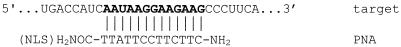Abstract
Peptide nucleic acid (PNA) is a synthetic DNA analogue that is resistant to nucleases and proteases and binds with exceptional affinity to RNA. Because of these properties PNA has the potential to become a powerful therapeutic agent to be used in vivo. Until now, however, the use of PNA in vivo has not been much investigated. Here, we have attempted to reduce the expression of the bcr/abl oncogene in chronic myeloid leukaemia KYO-1 cells using a 13mer PNA sequence (asPNA) designed to hybridise to the b2a2 junction of bcr/abl mRNA. To enhance cellular uptake asPNA was covalently linked to the basic peptide VKRKKKP (NLS-asPNA). Moreover, to investigate the cellular uptake by confocal microscopy, both PNAs were linked by their N-terminus to fluorescein (FL). Studies of uptake, carried out at 4 and 37°C on living KYO-1 cells stained with hexidium iodide, showed that both NLS-asPNA-FL and asPNA-FL were taken up by the cells, through a receptor-independent mechanism. The intracellular amount of NLS-asPNA-FL was about two to three times higher than that of asPNA-FL. Using a semi-quantitative RT– PCR technique we found that 10 µM asPNA and NLS-asPNA reduced the level of b2a2 mRNA in KYO-1 cells to 20 ± 5% and 60 ± 10% of the control, respectively. Western blot analysis showed that asPNA promoted a significant inhibition of p210BCR/ABL protein: residual protein measured in cells exposed for 48 h to asPNA was ∼35% of the control. Additionally, asPNA impaired cell growth to 50 ± 5% of the control and inhibited completion of the cell cycle. In summary, these results demonstrate that a PNA 13mer is taken up by KYO-1 cells and is capable of producing a significant and specific down-regulation of the bcr/abl oncogene involved in leukaemogenesis.
INTRODUCTION
Over the past few years, peptide nucleic acids (PNAs) have emerged as one of the most promising new types of therapeutic oligonucleotides in the form of antisense and anti-gene molecules (1). From the chemical point of view, PNA is both a peptide and a nucleic acid, since it is a hybrid molecule consisting of N-(2-aminoethyl)glycine monomers linked by amide bonds (2,3). The purine (A and G) and pyrimidine (C and T) nucleobases are attached to this polyamide backbone via a methylene carbonyl linkage. As PNAs are not charged at physiological pH, they bind to complementary nucleic acid strands with higher affinity than do natural nucleic acids (4). Although hybridisation of PNAs to complementary DNA and RNA obeys the Watson–Crick rules, both parallel and antiparallel duplexes can be formed. The N-terminus corresponds to the 5′ end and the C-terminus to the 3′ end of a normal oligonucleotide (4–6). However, antiparallel PNA–DNA and PNA–RNA duplexes are more stable and form more rapidly than the corresponding parallel duplexes (7). The PNA sequences used in this study are shown in Figure 1. They are designed to reduce the expression of the bcr/abl oncogene in chronic myelogenous leukaemia (CML) cells (8–10). In most CML patients, leukaemic cells contain the Philadelphia (Ph) chromosome, which derives from a reciprocal translocation of chromosomes 9q and 22q. Such a translocation generates a fused bcr/abl oncogene (with either a b2a2 or b3a2 junction) encoding a protein of 210 kDa (p210BCR/ABL) which is involved in leukaemogenesis (11,12). Protein p210BCR/ABL has a higher tyrosine phosphokinase activity than the normal ABL protein and its structure allows multiple protein–protein interactions that link p210BCR/ABL to several intracellular pathways, including the ras pathway, which regulates cell proliferation and differentiation (8–10). The presence of a unique sequence at b2a2 and b3a2 mRNA junctions and the observation that CML cells require p210BCR/ABL to survive make the bcr/abl gene an attractive target for antisense molecules. Attempts to reduce the intracellular level of bcr/abl mRNA in different cell lines by means of antisense oligonucleotides have been reported (13). However, the results have not been very satisfactory, because normal and thioate antisense oligonucleotides displayed a number of limits, including nuclease degradation, non-specific effects and insufficient target affinity. PNAs are not susceptible to degradation by endogenous nucleases and proteases and bind to RNA targets with exceptionally high affinity and in a sequence-specific manner. Because of these favourable properties, PNAs are potentially more powerful antisense agents than phosphorothioate oligonucleotides. To date, only a few reports have shown that PNAs, either free or conjugated, produce an antisense or antigene effect in vivo (14–19). In the study presented herein we have explored the capacity of 13mer antisense PNAs to inhibit the expression of bcr/abl in leukaemic KYO-1 cells. The PNAs were designed to bind to the b2a2 junction of bcr/abl mRNA and thus block its translation sterically. We found that the designed b2a2 antisense PNAs: (i) are taken up by KYO-1 cells; (ii) specifically reduce the level of p210BCR/ABL; (iii) inhibit completion of the cell cycle; (iv) impair cell growth to 50% of the control.
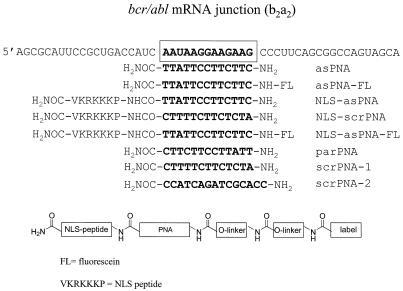
Figure 1.
