disappear in less than 4 h. The greater than 4 h time span for which γδ resolvase remains active is longer than six cell lives. If res sites occupy the same domain even briefly during this period, a resolution product could be produced.
To compare the efficiency of resolvases that span the widest range in time, nine strains (Higgins et al., 1996) were used with res site distances varying from 14 to 90 kb. Each strain was tested with the γδRes and the γδRes-SsrA proteins. The slope of the first-order decay curve gives the half-distance value (1/2D), which is the interval of separation that results in a 50% drop in recombination efficiency. Data from at least three independent experiments for each point are plotted (Fig. 3, Table 2). The 1/2D of γδRes protein was 36 kb whereas the γδRes-SsrA protein value was 9 kb (Fig. 3A). The correlation coefficient was good for both curves (R2 = 0.841 and 0.974 respectively). This change in slope for the WT and γδRes-SsrA resolvases shows that a stable resolvase assay would underestimate the domain number by more than twofold.
Fig. 3.
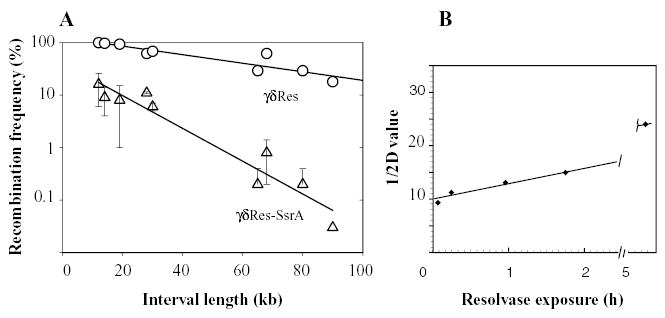
A. Resolution assays for nine intervals ranging in size from 12 to 90 kb in the his-cob region. Three independent experiments were averaged for each time point, and at least 300 colonies were counted for each time point. The distance penalty for recombination (the slope of the curve) is much lower for the WT γδ resolvase than for the short-lived, ssrA-tagged resolvase. The half-distance value calculated for the wild-type resolvase was 36 kb, and for the ssrA-tagged resolvase was 9 kb.
B. The bacterial chromosome changes over time. To illustrate the change, the first-order decay constants for different resolvases in Table 2 were plotted against the time that each enzyme persists in the cell. The resolvase exposure time is defined as the period it takes each type of resolvase to decay so that the resolution activity is half the maximum amount. Note that this value is not equivalent to the half-life for stable enzymes because saturating levels of resolvase persist for periods longer than the cell division time. The enzymes arranged in increasing exposure levels are γδRes-SsrA (1/2D = 9 kb), γδRes-SsrA-A8D (1/2D = 12 kb), γδRes-SsrA-L9D (1/2D = 13 kb), Tn3 Res (1/2D = 15 kb) and γδRes-SsrA-DD (1/2D = 23 kb). Data for the γδRes protein (1/2D = 36 kb) is not included because it remains at saturating levels for more than 6 h after induction.
Table 2.
Log-phase resolution data for nine intervals in the his-cob region (43–45 min) of the Salmonella typhimurium chromosome.
| Interval size (kb) | γδRes A1/2 > 4 h | γδRes-SsrA-DD A1/2 > 4 h | Tn3 Res A1/2 = 2 h | γδRes-SsrA-L9D A1/2 = 1 h | γδRes-SsrA-A8D A1/2 = 15 m | γδRes-SsrA A1/2 < 5 m |
|---|---|---|---|---|---|---|
| 12 | 99 ± 1 | 97 ± 1 | 81 ± 7 | 35 ± 4 | 39 ± 10 | 16 ± 10 |
| 14 | 97 ± 1 | 99 ± 1 | 81 ± 10 | 40 ± 6 | 26 ± 9 | 9 ± 5 |
| 19 | 93 ± 1 | 89 ± 6 | 49 ± 17 | 19 ± 3 | 18 ± 10 | 7 ± 7 |
| 28 | 62 ± 2 | 57 ± 3 | 44 ± 11 | 17 ± 3 | 14 ± 15 | 10 ± 1 |
| 30 | 68 ± 6 | 53 ± 6 | 36 ± 14 | 24 ± 3 | 21 ± 4 | 6 ± 1 |
| 65 | 29 ± 5 | 16 ± 2 | 4 ± 3 | 3 ± 1 | 1 ± 1.4 | 0.2 ± 0.2 |
| 68 | 62 ± 7 | 30 ± 2 | 14 ± 3 | 3 ± 1 | 4 ± 2 | 0.8 ± 0.6 |
| 80 | 29 ± 5 | 22 ± 3 | 1 ± 1 | 1 ± 1 | 1 ± 1 | <0.1 |
| 90 | 18 ± 1 | 6 ± 1 | 2 ± 2 | 1 ± 1 | 0.3 | <0.1 |
Recombination assays from at least three independent experiments were used to calculate a mean with one standard deviation. The A1/2 represents the time required for cellular resolution activity to fall to half-maximum values. Note that this does not reflect a half-life for the long-lived enzymes because cells remain saturated for resolvase for different lengths of time before a first-order decay can be detected.
Once cells are shifted from 30°C to 42°C to induce resolvase expression, about 6 min are required to detect resolution products for all resolvases (data not shown). During this 6 min interval transcription and translation must generate sufficient enzyme to saturate the res sites and the four strand exchanges necessary for recombination must be completed. Cellular persistence of γδRes-SsrA protein lasted for 6–10 min after induction (Fig. 1). Therefore, the γδRes-SsrA enzyme detects chromosome dynamics over a 10 min time span.
How do the intermediate half-life resolvases behave? Strains carrying the plasmids pγδRes-SsrA-A8D, pγδRes-SsrA-L9D and pγδRes-SsrA-DD were also tested for the same nine intervals. The 1/2D values were 12 kb, 13 kb and 23 kb, respectively, for these enzymes (Fig. 3B). The number of barriers detected in WT cells decreased gradually over time periods from 10 to 40 min, extending up to 5 h. In our previous experiments, the Tn3 resolvase would have catalysed resolution after cells begin to enter stationary phase. In stationary-phase domain barriers disappear (Staczek and Higgins, 1998). Assuming a Poisson distribution of domain barriers, our new data with resolvases having half-lives of 30 min or less converge on a median domain in a WT bacterial chromosome of 13 kb. One genome equivalent of Salmonella DNA would contain from 350 to 450 domains. This more than doubles the prior estimate of 150 supercoil domains (Higgins, 1999; Scheirer and Higgins, 2001).
The gyrase impact on domain number
Gyrase and TopoIV mutations influence domain structure (Staczek and Higgins, 1998). Gyrase mutants vary from exerting a mild impact on resolution for the gyrA205 allele to severe effects for the gyrB652 and gyrB1820 alleles. Using the Tn3 resolvase to probe genome structure, some severe gyrase mutants had approximately twice as many domains as WT (Staczek and Higgins, 1998). To confirm the results obtained with Tn3 resolvase using a tagged enzyme, experiments were carried out with the γδRes-SsrA-DD protein (Fig. 4). Intervals ranging from 14 to 90 kb were tested in strains with the WT gyrase, with the mild gyrA209 gene, and with the severe gyrB1820 allele (Fig. 4). Strains with WT gyrase had 1/2D of 23 kb (R2 = 0.91), the gyrA209 allele had a 1/2D of 18 (R2 = 0.91), while the gyrB1820 mutation gave a 1/2D of 12 (R2 = 0.89). These results confirmed our previous conclusions.
Fig. 4.