Sequence of the b2a2 junction of bcr/abl mRNA expressed in KYO-1 cells. The structures of the antisense and control PNAs used in this study are also shown.
MATERIALS AND METHODS
PNA synthesis
The PNAs used in this study were made by solid phase synthesis using a peptide synthesiser (Applied Biosystems 433A peptide synthesiser) and PNA synthesiser (Perspective Biosystems Expedite Nucleic Acid Synthesis System). All solvents (Biosolve) were used as received. The solid phase syntheses were performed on PEG-PS beads as solid support with rink-amide as linker (loading 0.17 µmol/mg). For PNAs conjugated to NLS, first the peptide part was assembled on the peptide synthesiser on the 20 µmol scale using protected amino acids [5 equivalents, Fmoc-Val-OH, Fmoc-Lys(Boc)-OH, Fmoc-Arg(Pbf)-OH and Fmoc-Pro-OH] under the agency of BOP/HOBt as the coupling agent. Only the first coupling, that of valine, was double, and after the synthesis was complete the final Fmoc was left on. The loading was determined by deprotection of a small amount of the resin and measuring the absorbance of the liberated Fmoc protecting group (loading 0.13 µmol/mg, efficiency 98%). Assembly of the PNAs was established using the standard protocol described in the synthesiser manual, on the 2 µmol scale using Fmoc chemistry and monomers in which the exocyclic amines were Bhoc protected (Perspective Biosystems). As solid support either PRG-PS beads with rink-amide as the linker (loading 0.17 µmol/mg) or NLS-functionalised resin (loading 0.13 µmol/mg) was used. After removal of the final Fmoc protecting group the oligomer was either cleaved from the resin and deprotected under acidic conditions (TFA/TIS/ H2O, 9:1:1 v/v/v) or first coupled twice with the O-linker [FmocHN(CH2)2O(CH2)OCH2CO2H], then reacted with 5(6)-carboxyfluorescein succinimide ester (Molecular Probes) and subsequently cleaved with concomitant deprotection. RP-HPLC purification and analysis were carried out on a JASCO HPLC system equipped with an Altima C18 column (10 × 250 mm). Gradient elution was performed at 40°C by building up a gradient starting with buffer A (0.1% TFA in water) and applying buffer B (0.1% TFA in acetonitrile/water, 3:1 v/v) with a flow rate of 4 ml/min. The PNAs obtained were lyophilised and characterised by matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF MS) and RP-HPLC.
Cell cultures
CML cell lines K562 (a gift of Dr M. Giunta, Bone Marrow Transplantation Unit, Medical School, University of Udine, Italy) and KYO-1 (a gift of Dr C. M. Broughton, Department of Hematology, University of Liverpool, UK) were maintained in exponential growth in RPMI 1640 medium containing 100 U/ml penicillin, 100 mg/ml streptomycin, 200 mM l-glutamine and 10% fetal bovine serum (FBS) (Celbio, Milan, Italy), which was heat inactivated at 56°C for 30 min.
Cell cycle analysis
Cell cycle analysis was performed on KYO-1 cells growing in the presence of 10 µM PNA for 30 h. Briefly, KYO-1 cells were seeded in a 24-well plate at a cell density of 105 cells/ml, 24 h before PNA treatment. Cells (2 × 105) were harvested (240 g, 4°C), resuspended in 0.5 ml of PBS and washed twice. While vortexing, 1 ml of ice-cold 70% ethanol was added dropwise to the pellet and the cells were allowed to fix overnight at 4°C. Prior to FACS analysis (Becton Dickinson) the cells were stained with a solution containing 0.05 mg/ml propidium iodide and 0.5 mg/ml RNase A. The cells were centrifuged and the supernatant poured off. The cells were vortexed in the residual ethanol and then 0.5 ml of propidium iodide was added to each sample tube. The cells were incubated for 30 min at 37°C and then analysed by FACS.
Proliferation assay in liquid culture
Aliquots of 2 × 104 exponentially growing KYO-1 cells in liquid culture were seeded in 200 µl of complete medium in 96-well microtitre plates and treated with the PNAs. The PNAs were added to the medium 24 h after seeding. Cellular growth was measured at 24, 48 and 72 h using 0.4 mg/ml 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (20). The absorbance was read at 540 and 690 nm using a microtitre plate reader (Titertek Multiscan MCC; Labsystems). The data are expressed in terms of T/C (OD of PNA-treated cells/OD of control cells) as a measure of cell viability and survival in the presence of test PNAs.
Confocal microscopy
KYO-1 cells were seeded in 12-well plates at a density of 0.8 × 105 cells in 600 µl of RPMI medium supplemented with 10% heat-inactivated FBS. After 24 h the cells were exposed for a certain time to fluorescein-labeled non-conjugated PNA (asPNA-FL) and PNA conjugated to the basic peptide VKRKKKP (NLS-asPNA-NLS). The cells were centrifuged, washed twice with PBS, spun on a glass slide and fixed with 3% paraformaldehyde (PFA) in PBS for 20 min. After washing with 0.1 M glycine, containing 0.02% sodium azide in PBS, to remove PFA, and Triton X-100 (0.1% in PBS), the cells were incubated with 24 µg/ml propidium iodide and 0.4 mg/ml RNase A for 30 min at 37°C in order to stain the nuclei. Then coverslips were mounted on the glass slide with mowiol 4-88 and DABCO (2.5%). The cells were analysed using a Leica DM IRBE confocal imaging system. Diaphragm and fluorescence detection levels were adjusted to minimise any interference between the fluorescein and propidium iodide channels. Hexidium iodide (Molecular Probes) was dissolved in DMSO and 10 mM stock solutions were stored at –20°C.
RNA purification and reverse transcription–polymerase chain reaction (RT–PCR) analysis
RNA purification. Total cellular RNA was extracted from K562 and KYO-1 cells using RNAqueous-4 PCR (Ambion). This method is based on cell lysis in a solution containing guanidinium thiocyanate. The RNA precipitate was treated with ultrapure DNase I to remove DNA traces and the RNA solutions were stored at –80°C.
cDNA synthesis. A volume of 5 µl of RNA solution in diethylpyrocarbonate (DEPC)-treated water was heated at 70°C and placed in ice. The solution was added to 16.6 µl of cDNA mix [10 µl of 5× buffer, 5 µl of 0.1 M DTT (Life Technologies), 2 µl of a 12.5 pmol primer AZ (5′-CCA TTTTTGGTTTGGGCTTCA-3′; MWG Biotech), 2 µl of a 5 mM solution containing equimolar amounts of dATP, dCTP, dGTP and dTTP (Amersham Pharmacia Biotech), 0.6 µl of 40 U/ml RNase inhibitor (Life Technologies) and 1 µl of 200 U M-MLV reverse transcriptase (Life Technologies)]. The reactions (50 µl/tube) were incubated for 1 h at 37°C. As a negative control the reverse transcription reaction was performed with 5 µl of DEPC-treated water. The cDNA was stored at –20°C.
Polymerase chain reaction. A volume of 10 µl of cDNA, heated at 95°C for 5 min, was mixed with 16.5 µl of BCR/ABL mix [10 µl of 10× Taq polymerase buffer (Boehringer), 3 µl of 25 mM MgCl2, 2 µl of 12.5 pmol/µl primer EA500 (5′-TGTGATTATAGCCTAAGACCC-3′; MWG Biotech), 2 µl of 12.5 pmol/µl primer EA122 (5′-GTTTCAG AAGCTTCTCCCTG-3′; MWG-Biotech), 2 µl of 5 mM dNTPs with equimolar amounts of dTTP, dCTP, dATP and dGTP (Amersham Pharmacia Biotech), 0.5 µl of 2.5 U/ml AmpliTaq DNA polymerase (Perkin Elmer)]. Amplification was carried out on an automated DNA thermal cycler (Progene) as follows: 35 cycles of denaturation (94°C for 30 s), annealing (60°C for 30 s) and extension (72°C for 30 s). Abl amplification (annealing at 55°C) was carried out with primers EA500 and ABL1A (5′-CCTCTCGCTGGA CCCAGTGA). Two positive cDNA controls were used. First, RNA extracted only from K562 cells was used to amplify the b3a2 bcr/abl mRNA. Secondly, the RNA extracted only from KYO-1 cells was used to amplify the b2a2 bcr/abl mRNA. Moreover, as a negative control we tried to amplify the controls of the RNA extraction and the control of the cDNA reaction.
Western blots
Aliquots of 2 × 105 KYO-1 cells were lysed in 2× Laemmli sample buffer (3.3% SDS, 22% glycerol, 1.1 M Tris–HCl, pH 6, 0.001% bromophenol blue, 10% β-mercaptoethanol). Samples were heated at 95°C for 10 min and loaded (70 µl) in either 7 or 12% resolving, 5% stacking SDS–polyacrylamide gel. Electrophoresis was carried out at 100 and 40 V for 45 min and 18 h, respectively. The relative concentration of the protein lysates was estimated electrophoretically and equal amounts of lysates were transferred to a nitrocellulose membrane (Sartorius) using a Multiphor II Novablot Transfer Unit (Amersham Pharmacia Biotech). The membranes were then cut at the 45 and 210 kDa molecular weight levels. Both membranes were used to detect the BCR/ABL and β-actin. A double antibody procedure was used to detect the proteins. The membranes were incubated under agitation for 1 h at room temperature in blocking solution (PBS containing 5% dry milk and 0.1% Tween 20), then at room temperature for 2 h in c-ABL (Ab-3) (1:50) and β-actin monoclonal antibodies (1:20 000) (Calbiochem). The antibody solution was removed and the membrane was washed three times with PBS containing 0.1% Tween 20. The membranes were then incubated (1 h at room temperature under agitation) with horseradish peroxidase-conjugated goat anti-mouse IgG (1:1000) (Sigma) for the Ab-3 and goat anti-mouse IgM (1:2000) (Calbiochem) for the β-actin antibody. Chemiluminescence was detected immediately as described by the manufacturer (Super Signal West Pico Trial; Pierce). Films were exposed for ∼15 min for BCR/ABL and 1 min for β-actin.