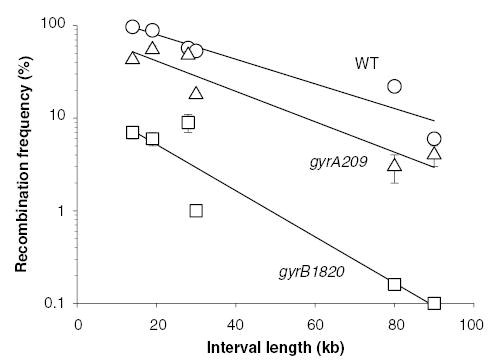
Resolution assays with the γδRes-SsrA-DD protein in the presence of wild-type (WT) gyrase (open circles) gyrA209 (open triangles) and gyrB1820 (open squares). The 1/2D values for recombination calculated for each curve were: 23 kb for the WT strain, 18 kb for a strain with gyrA209 and 12 kb for a strain with the gyrB1820 gene.
If the median domain in a WT cell is 13 kb when assayed with a time-sensitive resolvase, how many domains exist in a gyrB1820 mutant? Doubling the domain number should cause the fraction of cells capable of resolving a 14 kb interval to be small. The resolution of a 14 kb interval was tested with the γδRes-SsrA, γδRes-SsrA-L9D, γδRes-SsrA-A8D or γδRes-SsrA-DD proteins. Each enzyme was tested in combination with one of the five Ts gyrase alleles (Fig. 5). Gyrase had a dramatic impact on 14 kb resolution reactions. Even rather weak gyrA alleles showed a significant decline in resolution efficiency with enzymes having half-lives of 30 min or less. For example, resolution with the gyrA213 allele decreased fivefold relative to a WT strain whereas the gyrA209 allele depressed recombination by 10-fold. The most dramatic effect was obtained with the gyrB1820 mutation where resolution efficiency fell to less than 1% of WT levels using all three time-restricted proteins.
Fig. 5.
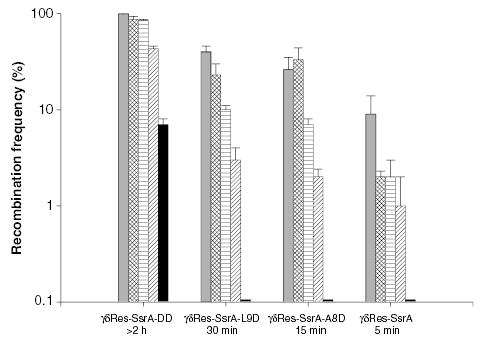
Resolution efficiency of a 14 kb domain in strains carrying different DNA gyrase mutations. A 14 kb interval was examined with resolvases having the indicated half-life. The recombination efficiency of each resolvase is reported for: wild-type gyrase, grey bars; gyrA205, hatched bars; gyrA213, horizontally striped bars; gyrA209, diagonally striped bars, and gyrB1820, black bars.
The rank for each gyrase mutant from mild to severe for the effect on the change in the distance rule correlated well with its effect on the efficiency of producing a 14 kb deletion. We previously interpreted this correlation to indicate that supercoil movement is more restricted in the gyrase mutants. However, one class of promoters responds to altered gyrase activity with lower transcriptional output. If synthesis from the λ PL promoter was strongly decreased by gyrase mutations, this could contribute to a decline in the resolution efficiency. To test how gyrase mutations affect the ability of Res protein to saturate res sites, a short interval containing only the tet gene in plasmid pRR51 was tested for resolution efficiency. Tet resistance could not be scored directly because all gyrase mutants are linked to a Tet marker in the chromosome. To circumvent this problem, cells harbouring pRR51 were induced for resolvase synthesis in gyrase mutants, and after 30 min plasmid was extracted from each strain and transformed into the LT2 strain. All enzymes resolved the plasmid interval at 65% efficiency or greater, except for the γδRes-SsrA protein (52%) (Table 3). Therefore, impact of gyrase mutants on resolution of the 14 kb chromosomal domain could have a small (50%) expression component, but most of the 10- to 100-fold effects on recombination efficiency resulted from inhibition of synaptic pairing of chromosomal sites. Another possibility is that the half-life of SsrA-tagged resolvases are changed in gyrase mutants. We tested resolvase activity in all four gyrase mutants used in this study, and resolvase half-life was unaltered (data not shown). Thus, in severe gyrase mutants, the chromosomes become balkanized into > 1000 domains per genome equivalent.
Table 3.
Efficiency of time-sensitive resolvases in gyrase mutants.
| γδRes-SsrA-DD | γδRes-SsrA-L9D | γδRes-SsrA-A8D | γδRes-SsrA | |
|---|---|---|---|---|
| Wild-type gyrase | 99 | 99 | 99 | 52 ± 19 |
| gyrA205 | 98 | 99 | 99 | 19 ± 10 |
| gyrA213 | 99 | 96 ± 2 | 95 ± 2 | 6 ± 4 |
| gyrA209 | 98 | 68 ± 8 | 76 ± 22 | 7 ± 1 |
| gyrB1820 | 80 ± 10 | 64 ± 4 | 69 ± 26 | 7 ± 1 |
Discussion
Time-sensitive protein expression
To develop a new tool for measuring chromosome dynamics within specific windows of time, we designed proteins with cellular half-lives varying from 5 min to several hours by attaching different 11-amino-acid tags to the C-terminus of the γδ resolvase. Using a thermoinducible λ regulatory system cloned on a low-copy plasmid and various resolvase derivatives, this system allowed us to synchronously express resolvases in cell populations of E. coli and Salmonella and to measure supercoil dynamics for intervals varying from 10 min to 6 h. Compared with other induction systems that we’ve tested, the λ control system is exceptional both for the level and uniformity of protein expressed in a short time and for the tightness of repression at 30°C. The cI857 repressor dissociates from DNA at 42°C, but re-folds rapidly and binds DNA when cells are returned to 30°C (Gaitanaris et al., 1990). This performance is superior to systems we’ve developed using lac and ara regulatory elements. One difference may have to do with the simplicity and ease of temperature shift versus complications involved in inducer transport and added effects of c-AMP on arabinose-modulated expression. We suggest that this general strategy is ideal for analysing temporal effects of many cellular proteins that can be modified at the C-terminus without altering protein function.
Many experiments have been designed to study the structure of DNA inside living cells. Assays include gel analysis of plasmid linking number (Wang, 1986; Vologodskii, 1992), formation of left-handed Z-DNA conformation (Klysik et al., 1982; Peck et al., 1982; Mirkin et al., 1987), extrusion of cruciforms in special plasmids (Murchie and Lilley, 1987; McClellan et al., 1990; Dayn et al., 1991), quantitative measurements of trimethylpsoralen intercalation (Sinden et al., 1980; Sinden and Pettijohn, 1981; Mojica and Higgins, 1997) and measures of transcription from promoters that respond to superhelical tension (Miller and Simons, 1993; Pavitt and Higgins, 1993; Figueroa-Bossi et al., 1998). Although these experiments yield important information about DNA conformation, they are largely uninformative about DNA structure in many specific segments of the bacterial chromosome. The advantage of the Tn3 and γδ resolution assay is that the technique is non-invasive, requires a minimum level of supercoiling and can be carried out for almost any segment of the chromosome (Deng et al., 2004).
Previous work from this lab showed that for about 5% of the Salmonella genome, the efficiency of recombination is a first-order function of physical distance (Higgins et al., 1996). The probability of two sites forming a plectonemic synapse behaves in a stochastic manner, i.e. barriers to supercoil diffusion vary in position from cell to cell and can change within a single cell over time. Like other first-order functions (radioactive decay) the slope of resolution efficiency curves gives a number that we call the 1/2D, which is the distance that causes resolution efficiency to fall by 50%. For the Tn3 resolvase, which is mistakenly called γδ resolvase in previous works (Higgins et al., 1996; Staczek and Higgins, 1998; Scheirer and Higgins, 2001), this distance was about 15 kb.