RESULTS
Target and antisense PNA molecules
Ph-positive KYO-1 cells express a bcr/abl mRNA which is characterised by a bcr exon 2/abl exon 2 (b2a2) junction. As this junction is a unique sequence in CML cells, it has been chosen as the target for the antisense PNA molecules used in this study (Fig. 1). According to previous observations, free PNAs do not penetrate or hardly penetrate cell membranes in the absence of a specific carrier (21,22). So, to enhance cellular uptake, the designed antisense PNA was covalently bound at the C-terminus (3′) to a short peptide rich in basic amino acids, having the sequence of the SV40 nuclear localisation signal peptide (NLS) (HO2C-VKRKKKP-NH2). Moreover, to perform uptake studies by flow cytometry and confocal microscopy, both free and NLS-conjugated PNAs were covalently linked at their N-terminus (5′) to 5(6)-carboxyfluorescein via an amide bond. The PNA 13mers shown in Figure 1, designed with a sequence complementary to the bcr/abl mRNA b2a2 junction, were assembled following a well-established solid phase protocol (23). Sequential elongation of the rink-amide-functionalised solid support, either with or without the NLS peptide present, using Fmoc/Bhoc-protected monomers under the agency of the coupling reagent HATU, gave the immobilised constructs. At this stage, the label could be introduced by reacting the deprotected N-terminus with the succinimidyl ester of 5(6)-carboxyfluorescein. Removal of the N-Bhoc protecting groups and concomitant release of the support was affected under acidic conditions, to give, after RP-HPLC purification, the PNA conjugates. The homogeneity and identity of these constructs were established by RP-HPLC and MALDI-TOF MS. Antisense PNA oligomers were synthesised with (NLS-asPNA) and without (asPNA) NLS peptide and both of them also with fluorescein (asPNA-FL and NLS-asPNA-FL). As a control we designed parPNA, a PNA with an inverted sequence compared to asPNA, and two PNAs with scrambled sequences, scrPNA-1 and scrPNA-2 (Fig. 1).
PNA uptake: confocal microscopy and FACS
It has recently been reported that unmodified, carrier-free PNA injected into rats either directly into the brain or i.p. was able to enter neuronal cells and specifically reduce gene expression (18). Other recent studies show that unmodified antisense PNA molecules are taken up by protozoa (Entameba hystolitica) (21) and by neurons in culture (17). These data suggest that a certain amount of PNA is intrinsically taken up by the cells, in contrast to the proposal that PNAs do not pass or hardly pass through the membranes (22,24–26). Thus, we wished to investigate the uptake of free asPNA-FL and peptide-conjugated NLS-asPNA-FL by KYO-1 cells using confocal microscopy. Two cell staining procedures were followed. In one case, after incubation with free asPNA-FL or NLS-asPNA-FL, the cells were fixed on a glass slide and exposed to 0.1% Triton X-100 and propidium iodide. As Triton X-100 permeabilises the cell membrane, it is possible that some of the PNA taken up by the cells is lost during the washing steps. In order to avoid this we also stained the cells directly, i.e. without fixing them on a glass plate, with 3 µM hexidium iodide, a dye that binds to DNA and RNA in living cells and displays a red fluorescence (27). KYO-1 cells were treated for 2 h with hexidium iodide, transferred to growth medium containing either free asPNA-FL or NLS-asPNA-FL for a certain time and then directly analysed with a confocal microscope. Figures 2A and B and 3 show images obtained by both staining methods. PNA-treated KYO-1 cells fixed on glass and stained with propidium iodide are shown in Figure 2A. Figure 2A shows the nuclei of KYO-1 cells stained in red by propidium iodide, while Figure 2B shows the green fluorescent light emitted by asPNA-FL (incubation time 24 h). It is clear from these pictures that asPNA-FL is taken up by the cells. A superimposed view of Figure 2A and B is reported in Figure 2C. This image clearly shows that asPNA-FL is partly located in the nuclei and partly in the cytoplasm (green crown surrounding the nucleus, Fig. 2D). The intracellular localisation of asPNA-FL was also confirmed by xz-planes taken at the level of the arrow (Fig. 2E). The uptake of NLS-asPNA-FL by KYO-1 cells is shown in Figure 2B. NLS-asPNA-FL is localised in both the cytoplasm and nucleus, where it appears mainly confined to the nucleoli. Figure 3 shows the PNA uptake in living KYO-1 cells stained with hexidium iodide. The images show in an unambiguous way that both asPNA-FL and NLS-asPNA-FL are taken up by the cells. To investigate the mechanism by which PNA is internalised in KYO-1 cells, experiments were performed at 4°C. Basically, the cells were first stained with hexidium iodide, cooled to 4°C over 1 h and then exposed to 10 µM asPNA-FL or NLS-asPNA-FL for a further 5 h at 4°C. After incubation the cells were immediately analysed with a confocal microscope. We observed that both free and NLS-conjugated PNAs were also detected in the cells at low temperature (not shown).
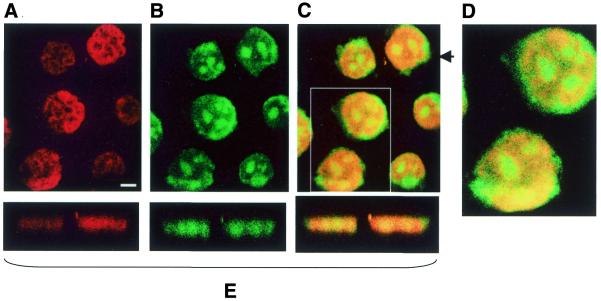
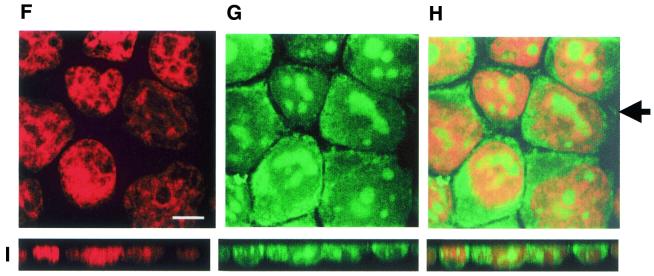
Figure 2.