Domain numbers estimates
Two significant findings about chromosome structure are reported in this work. First, individual cells of WT strains of Salmonella have more than 400 domains that constrain negative supercoiling for each genome equivalent of DNA. This doubles the domain number we estimated from previous experiments employing the WT Tn3 resolvase (Higgins et al., 1996; Scheirer and Higgins, 2001). Two factors contributed to this revision and to the need for time-sensitive enzymes in evaluating genome structure. (i) The WT resolvase from Tn3 or γδ transposons remains in cells at recombination-active concentrations for up to 6 h after expression (Fig. 1 and R.A. Stein, unpublished data). In stationary cells, supercoil domains expand (Staczek and Higgins, 1998). Because cells enter stationary phase before the resolvase activity disappears, a significant amount of recombination can occur in stationary phase. The differences in 1/2D for the γδRes, γδRes-SsrA-DD and Tn3 resolvases (36, 23 and 15 kb respectively) reflect how long these proteins persist as cells enter stationary phase (data not shown.) (ii) Recent work from this lab showed that the induction of transcription from a strong promoter causes a new domain to appear in the bacterial chromosomes (Deng et al., 2004). Transcription-induced domains can appear and disappear over a 20 min time span. The fact that resolvases with half-lives of 5–30 min all give similar 1/2D value (9–13 kb) indicates that barriers are relatively stable over this reference frame, at least in the intervals that were studied in this work.
A 400-domain chromosome agrees with new measurements made by independent techniques in E. coli (Postow et al., 2004). Postow and co-workers introduced chromosome breaks using controlled expression of rare-cutting restriction enzymes. By measuring the distance from a break to a supercoil-responsive promoter, they used genome-wide microarray technology to measure supercoil diffusion. They also analysed the loop sizes of nucleoids spread on electron microscope grids. The model of chromosome structure that best fit their data and Monte Carlo computer simulations was a family of loops with a median size of 10 kb having variable length and sequence determinants. These measurements are in close agreement with the conclusions we reached with tagged resolvases.
Controlling domain number
Gyrase mutants cause the number of independent domains to increase, exceeding 1000 domains per genome equivalent in rare severe gyrase mutants (Fig. 5). This trend was seen previously in assays using Tn3 resolvase (Staczek and Higgins, 1998), but time-sensitive enzymes made the effect more obvious. With time-shortened enzymes, nearly all gyrase mutants increased the number of domains. The impact of gyrase on domain size was more striking than the impact of these same mutations on superhelix density monitored by plasmid linking numbers (Staczek and Higgins, 1998) (Fig. 5). Most models of chromosome structure focus on torsional aspects of negative supercoiling. These models consider the impact of gyrase on the helical repeat, which can influence protein binding to complex sites, the influence on helix melting, which can alter initiation kinetics, and the waves of negative and positive supercoils that attend processes like replication and transcription (see Higgins and Vologodskii, 2004).
We propose that the domain number is a critical factor in prokaryotic genome structure. If domains are too large, DNA compaction and efficient segregation is at risk; if there are too many domains problems might arise during transcription and replication. Three observations are consistent with this view. First, genetic selections to find genes involved in chromosome condensation and partitioning uncovered proteins closely related to eukaryotic cohesins and condensins (Hiraga et al., 1989; Hiraga, 1992). In E. coli the gene is mukB and in Bacillus subtilis and Caulobacter crescentus the gene is called smc. Defects in these proteins can cause a diffuse appearance of cellular nucleoids (Hiraga et al., 1989; Hiraga, 1992), influence supercoiling (den Blaauwen et al., 2001; Lindow et al., 2002; Adachi and Hiraga, 2003; Case et al., 2004) and result in poor segregation of chromosomes between dividing sister cells (Weitao et al., 2000). Second, a new technique for gene localization is based on ‘tagging’ chromosomes with either lac-operator or tet-operator modules (Lau et al., 2003). When such green fluorescent protein (GFP) or CFP-fused repressors are expressed in cells with tagged chromosomes, a focus is observed that localizes one particular genetic region. Viollier et al. (2004) measured GFP or CFP-tagged repressor foci in 112 tagged strains of C. crescentus, scoring thousands of images for each strain. Each tagged gene moved to a post-replication position that reflected its genetic linkage to the origin and terminus of the chromosome. Such high fidelity positioning has been explained by underpinning structure with many small domains (Breier and Cozzarelli, 2004).
Third, the down side of too many domains becomes obvious in severe gyrase mutants. Strains with the gyrB1820 allele have >1000 domains per genome equivalent. However, the phenotype is complex, including constitutive SOS induction, synthetic lethality with RecA and RecBC, and Recless DNA degradation of nascent DNA (Gari et al., 1996; 2001). These are all facets that suggest frequent replication fork collapse (Bidnenko et al., 2002; Grompone et al., 2003; Michel et al., 2004).
A complete description of chromosome structure remains elusive. A level of organization beyond a 10 kb domain pattern is suggested by several experiments. Using a system based on site-specific inversions catalysed by the phage λ Int and Xis proteins, Garcia-Russell showed that some segments of chromosome are unable to make intimate contact with each other (Garcia-Russell et al., 2004). In a similar study, Valens et al. (2004) found a distance rule for lambda excisive recombination indicating colocalization of many different regions. Bacteriophage Mu, which is 37 kb, contains a central strong gyrase site that functions in an early stage of transposition (Pato et al., 1990; Pato and Karlock, 1994). Evidence points to the critical step being formation of a functional synapse (Pato et al., 1995; Pato and Banerjee, 1996; 1999), but the detailed structure of a Mu synapse (a five-noded plectoneme; Pathania et al., 2002) suggests an end-pairing mechanism more complex than slithering.
Finally, the similarity in domain structure between E. coli and Salmonella typhimurium may indicate that a common set of proteins and mechanisms contribute to domain structure in two organisms separated from a common ancestor for 140 million years. However, significant differences are also apparent. The deletion of mukB leads to a much more severe phenotype in Salmonella than it does in E. coli (N.P. Higgins, unpublished). Deletions of hupA and hupB in Salmonella have different effects on phage Mu biology than in E. coli (Hillyard et al., 1990) and specific gyrase mutations that are viable in Salmonella are lethal in E. coli (K.M. Champion and N.P. Higgins, unpublished data) Developing a model of how the small DNA-binding proteins like HU, H-NS, IHF, STPA, FIS, the ATP-requiring type II topoisomerases, the type I topoisomerases and the cohensin-related proteins organize bacterial chromatin remains a tough challenge, but the comparative approach involving E. coli and Salmonella promises to be very useful.
Experimental procedures
Bacterial strains
Derivatives of S. typhimurium LT2 used in the study are described in Table 4. Gene transduction with bacteriophage P22 was performed as previously described (Higgins et al., 1996), and single colonies were re-streaked on green plates (Smith and Levine, 1967) to identify phage-free transductants.
Table 4.