Confocal microscopy images of PNA-treated KYO-1 cells fixed on glass slides and stained with propidium iodide. (A–D) Cells incubated for 24 h with 10 µM asPNA-FL. (E) xz-Planes taken at the level of the arrow. (F–H) Uptake of NLS-asPNA-FL in KYO-1 cells stained with propidium iodide. (I) xz-Sections taken at the level of the arrow. Experiments were performed at 37°C. Bar 5 µm.
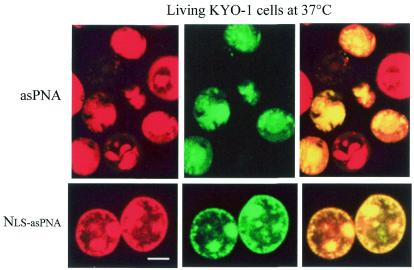
Figure 3.
Confocal microscopy of living KYO-1 cells treated for 12 h with asPNA-FL or NLS-asPNA-FL and stained with the vital dye hexidium iodide. (Top) Uptake at 37°C by KYO-1 cells treated with 10 µM asPNA-FL. (Bottom) Uptake by KYO-1 cells treated with 10 µM NLS-asPNA-FL. Bar 5 µm.
An estimate of the fluorescence intensity associated with the KYO-1 cells treated with asPNA-FL and NLS-asPNA-FL was obtained by flow cytometry. The average fluorescence intensity associated with the cells treated with 10 µM asPNA-NLS-FL was 210 ± 20 units, while the fluorescence of the cells treated with asPNA-FL was about three times lower, i.e. 70 ± 15 units. These values did not significantly change over incubation periods up to 40 h.
The designed antisense PNA binds the b2a2 mRNA target in vitro
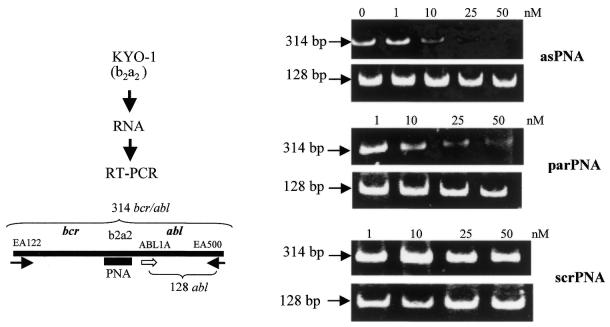
In order to demonstrate that the designed antisense PNAs stably hybridise to their natural target, i.e. the b2a2 mRNA junction, asPNA, asPNA-NLS, parPNA and scrPNA were incubated separately with total RNA extracted from KYO-1 cells, at concentrations increasing from 1 to 50 nM. Both asPNA and NLS-asPNA are complementary to a 13mer stretch of the b2a2 junction of bcr/abl mRNA, with which they can form the antiparallel PNA–RNA heteroduplex shown in Scheme 1, which could sterically block reverse transcription. In contrast, parPNA is expected to hybridize to the b2a2 junction with a parallel orientation, forming a PNA–DNA duplex which is expected to be less stable than that formed by asPNA (3,4). After RT–PCR, carried out using the primers EA122 and EA500, the reaction products were separated by polyacrylamide electrophoresis (Fig. 4). It can be seen that asPNA inhibits reverse transcription in a dose-dependent manner, completely blocking the enzymatic activity at a concentration as low as 50 nM. Surprisingly, parPNA too showed a remarkable ability to inhibit reverse transcription, under the experimental conditions adopted. In contrast, samples incubated either in the absence of antisense PNA or the presence of srcPNA (a PNA with a scrambled sequence) did not show reverse transcription inhibition. The efficiency of reverse transcriptase was checked by amplifying a 128 nt stretch of the abl gene, using primers ABL1A and EA500, in each reaction tube. It can be seen that in all samples the expected abl fragment was obtained. These results demonstrate that the designed antisense PNAs are able to bind at nanomolar concentrations to the b2a2 mRNA junction with high affinity and completely block the catalytic activity of reverse transcriptase. We also performed RT–PCR experiments with NLS-asPNA and have observed that it produces an inhibitory effect roughly similar to that obtained with the unconjugated asPNA (not shown). The thermodynamic stability of parallel and antiparallel PNA–RNA hybrids was predicted using the nearest neighbour stacking energy parameters reported by Santa Lucia et al. (28), which allows Tm calculation for DNA–DNA duplexes. Taking into account that PNA–DNA duplexes are more stable than the corresponding DNA–DNA duplexes by ∼1.0 K/base (3) and that PNA–DNA duplexes are less stable than PNA–RNA by ∼4°C (29), we calculated for the antiparallel asPNA–b2a2 mRNA heteroduplex a Tm of 68°C at 1 µM, 52°C at 10 nM and 42°C at 10 pM. As for the parallel heteroduplex formed by parPNA with the b2a2 target, its stability should be comparable to that of the corresponding DNA–DNA duplex, i.e. 52°C at 1 µM, 37°C at 10 nM and 26°C at 10 pM (3,27).