Bacterial strains used in this study.
| Strain | Interval size (kb) | Genotype | Plasmid |
|---|---|---|---|
| NH0185 | hsdL6 hsdSA29(rLT− mlT+ rs− ms+) metA22 metE551 ilv-452 trpB2 xyl404 rpsL120(Strr) galE496 | ||
| NH3568 | LT2 | pJBRescI (Tn3) | |
| NH3645 | LT2 | pγδRes | |
| NH3546 | LT2 | pγδRes-SsrA | |
| NH3578 | LT2 | pγδRes-SsrA-A8D | |
| NH3643 | LT2 | pγδRes-SsrA-DD | |
| NH3644 | LT2 | pγδRes-SsrA-L9D | |
| NH3566 | clpP::kan | pγδRes-SsrA | |
| NH3569 | clpP::kan | pJBRescI | |
| NH2124 | 12 | cobT714::MudJr2 zea-3777::Tn10dTc cobU713::Tn10dGn | pJBRescI |
| NH3618 | 12 | cobT714::MudJr2 zea-3777::Tn10dTc cobU713::Tn10dGn | pγδRes |
| NH3561 | 12 | cobT714::MudJr2 zea-3777::Tn10dTc cobU713::Tn10dGn | pγδRes-SsrA |
| NH3580 | 12 | cobT714::MudJr2 zea-3777::Tn10dTc cobU713::Tn10dGn | pγδRes-SsrA-A8D |
| NH3573 | 12 | cobT714::MudJr2 zea-3777::Tn10dTc cobU713::Tn10dGn | pγδRes-SsrA-L9D |
| NH3592 | 12 | cobT714::MudJr2 zea-3777::Tn10dTc cobU713::Tn10dGn | pγδRes-SsrA-DD |
| NH2118 | 14 | cobT714::MudJr2 zea-3777::Tn10dTc cobP712::Tn10dGn | pJBRescI |
| NH3617 | 14 | cobT714::MudJr2 zea-3777::Tn10dTc cobP712::Tn10dGn | pγδRes |
| NH3638 | 14 | cobT714::MudJr2 zea-3777::Tn10dTc cobP712::Tn10dGn | pγδRes-SsrA |
| NH3552 | 14 | cobT714::MudJr2 zea-3777::Tn10dTc cobP712::Tn10dGn | pγδRes-SsrA-A8D |
| NH3572 | 14 | cobT714::MudJr2 zea-3777::Tn10dTc cobP712::Tn10dGn | pγδRes-SsrA-L9D |
| NH3594 | 14 | cobT714::MudJr2 zea-3777::Tn10dTc cobP712::Tn10dGn | pγδRes-SsrA-DD |
| NH2142 | 19 | cobT714::MudJr1 zea-3777::Tn10dTc cobJ709::Tn10dGn | pJBRescI |
| NH3616 | 19 | cobT714::MudJr1 zea-3777::Tn10dTc cobJ709::Tn10dGn | pγδRes |
| NH3560 | 19 | cobT714::MudJr1 zea-3777::Tn10dTc cobJ709::Tn10dGn | pγδRes-SsrA |
| NH3579 | 19 | cobT714::MudJr1 zea-3777::Tn10dTc cobJ709::Tn10dGn | pγδRes-SsrA-A8D |
| NH3577 | 19 | cobT714::MudJr1 zea-3777::Tn10dTc cobJ709::Tn10dGn | pγδRes-SsrA-L9D |
| NH3587 | 19 | cobT714::MudJr1 zea-3777::Tn10dTc cobJ709::Tn10dGn | pγδRes-SsrA-DD |
| NH2119 | 28 | cobT714::MudJr2 zea-3777::Tn10dTc cob-708::Tn10dGn | pJBRescI |
| NH3614 | 28 | cobT714::MudJr2 zea-3777::Tn10dTc cob-708::Tn10dGn | pγδRes |
| NH3556 | 28 | cobT714::MudJr2 zea-3777::Tn10dTc cob-708::Tn10dGn | pγδRes-SsrA |
| NH3551 | 28 | cobT714::MudJr2 zea-3777::Tn10dTc cob-708::Tn10dGn | pγδRes-SsrA-A8D |
| NH3575 | 28 | cobT714::MudJr2 zea-3777::Tn10dTc cob-708::Tn10dGn | pγδRes-SsrA-L9D |
| NH3588 | 28 | cobT714::MudJr2 zea-3777::Tn10dTc cob-708::Tn10dGn | pγδRes-SsrA-DD |
| NH3590 | 30 | cobT714::MudJr1 zea-3777::Tn10dTc pduF358::Tn10dGn | pγδRes-SsrA-DD |
| NH3642 | 30 | cobT714::MudJr1 zea-3777::Tn10dTc pduF358::Tn10dGn | pγδRes-SsrA-L9D |
| NH3606 | 30 | cobT714::MudJr1 zea-3777::Tn10dTc pduF358::Tn10dGn | pγδRes-SsrA-A8D |
| NH3621 | 30 | cobT714::MudJr1 zea-3777::Tn10dTc pduF358::Tn10dGn | pγδRes |
| NH3639 | 30 | cobT714::MudJr1 zea-3777::Tn10dTc pduF358::Tn10dGn | pγδRes-SsrA |
| NH2075 | 30 | cobT714::MudJr1 zea-3777::Tn10dTc pduF358::Tn10dGn | pJBRescI |
| NH2453 | 65 | cobT714::MudJr2 zea-3777::Tn10dTc zec-8253::Tn10dGn | pJBRescI |
| NH3613 | 65 | cobT714::MudJr2 zea-3777::Tn10dTc zec-8253::Tn10dGn | pγδRes |
| NH3557 | 65 | cobT714::MudJr2 zea-3777::Tn10dTc zec-8253::Tn10dGn | pγδRes-SsrA |
| NH3558 | 65 | cobT714::MudJr2 zea-3777::Tn10dTc zec-8253::Tn10dGn | pγδRes-SsrA-A8D |
| NH3574 | 65 | cobT714::MudJr2 zea-3777::Tn10dTc zec-8253::Tn10dGn | pγδRes-SsrA-L9D |
| NH3591 | 65 | cobT714::MudJr2 zea-3777::Tn10dTc zec-8253::Tn10dGn | pγδRes-SsrA-DD |
| NH2223 | 68 | cobT714::MudJr2 zea-3777::Tn10dTc phs-209::Tn10dGn | pJBRes cI |
| NH3619 | 68 | cobT714::MudJr2 zea-3777::Tn10dTc phs-209::Tn10dGn | pγδRes |
| NH3640 | 68 | cobT714::MudJr2 zea-3777::Tn10dTc phs-209::Tn10dGn | pγδRes-SsrA |
| NH3549 | 68 | cobT714::MudJr2 zea-3777::Tn10dTc phs-209::Tn10dGn | pγδRes-SsrA-A8D |
| NH3571 | 68 | cobT714::MudJr2 zea-3777::Tn10dTc phs-209::Tn10dGn | pγδRes-SsrA-L9D |
| NH3589 | 68 | cobT714::MudJr2 zea-3777::Tn10dTc phs-209::Tn10dGn | pγδRes-SsrA-DD |
| NH2149 | 80 | cobT714::MudJr2 zea-3777::Tn10dTc hisD10201::Tn10dGn | pJBRescI |
| NH3620 | 80 | cobT714::MudJr2 zea-3777::Tn10dTc hisD10201::Tn10dGn | pγδRes |
| NH3641 | 80 | cobT714::MudJr2 zea-3777::Tn10dTc hisD10201::Tn10dGn | pγδRes-SsrA |
| NH3555 | 80 | cobT714::MudJr2 zea-3777::Tn10dTc hisD10201::Tn10dGn | pγδRes-SsrA-A8D |
| NH3570 | 80 | cobT714::MudJr2 zea-3777::Tn10dTc hisD10201::Tn10dGn | pγδRes-SsrA-L9D |
| NH3593 | 80 | cobT714::MudJr2 zea-3777::Tn10dTc hisD10201::Tn10dGn | pγδRes-SsrA-DD |
| NH2226 | 90 | cobT714::MudJr1 zea-3777::Tn10dTc zee-8251::Tn10dGn | pJBRescI |
| NH3615 | 90 | cobT714::MudJr1 zea-3777::Tn10dTc zee-8251::Tn10dGn | pγδRes |
| NH3559 | 90 | cobT714::MudJr1 zea-3777::Tn10dTc zee-8251::Tn10dGn | pγδRes-SsrA |
| NH3554 | 90 | cobT714::MudJr1 zea-3777::Tn10dTc zee-8251::Tn10dGn | pγδRes-SsrA-A8D |
| NH3576 | 90 | cobT714::MudJr1 zea-3777::Tn10dTc zee-8251::Tn10dGn | pγδRes-SsrA-L9D |
| NH3586 | 90 | cobT714::MudJr1 zea-3777::Tn10dTc zee-8251::Tn10dGn | pγδRes-SsrA-DD |
| NH3622 | 14 | cobT714::MudJr2, cobP712::Tn10dGn zeh-754::Tn10 gyrA209ts | pγδRes-SsrA |
| NH3623 | 14 | cobT714::MudJr2, cobP712::Tn10dGn zeh-754::Tn10 gyrA209ts | pγδRes-SsrA-A8D |
| NH3624 | 14 | cobT714::MudJr2, cobP712::Tn10dGn zeh-754::Tn10 gyrA209ts | pγδRes-SsrA-L9D |
| NH3625 | 14 | cobT714::MudJr2, cobP712::Tn10dGn zeh-754::Tn10 gyrA209ts | pγδRes-SsrA-DD |
| NH3626 | 14 | cobT714::MudJr2, cobP712::Tn10dGn zib-748::Tn10 gyrB1820ts | pγδRes-SsrA |
| NH3627 | 14 | cobT714::MudJr2, cobP712::Tn10dGn zib-748::Tn10 gyrB1820ts | pγδRes-SsrA-A8D |
| NH3628 | 14 | cobT714::MudJr2, cobP712::Tn10dGn zib-748::Tn10 gyrB1820ts | pγδRes-SsrA-L9D |
| NH3629 | 14 | cobT714::MudJr2, cobP712::Tn10dGn zib-748::Tn10 gyrB1820ts | pγδRes-SsrA-DD |
| NH3630 | 14 | cobT714::MudJr2, cobP712::Tn10dGn zeh-754::Tn10 gyrA205ts | pγδRes-SsrA |
| NH3631 | 14 | cobT714::MudJr2, cobP712::Tn10dGn zeh-754::Tn10 gyrA205ts | pγδRes-SsrA-A8D |
| NH3632 | 14 | cobT714::MudJr2, cobP712::Tn10dGn zeh-754::Tn10 gyrA205ts | pγδRes-SsrA-L9D |
| NH3633 | 14 | cobT714::MudJr2, cobP712::Tn10dGn zeh-754::Tn10 gyrA205ts | pγδRes-SsrA-DD |
| NH3634 | 14 | cobT714::MudJr2, cobP712::Tn10dGn zeh-754::Tn10 gyrA213ts | pγδRes-SsrA |
| NH3635 | 14 | cobT714::MudJr2, cobP712::Tn10dGn zeh-754::Tn10 gyrA213ts | pγδRes-SsrA-A8D |
| NH3636 | 14 | cobT714::MudJr2, cobP712::Tn10dGn zeh-754::Tn10 gyrA213ts | pγδRes-SsrA-L9D |
| NH3637 | 14 | cobT714::MudJr2, cobP712::Tn10dGn zeh-754::Tn10 gyrA213ts | pγδRes-SsrA-DD |
Plasmid construction
Plasmid pJBRescI was originally derived from pACYC184 (Bliska and Cozzarelli, 1987). In this plasmid, the tnpR gene encoding Tn3 resolvase is regulated by a temperature-sensitive (Ts) λ repressor, cI857. Due to a strain transfer recording error, we believed that we were using a plasmid with a copy of γδ resolvase (see Fig. 1). At permissive temperature (30°C) resolvase transcription is tightly repressed. Incubation at 42°C for 10 min results in reversible denaturation of the repressor (Gaitanaris et al., 1990) and a burst of the resolvase protein expression. Plasmid pJBRescI was digested with BamHI and BsaHI to remove a 600 bp fragment containing the Tn3 tnpR gene. Digested plasmid was subjected to electrophoresis on a 1% agarose gel, and after extraction followed by nucleotide end-filling with T7 DNA polymerase (Promega), the product was blunt-end ligated to create the empty vector pJB.
The γδ tnpR gene was amplified from the F factor by carrying out PCRs using oligonucleotides RS01 (5′-GCTAGCCTTGGATCCGATAATAAAGAAGGAGATATCATAT GCGACTTTTTGGTTACGC-3′) and RS02 (5′-GTCCGCATCGGATCCGCTAACGTAATTAGTTGCTTTCATT-3′). PCR products were precipitated with ethanol, resuspended in enzyme buffer, digested with BamHI (which cuts in the underlined sequence shown above), ligated to BamHI-linearized pJB and transformed into chemically competent Top 10 E. coli cells (Invitrogen). Individual clones were sequenced to confirm proper orientation and sequence and then introduced into the restriction-defective S. typhimurium strain NH0185 by electroporation. Plasmids isolated from NH0185 can be efficiently moved into any S. typhimurium strain derived from LT2.
To create pγδRes-SsrA, the γδ tnpR gene was first amplified with oligonucleotides RS03 (ATGCGACTTTTTGGTTA CGC) and RS04 (TTAAGCTGCTAAAGCGTAGTTTTCGTC GTTTGCTGCGTTGCTTTCATTTATTAC) where the segment that encodes the ssrA tag is italicized. The product was cloned into the TopoTA vector, yielding TopoTA-tnpR-ssrA plasmid encoding a resolvase tagged at its C-terminus with the signal for rapid degradation by the ClpXP proteosome (Levchenko et al., 2000; Joshi et al., 2004). Oligonucleotides RS01 and RS05 (5′-GTCCGCATCGGATCCGCTAACGTAAT TAAGCTGCTAAAGCGTAGTT-3′) were then used to PCR amplify a 580 bp tnpR-ssrA DNA fragment with BamHI cleavage sites (underlined) at both ends. After BamHI digestion and gel purification (Qiagen), the PCR product was ligated into pJB vector linearized with BamHI. Clones were screened for inserts in the correct orientation and then subjected to sequence analysis to confirm the expected sequence.
To make pγδRes-SsrA-L9D, the tnpR-ssrA fragment was amplified from the Topo-tnpR-ssrA template by using oligonucleotides RS01 and RS06 (5′-GTCCGCATCGGAT CCGCTAACGTAATTAAGCTGCGTCAGCGTAGTT-3′) (L9D replacement indicated with bold characters). pγδRes-SsrA-A8D was generated with oligonucleotides RS01 and RS07 (5′-GTCCGCATCGGATCCGCTAACGTAATTAAGCTGCTAAGTCGTAGTTTTC-3′) using the strategy described above. To make a plasmid that is immune to ClpXP degradation, the same procedure was carried out with RS08 (5′-GTCCG CATCGGATCCGCTAACG TAATTAGTCGTCTTAAAGCGTA GTTTTC-3′) where the A10A11 and D10D11 mutations are indicated by bold type. DNA sequence analysis confirmed that each new encoded proper C-terminal tags: γδ-ssrA gene encoded AANDENYALAA, γδ-ssrA-L9D encoded AANDEN YADAA, γδ-ssrA-A8D encoded AANDENYDLAA and γδ-ssrA-DD encoded AANDENYALDD.