Scheme 1.
Figure 4.
In vitro inhibition of reverse transcription by PNA. Total RNA extracted from KYO-1 (b2a2 junction) cells was incubated overnight with control or antisense PNAs at increasing concentrations (0–50 nM). The RNA samples were subjected to RT–PCR, using bcr/abl primers EA122 and EA500, and the products separated in a 12% polyacrylamide gel. The cDNA amplification with primers EA122 and EA500 provides a 314 bp band. RT–PCR is inhibited by asPNA and parPNA, which bind to RNA and block reverse transcription, but not srcPNA, which does not bind to RNA. As a control, the amplification of a 128 bp DNA fragment of the abl gene was amplified.
Antisense PNA binds to the b2a2 junction in vivo and reduces bcr/abl mRNA
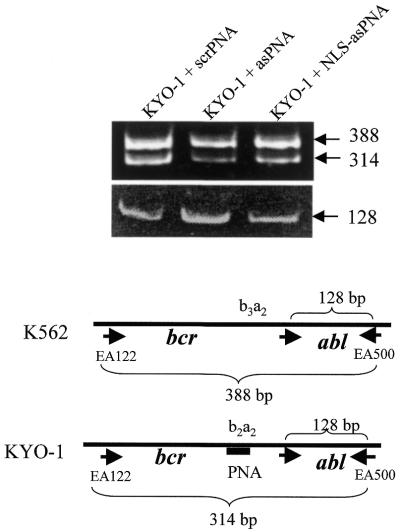
In contrast to phosphate and phosphorothioate oligonucleotides, antisense PNAs do not induce degradation of PNA–RNA complexes by RNase H (30). Despite this, it has been found that antisense PNA depressed the amount of mRNA coding for prepro-oxytocin in neurons (17). We therefore measured the relative level of bcr/abl mRNA from PNA-treated KYO-1 cells following a competitive PCR strategy as previously described (31,32). The total RNA extracted from a nearly 1:1 mixture of PNA-treated KYO-1 cells (containing the target b2a2 junction) and untreated K562 cells (containing a b3a2 junction which is not recognised by the PNA, competitor) was transformed into cDNA and amplified by PCR. Target and competitor mRNA share the primer recognition sites, but differ in size by 74 bp. Running the RT–PCR products in a polyacryalmide gel, two bands were obtained: one of 388 bp from K562, the other of 314 bp from KYO-1 cells. The ratio between b2a2 mRNA from PNA-treated KYO-1 cells (314 bp) and b3a2 mRNA from untreated K562 cells (388 bp) provides an estimate of the relative amount of target mRNA compared to the control mRNA from untreated cells. In the presence of 10 µM asPNA, the b2a2/b3a2 ratio was reduced to 20 ± 5% of the control, while 10 µM NLS-asPNA reduced the b2a2/b3a2 ratio to only 60 ± 10% of the control (Fig. 5). The efficiency of the RT reaction in each reaction tube was checked by amplifying a 128 bp fragment of abl.
Figure 5.
Effect of b2a2 antisense PNA on the level of bcr/abl mRNA in KYO-1 cells quantified by RT–PCR. The RNA from an equal number of PNA-untreated K562 (competitor) and PNA-treated KYO-1 cells was extracted. The extracted RNA was subjected to RT–PCR. Using primers EA122 and EA500, two amplified bcr/abl bands of 388 and 314 bp were obtained from K562 (b3a2 junction) and KYO-1 cells (b2a2 junction), respectively. When the KYO-1 cells were treated with asPNA or NLS-asPNA, the intensity of the 314 bp band, but not that of the 388 bp band, was reduced, compared to the band obtained by treating KYO-1 cells with scrPNA, a control PNA which is unable to bind to bcr/abl mRNA. As a control we also amplified a 128 bp DNA fragment of abl in each reaction tube, using primers ABL1A and EA500. As expected, this amplification is not influenced by PNA.
Depression of p210BCR/ABL level by antisense PNA
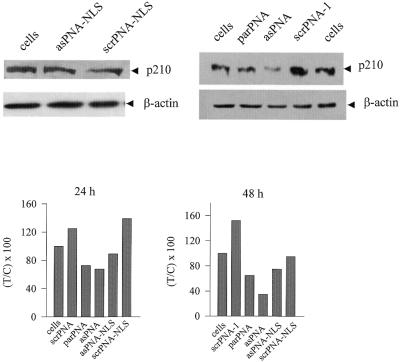
The capacity of antisense PNA to reduce the expression of bcr/abl in KYO-1 cells was measured by immunoblotting. KYO-1 cells were cultured in the absence and presence of antisense or control PNAs for 24, 48 and 72 h. After incubation, the amount of protein in untreated and PNA-treated KYO-1 cells was determined by western blot using an anti-BCR/ABL monoclonal antibody. To demonstrate gene specificity, the level of β-actin was also measured in all the samples. Figure 6 shows typical western blots at 48 h. It can be noted that after 48 h incubation with 10 µM asPNA the level of p210BCR/ABL protein is reduced to ∼35% compared to that in untreated cells. Western blots were also carried out at 24 h and the level of p210BCR/ABL compared to that of untreated cells is reported in the enclosed histograms. It can be seen that the maximum reduction of p210BCR/ABL level is observed in the cells exposed to asPNA for 48 h. The data show that 10 µM NLS-asPNA induces at 48 h an inhibition of bcr/abl expression much lower than that observed with asPNA. An antisense effect was also observed with parPNA, as this molecule forms a parallel hybrid with the mRNA target (5). However, as parPNA has a lower affinity for bcr/abl mRNA, it promotes a weak inhibitory effect at the times considered: residual p210BCR/ABL is 73 and 65% at 24 and 48 h, respectively. We also observed that exposing the cells to the PNAs for 72 h resulted in a non-significant reduction of the level of p210BCR/ABL (not shown).