Media
Bacteria were grown in complex LB medium (5 g of NaCl, 10 g of bactotryptone, 5 g of yeast extract per litre). Plates used for recombination assays were prepared by using NCE salts (Davis et al., 1980) and casamino acids at 0.2% with X-Gal at a final concentration of 40 μg ml−1. Antibiotic concentrations were 50 μg ml−1 for kanamycin, 20 μg ml−1 for chloramphenicol, 15 μg ml−1 for gentamicin and 10 μg ml−1 for tetracycline.
Polymerase chain reaction (PCR)
Recombinant Taq DNA polymerase and DNA extender (Fisher) were used in an equimolar ratio in thermocycling reactions. Reactions were performed in an air thermocycler (Idaho Technologies) (15 s denaturation at 94°C, 15 s annealing at 55°C, 45 s synthesis at 72°C, 30 cycles, S = 6).
Resolution assays
Recombination assays were performed as described previously (Higgins et al., 1996) with one modification to decrease the number of sectored colonies. Fresh colonies from an LB plate containing chloramphenicol, kanamycin and gentamicin were inoculated in 2 ml of LB medium containing chloram-phenicol and kanamycin and incubated with shaking at 30°C overnight. The next morning, 100 μl of aliquots were inoculated into 10 ml of LB with chloramphenicol and kanamycin in 125 ml Nephlo culture flasks (Bellco Biotechnology) and grown with shaking at 30°C. When cultures reached Klett 50, 500 μl were placed into a sterile glass tube and incubated for 10 min in a 42°C shaking water bath. Twenty microlitres of each induced and uninduced culture were added to 180 μl of LB and incubated at 30°C overnight in 96-well microtitre plates (Greiner BioOne). The next day, serial 10-fold dilutions were made and cells from the 10−4 and 10−5 dilution were spread on plates containing chloramphenicol and X-Gal. To score resolution frequencies, the recombinant (white) and non-recombinant (blue) colonies were counted after plates had been incubated for several days at 30°C.
Measuring resolvase half-life
The half-life of different resolvase derivatives was measured using pRR51, a 5.9 kb plasmid with two directly repeated res sites flanking a gene encoding Tet resistance plus an Amp resistance gene linked to the replication origin (Reed, 1981). Strain NH3568 contains the compatible plasmid pJBRescI encoding Tn3 resolvase. NH3568 was grown in LB with chloramphenicol at 30°C and at a density of Klett 50 (mid-log phase), the culture was shifted to 42°C for 10 min and then returned to 30°C. Aliquots of thermoinduced bacterial cultures were quickly (within 2 min) pelleted three times, resuspended in 10% ice-cold glycerol, and then plasmid pRR51 was introduced by electroporation. Culture was spread on LB plates with chloramphenicol and ampicillin and incubated overnight (without shaking) at 30°C. The next day, 500 single colonies for each time point were picked and patched on LB Tet-plates to measure the fraction of recombinant (Tet-sensitive) colonies.
Western blots
To measure γδRes-SsrA protein stability, strain NH3546 was grown at 30°C to mid-log phase (Klett 50) then shifted to 42°C for 10 min. Immediately after induction, a 1.5 ml sample was removed, centrifuged for 20 s and the pellet was fast-frozen in a dry ice-ethanol bath. The induced culture was placed back to 30°C and samples were similarly removed after different incubation times, pelleted and snap frozen. Frozen pellets were resuspended in 50 μl of SDS sample buffer and boiled for 10 min in a water bath and centrifuged to remove insoluble cell debris. Independent samples (15 μl) were applied to a 12% SDS-PAGE gel and the protein bands were transferred to a PVDF membrane as previously described (Maniatis et al., 1982). PVDF membranes were processed by using the Western Breeze Chemoluminescence Detection Kit (Invitrogen) according to manufacturer’s instructions. Polyclonal antibodies against the ssrA tag, a gift from Tania Baker, were diluted to 1:1000 before use.
Gyrase mutants
The pRR51 reporter plasmid was used to examine the persistence of the various resolvases in the presence of the various gyrase alleles used in this study. WT LT2 and strains harbouring the gyrA 209, gyrA 213, gyrB 1820 or gyrA 205 alleles were transformed with the pRR51 reporter plasmid as well as the plasmid encoding various half-life resolvases (Table 4). Single colonies were grown up to stationary phase at 30°C overnight and the next morning cultures were diluted 1:100 and grown to Klett 50. After heat induction at 42°C for 10 min, plasmids were isolated from 1 ml of cultures (Promega), electroporated into LT2 strains and the cultures were then grown for several hours at 30°C and plated on Amp plates. The next day, individual colonies were patched on Amp and Tet plates and the deletion efficiencies were calculated (Table 3). Three independent experiments were performed for each strain.
Acknowledgments
We thank Tania Baker for the gift of SsrA tag-specific antisera, and Lisa Postow, Nicholas Cozzarelli and Christine Hardy for fruitful discussions and sharing preliminary data. We also thank Martin Pato for a thorough and thoughtful review. This work was supported by National Institutes of Health Grant GM33143.
References
- Adachi S, Hiraga S. Mutants suppressing novo-biocin hypersensitivity of a mukB null mutation. J Bacteriol. 2003;185:3690–3695. doi: 10.1128/JB.185.13.3690-3695.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin KR, Abola AP, Kanaar R, Cozzarelli NR. Contributions of supercoiling to Tn3 resolvase and phage Mu Gin site-specific recombination. J Mol Biol. 1996;256:50–65. doi: 10.1006/jmbi.1996.0067. [DOI] [PubMed] [Google Scholar]
- Bidnenko V, Ehrlich SD, Michel B. Replication fork collapse at replication terminator sequences. EMBO J. 2002;21:3898–3907. doi: 10.1093/emboj/cdf369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Blaauwen T, Lindqvist A, Lowe J, Nanninga N. Distribution of the Escherichia coli structural maintenance of chromosomes (SMC)-like protein MukB in the cell. Mol Microbiol. 2001;42:1179–1188. doi: 10.1046/j.1365-2958.2001.02691.x. [DOI] [PubMed] [Google Scholar]
- Bliska JB, Cozzarelli NR. Use of site-specific recombination as a probe of DNA structure and metabolism in vivo. J Mol Biol. 1987;194:205–218. doi: 10.1016/0022-2836(87)90369-x. [DOI] [PubMed] [Google Scholar]
- Breier AM, Cozzarelli NR. Linear ordering and dynamic segregation of the bacterial chromosome. Proc Natl Acad Sci USA. 2004;101:9175–9176. doi: 10.1073/pnas.0403722101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case RB, Chang YP, Smith SB, Gore J, Cozzarelli NR, Bustamante C. The bacterial condensin MukBEF compacts DNA into a repetitive, stable structure. Science. 2004;305:222–227. doi: 10.1126/science.1098225. [DOI] [PubMed] [Google Scholar]
- Davis, R.W., Botstein, D., and Roth, J.R. (1980) A Manual for Genetic Engineering. Advanced Bacterial Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Dayn A, Malkhosyan S, Duzhy D, Lyamichev V, Panchenko Y, Mirkin S. Formation of (dA-dT)n cruciforms in Escherichia coli cells under different environmental conditions. J Bacteriol. 1991;173:2658–2664. doi: 10.1128/jb.173.8.2658-2664.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delius H, Worcel A. Electron microscopic studies on the folded chromosome of Escherichia coli. Cold Spring Harb Symp Quant Biol. 1973;38:53–58. doi: 10.1101/sqb.1974.038.01.008. [DOI] [PubMed] [Google Scholar]
- Deng S, Stein RA, Higgins NP. Transcription-induced barriers to supercoil diffusion in the Salmonella typhimurium chromosome. Proc Natl Acad Sci USA. 2004;101:3398–3403. doi: 10.1073/pnas.0307550101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Bossi N, Guerin M, Rahmouni R, Leng M, Bossi L. The supercoiling sensitivity of a bacterial tRNA promoter parallels its responsiveness to stringent control. EMBO J. 1998;17:2359–2367. doi: 10.1093/emboj/17.8.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Seidel M, Wickner SH, Sauer RT, Baker TA. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc Natl Acad Sci USA. 2001;98:10584–10589. doi: 10.1073/pnas.191375298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitanaris GA, Papavassiliou AG, Rubock P, Silverstein SJ, Gottesman ME. Renaturation of denatured 1 repressor requires heat shock proteins. Cell. 1990;61:1013–1020. doi: 10.1016/0092-8674(90)90066-n. [DOI] [PubMed] [Google Scholar]
- Garcia-Russell N, Harmon TG, Le TQ, Amaladas NH, Mathewson RD, Segall AM. Unequal access of chromosomal regions to each other in Salmonella: probing chromosome structure with phage λ integrase-mediated long-range rearrangements. Mol Microbiol. 2004;52:329–344. doi: 10.1111/j.1365-2958.2004.03976.x. [DOI] [PubMed] [Google Scholar]
- Gari E, Figueroa-Bossi N, Blanc-Potard AB, Spirito F, Schmid MB, Bossi L. A class of gyrase mutants of Salmonella typhimurium show quinolone-like lethality and require Rec functions for viability. Mol Microbiol. 1996;21:111–122. doi: 10.1046/j.1365-2958.1996.6221338.x. [DOI] [PubMed] [Google Scholar]
- Gari E, Bossi L, Figueroa-Bossi N. Growth-dependent DNA breakage and cell death in a gyrase mutant of Salmonella. Genetics. 2001;159:1405–1414. doi: 10.1093/genetics/159.4.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser P, Sharpe ME, Raether B, Perego M, Ohlsen K, Errington J. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes Dev. 1997;11:1160–1168. doi: 10.1101/gad.11.9.1160. [DOI] [PubMed] [Google Scholar]
- Gordon S, Rech J, Lane D, Wright A. Kinetics of plasmid segregation in Escherichia coli. Mol Microbiol. 2004;51:461–469. doi: 10.1046/j.1365-2958.2003.03837.x. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Roche E, Zhou Y, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith JD. Visualization of prokaryotic DNA in a regularly condensed chromatin like fiber. Proc Natl Acad Sci USA. 1976;73:563–567. doi: 10.1073/pnas.73.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grompone G, Ehrlich SD, Michel B. Replication restart in gyrB Escherichia coli mutants. Mol Microbiol. 2003;48:845–854. doi: 10.1046/j.1365-2958.2003.03480.x. [DOI] [PubMed] [Google Scholar]
- Hayes CS, Bose B, Sauer RT. Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. J Biol Chem. 2002a;277:33825–33832. doi: 10.1074/jbc.M205405200. [DOI] [PubMed] [Google Scholar]
- Hayes CS, Bose B, Sauer RT. Stop codons preceded by rare arginine codons are efficient determinants of SsrA tagging in Escherichia coli. Proc Natl Acad Sci USA. 2002b;99:3440–3445. doi: 10.1073/pnas.052707199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, N.P. (1999) DNA supercoiling and its consequences for chromosome structure and function. In Organization of the Prokaryotic Genome, Vol. 1. Charlebois, R.L. (ed). Washington, DC: American Society for Microbiology Press, pp. 189–202.
- Higgins, N.P., and Vologodskii, A. (2004) Topological behavior of plasmid DNA. In Plasmid Biology. Phillips, G., and Funnell, B. (eds). Washington, DC: American Society for Microbiology Press, pp. 181–201.
- Higgins NP, Yang X, Fu Q, Roth JR. Surveying a supercoil domain by using the γδ resolution system in Salmonella typhimurium. J Bacteriol. 1996;178:2825–2835. doi: 10.1128/jb.178.10.2825-2835.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillyard D, Edlun M, Hughes K, Marsh M, Higgins NP. Subunit-specific phenotypes of Salmonella typhimurium HU mutants. J Bacteriol. 1990;172:5402–5407. doi: 10.1128/jb.172.9.5402-5407.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga, S. (1992) Chromosome and plasmid partition in Escherichia coli. In Annual Review of Biochemistry, Vol. 61. Richardson, C., Abelson, J., Meister, A., and Walsh, C. (eds). Palo Alto: Annual Reviews, pp. 283–306. [DOI] [PubMed]
- Hiraga S, Niki H, Ogura T, Ichinose D, Mori H, Ezaki B, Jaffe A. Chromosome partitioning in Escherichia coli: novel mutants producing anucleate cells. J Bacteriol. 1989;171:1496–1505. doi: 10.1128/jb.171.3.1496-1505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi SA, Hersch GL, Baker TA, Sauer RT. Communication between ClpX and ClpP during substrate processing and degradation. Nat Struct Biol. 2004;11:404–411. doi: 10.1038/nsmb752. [DOI] [PubMed] [Google Scholar]
- Kavenoff R, Bowen B. Electron microscopy of membrane-free folded chromosomes from Escherichia coli. Chromosoma. 1976;59:89–101. doi: 10.1007/BF00328479. [DOI] [PubMed] [Google Scholar]
- Kavenoff R, Ryder O. Electron microscopy of membrane-associated folded chromosomes of Escherichia coli. Chromosoma. 1976;55:13–25. doi: 10.1007/BF00288323. [DOI] [PubMed] [Google Scholar]
- Keiler KC, Waller PRH, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- Klysik J, Stirdivant SM, Wells RD. Cloning, characterization, and instability of inserts containing different lengths of (dC-dG) in Escherichia coli. J Biol Chem. 1982;257:10152–10158. [PubMed] [Google Scholar]
- Krasnow MA, Cozzarelli NR. Site-specific relaxation and recombination by the Tn3 resolvase: recognition of the DNA path between oriented res sites. Cell. 1983;32:1313–1324. doi: 10.1016/0092-8674(83)90312-4. [DOI] [PubMed] [Google Scholar]
- Lau IF, Filipe SR, Soballe B, Okstad OA, Barre FX, Sherratt DJ. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol Microbiol. 2003;49:731–743. doi: 10.1046/j.1365-2958.2003.03640.x. [DOI] [PubMed] [Google Scholar]
- Levchenko I, Seidel M, Sauer RT, Baker TA. A specificity-enhancing factor for the ClpXP degradation machine. Science. 2000;289:2354–2356. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- Lindow JC, Tritton RA, Grossman AD. Structural maintenance of chromosomes protein of Bacillus subtilis affects supercoiling in vivo. J Bacteriol. 2002;184:5317–5322. doi: 10.1128/JB.184.19.5317-5322.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan JA, Boublikova P, Palecek E, Lilley DMJ. Superhelical torsion in cellular DNA responds directly to environmental and genetic factors. Proc Natl Acad Sci USA. 1990;87:8373–8377. doi: 10.1073/pnas.87.21.8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T., Fritsch, E.F., and Sambrook, J. (1982) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Michel B, Grompone G, Flores MJ, Bidnenko V. Multiple pathways process stalled replication forks. Proc Natl Acad Sci USA. 2004;101:12783–12788. doi: 10.1073/pnas.0401586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WG, Simons RW. Chromosomal super-coiling in Escherichia coli. Mol Microbiol. 1993;10:675–684. doi: 10.1111/j.1365-2958.1993.tb00939.x. [DOI] [PubMed] [Google Scholar]