Figure 6.
Immunoblot of KYO-1 cell lysates using anti-ABL and anti-β-actin monoclonal antibodies. KYO-1 cells were incubated with 10 µM anti sense or control PNAs and the amount of protein p210BCR/ABL was evaluated by western blot analysis, after the cells were exposed to the PNAs for 24 and 48 h. The level of β-actin in the PNA-treated cells was also measured. A typical western blot analysis of cell lysates at 48 h after PNA treatment is shown in the figure. The levels of p210BCR/ABL in lysates obtained from KYO-1 cells treated with antisense and control PNAs at 24 and 48 h are shown in the enclosed histograms. The ordinate reports the residual BCR/ABL protein expressed as percent T/C, where T is the p210BCR/ABL/β-actin ratio of PNA-treated cells and C is the p210BCR/ABL/β-actin ratio of PNA-untreated cells. The uncertainty on each value is at most 20%.
Antiproliferative effect of antisense PNA
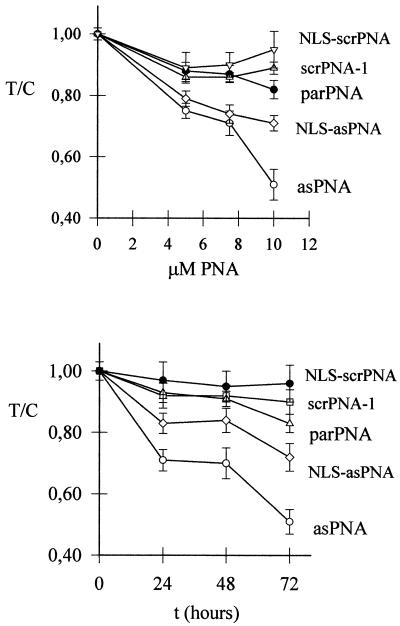
The effect on cell growth of the anti-b2a2 PNAs was investigated by the MTT assay. In a first set of experiments cell growth was measured at increasing PNA concentrations. Control and antisense PNAs were added to the culture medium at 5, 7.5 or 10 µM and the cells were grown for 72 h. The ratio T/C between treated (T) and untreated (C) cells as a function of PNA concentration is reported in Figure 7 (top). It can be seen that asPNA causes a strong antiproliferative effect compared to untreated cells (26, 28 and 50% at 5, 7.5 and 10 µM, respectively) after an incubation time of 72 h. NLS-asPNA was found to promote a weaker cell growth inhibition (21, 29 and 28%) at the same concentrations. In contrast, control molecules scrPNA, NLS-scrPNA and parPNA showed a weak inhibitory effect of ∼5–10% at 10 µM. The effect on cell growth as a function of time of 10 µM PNAs is shown in Figure 7 (bottom). It can be seen, in accordance with the data in Figure 7 (top), that the cell growth inhibition promoted by asPNA reaches a level of 50% at 72 h. After 72 h incubation, NLS-asPNA inhibits cell growth by ∼25%, while parPNA produced a smaller effect on cell growth.
Figure 7.
Antisense PNA inhibits the proliferation of KYO-1 cells in liquid culture. (Top) Cell growth in the presence of increasing PNA concentrations. The cells were grown for 72 h in the presence of the indicated PNAs, before a standard MTT assay was performed. (Bottom) Cell growth as a function of time in the presence of 10 µM PNA, measured by MTT assay. The data are reported as T/C (OD of PNA-treated cells/OD of PNA-untreated cells).
Cell cycle analysis of KYO-1 cells grown in the presence of antisense PNA
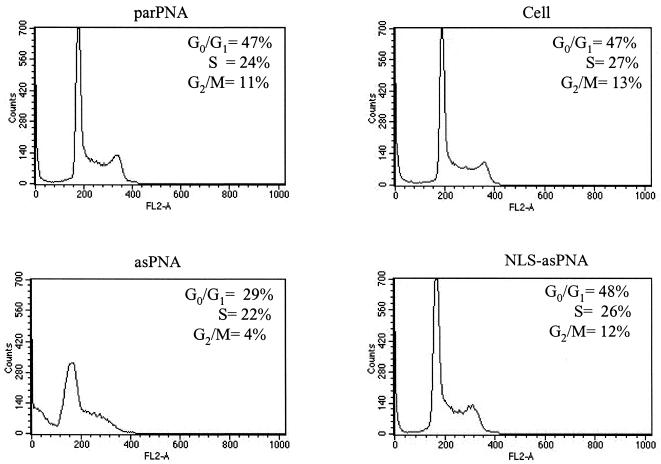
To further investigate the biological consequences of asPNA in KYO-1 cells, the cell cycle was assessed by FACS analysis, using propidium iodide staining of the DNA content. KYO-1 cells were grown in liquid culture for different times in the presence of 10 µM asPNA or control PNAs. Figure 8 shows that asPNA, the compound with the highest antisense activity, produced a significant effect on the cell cycle. After 30 h growth, untreated cells or cells treated with parPNA or NLS-asPNA exhibit a DNA content between 2N (∼47 ± 1% of cells) and 4N (12 ± 1% of cells), suggesting that the S phase is active (26 ± 2% cells). In contrast, the cell cycle profile of the cells treated with asPNA shows a reduction in the G2/M peak from 12 ± 1% to 3.7 ± 0.5%, suggesting that in the presence of asPNA completion of the cell cycle appears inhibited. According to the cell cycle analysis, a fraction (25 ± 4%) of KYO-1 cells treated with asPNA underwent apoptosis. Thus, it is possible that the inhibition of cell proliferation observed with the MTT assay results from the fact that asPNA induces apoptosis.