- Mirkin SM, Lyamichev VI, Kumarev VP, Kobzev VF, Nosikov VV, Vologodskii AV. The energetics of the B–Z transition in DNA. J Biomol Struct Dynamics. 1987;5:79–88. doi: 10.1080/07391102.1987.10506376. [DOI] [PubMed] [Google Scholar]
- Mojica FJM, Higgins CF. In vivo supercoiling of plasmid and chromosomal DNA in an Escherichia coli hns mutant. J Bacteriol. 1997;179:3528–3533. doi: 10.1128/jb.179.11.3528-3533.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie AIH, Lilley DMJ. The mechanism of cruciform formation in supercoiled DNA: initial opening of central basepairs in salt-dependent extrusion. Nucleic Acids Res. 1987;15:9641–9654. doi: 10.1093/nar/15.23.9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H, Yamaichi Y, Hiraga S. Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev. 2000;14:212–223. [PMC free article] [PubMed] [Google Scholar]
- Pathania S, Jayaram M, Harshey RM. Path of DNA within the Mu transpososome: transposase interactions bridging two Mu ends and the enhancer trap five DNA supercoils. Cell. 2002;109:425–436. doi: 10.1016/s0092-8674(02)00728-6. [DOI] [PubMed] [Google Scholar]
- Pato M, Banerjee M. The Mu strong gyrase-binding site promotes efficient synapsis of the prophage termini. Mol Microbiol. 1996;22:283–292. doi: 10.1046/j.1365-2958.1996.00115.x. [DOI] [PubMed] [Google Scholar]
- Pato ML, Banerjee M. Replacement of the bacteriophage Mu gyrase site and effect on Mu DNA replication. J Bacteriol. 1999;181:5783–5789. doi: 10.1128/jb.181.18.5783-5789.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pato ML, Karlock M. Central location of the Mu strong gyrase binding site is obligatory for optimal rates of replicative transposition. Proc Natl Acad Sci USA. 1994;91:7056–7060. doi: 10.1073/pnas.91.15.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pato M, Howe MM, Higgins NP. A DNA gyrase binding site at the center of the bacteriophage Mu genome required for efficient replicative transposition. Proc Natl Acad Sci USA. 1990;87:8716–8720. doi: 10.1073/pnas.87.22.8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pato ML, Karlock M, Wall C, Higgins NP. Characterization of Mu prophage lacking the central strong gyrase binding site: location of the block in replication. J Bacteriol. 1995;177:5937–5942. doi: 10.1128/jb.177.20.5937-5942.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavitt GD, Higgins CF. Chromosomal domains of supercoiling in Salmonella typhimurium. Mol Microbiol. 1993;10:685–696. doi: 10.1111/j.1365-2958.1993.tb00940.x. [DOI] [PubMed] [Google Scholar]
- Peck LJ, Nordheim A, Rich A, Wang JC. Flipping of cloned d(pCpG)n.d(pCpG)n DNA sequences from right- to left-handed helical structure by salt, Co(III), or negative supercoiling. Proc Natl Acad Sci USA. 1982;79:4560–4564. doi: 10.1073/pnas.79.15.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettijohn DE, Pfenninger O. Supercoils in prokaryotic DNA restrained in vivo. Proc Natl Acad Sci USA. 1980;77:1331–1335. doi: 10.1073/pnas.77.3.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L, Hardy CD, Arsuaga J, Cozzarelli NR. Topological domain structure of the Escherichia coli chromosome. Genes Dev. 2004;18:1766–1779. doi: 10.1101/gad.1207504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed RR. Transposon-mediated site-specific recombination: a defined in vitro system. Cell. 1981;25:713–719. doi: 10.1016/0092-8674(81)90178-1. [DOI] [PubMed] [Google Scholar]
- Scheirer K, Higgins NP. Transcription induces a supercoil domain barrier in bacteriophage Mu. Biochimie. 2001;83:155–159. doi: 10.1016/s0300-9084(00)01215-3. [DOI] [PubMed] [Google Scholar]
- Sinden RR, Pettijohn DE. Chromosomes in living Escherichia coli cells are segregated into domains of supercoiling. Proc Natl Acad Sci USA. 1981;78:224–228. doi: 10.1073/pnas.78.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden RR, Carlson JO, Pettijohn DE. Torsional tension in the DNA double helix measured with trimethylpsoralen in living E. coli cells: analogous measurements in insect and human cells. Cell. 1980;21:773–783. doi: 10.1016/0092-8674(80)90440-7. [DOI] [PubMed] [Google Scholar]
- Smith HO, Levine M. A phage P22 gene controlling integration of prophage. Virology. 1967;31:207–216. doi: 10.1016/0042-6822(67)90164-x. [DOI] [PubMed] [Google Scholar]
- Spirito F, Figueroa-Bossi N, Bossi L. The relative contributions of transcription and translation to plasmid DNA supercoiling in Salmonella typhimurium. Mol Microbiol. 1994;11:111–122. doi: 10.1111/j.1365-2958.1994.tb00294.x. [DOI] [PubMed] [Google Scholar]
- Staczek P, Higgins NP. DNA gyrase and Topoisomerase IV modulate chromosome domain size in vivo. Mol Microbiol. 1998;29:1435–1448. doi: 10.1046/j.1365-2958.1998.01025.x. [DOI] [PubMed] [Google Scholar]
- Teleman AA, Graumann PL, Lin DCH, Grossman AD, Losick R. Chromosome arrangement within a bacterium. Curr Biol. 1998;8:1102–1109. doi: 10.1016/s0960-9822(98)70464-6. [DOI] [PubMed] [Google Scholar]
- Tu GF, Reid GE, Zhang JG, Moritz RL, Simpson RJ. C-terminal extension of truncated recombinant proteins in Escherichia coli with a 10Sa RNA decapeptide. J Biol Chem. 1995;270:9322–9326. doi: 10.1074/jbc.270.16.9322. [DOI] [PubMed] [Google Scholar]
- Valens M, Penaud S, Rossignol M, Cornet F, Boccard F. Macrodomain organization of the Escherichia coli chromosome. EMBO J. 2004;23:4330–4341. doi: 10.1038/sj.emboj.7600434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci USA. 2004;101:9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vologodskii, A.V. (1992) Supercoiling, knotting, looping, and other large-scale conformational properties of DNA. In Topology and Physics of Circular DNA. Vologodskii, A.V. (ed.). Boca Raton: CRC Press, pp. 372–375.
- Wang, J.C. (1986) Circular DNA. In Cyclic Polymers. Semlyen, J.A. (ed.). London: Elsevier, pp. 225–260.
- Webb CD, Teleman A, Gordon S, Straight A, Belmont A, Lin DCH, et al. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- Weitao T, Dasgupta S, Nordstrom K. Role of the mukB gene in chromosome and plasmid partition in Escherichia coli. Mol Microbiol. 2000;38:392–400. doi: 10.1046/j.1365-2958.2000.02138.x. [DOI] [PubMed] [Google Scholar]
- Welty DJ, Jones JM, Nakai H. Communication of ClpXP protease hypersensitivity to bacteriophage Mu repressor isoforms. J Mol Biol. 1997;272:31–41. doi: 10.1006/jmbi.1997.1193. [DOI] [PubMed] [Google Scholar]