Figure 8.
Cell cycle analysis of PNA-treated KYO-1 cells. KYO-1 cells grown for 30 h in medium containing 10 µM PNA were subjected to cell cycle analysis. Cells were stained with propidium iodide and analysed by flow cytometry. Inset numbers indicate the percentage of cells in the phases of the cell cycle. The percentages of dead cells are 18 and 14% in parPNA- and NLS-asPNA-treated cells respectively, 13% in PNA-untreated cells and 45% in asPNA-treated cells; this high value may be due to apoptosis.
DISCUSSION
The results reported in this paper show that a 13mer antisense PNA targeted to the b2a2 mRNA junction in KYO-1 cells down-regulates bcr/abl gene expression up to ∼35% of that observed in untreated cells and inhibits cell growth by 50% in a 3 day liquid culture assay. Another finding of this study is that the major biological effect is observed with antisense PNA in the free form and not with its peptide-conjugated analogue. The biological activity in vivo of non-conjugated PNA is rather controversial because it is generally believed that this neutral molecule is unable to penetrate cell membranes. However, recent data disagree with this proposal, because: (i) free PNA administered i.p. was able to cross the blood–brain barrier and to reduce the expression of neurotensin (18) and mu (33) receptor genes; (ii) free PNA added to cells in culture and targeted to a particular region of the non-coding RNA Xist caused disruption of X-chromosome inactivation (34); (iii) free PNA administered i.v. in rat and targeted against the δ-opioid receptor gene reduced gene function (16). In addition to these studies, our own confocal and flow cytometry data show that a certain amount of free PNA can indeed penetrate the cell membrane of KYO-1 cells. We followed two staining procedures. In one case the cells were fixed on a glass slide, permeabilised with Triton X-100 and stained with propidium iodide. In the other case, unfixed cells were treated with hexidium iodide, a vital dye staining both the cytoplasm and nucleus of living cells. This last procedure avoids several washing steps and has the advantage of showing the uptake by intact and living cells. When the PNA was conjugated to NLS, its capacity to penetrate the membranes increased about three times, according to flow cytometry data, in keeping with previous findings (17). The observation that the uptake of NLS-asPNA was still detected at 4°C strongly suggests that this peptide-conjugated PNA is translocated into the cells through a receptor-independent mechanism, which might be based on direct fusion of the neutral PNA with the cell membrane. The cluster of positive charges on NLS should favour uptake by disrupting the structure of the outer membrane layer, acting on the negatively charged phospholipids (35). Moreover, since asPNA is also internalised in the cells at 4°C, its uptake should not be mediated by endocytosis, as occurs for phosphate and phosphorothioate oligonucleotides (36).
The capacity of the designed PNAs to hybridise to the b2a2 RNA target was checked in vitro by RT–PCR. The fact that asPNA (and NLS-asPNA) completely blocked reverse transcription suggests that these molecules can form very stable complexes with the b2a2 target. It is noteworthy that parPNA also forms a sufficiently stable parallel PNA–RNA duplex capable of inhibiting reverse transcription. If we assume that only a small fraction, for instance 10–3, of the PNA added to the culture medium (usually 10 µM) is taken up by the cells, the intracellular PNA concentration will be ∼10 nM. At this low concentration the antiparallel PNA–RNA hybrid should still be characterised by a Tm of ∼52°C, which is compatible with a strong biological activity. For instance, an antisense PNA injected i.p. into rats was found to reduce neurotensin receptor expression at concentrations as low as 2.4 ng/whole brain (18). In contrast, 10 nM parPNA, which forms a heteroduplex with a predicted Tm of ∼37°C with the b2a2 mRNA target, should produce a lower antisense effect, which is in keeping with the experimental results.
Another question about the use of antisense PNAs that remains to be clarified regards their mechanism of action. It is known that PNAs do not activate RNase H, so the antisense effect of this molecule should be ascribed to steric blocking of translation. It has been shown in a cell-free system that when the target is near the translation initiation site, hybridisation of the PNA to this target hinders the assembly of the translation initiation complex. When the target is far from the translation initiation site, as at the b2a2 junction, the PNA should induce production of a truncated protein (37). Interestingly, this and other studies (17) have reported that PNA can promote a reduction in mRNA. To rationalise this finding, Aldrian-Herrada et al. (17) have hypothesised that when mRNA is blocked by an antisense PNA, it may enter a metabolic pathway that leads to degradation. It is also possible, however, that the reduction in mRNA is simply a consequence of the fact that RNA was quantified by RT–PCR. In fact, since PNA–RNA hybrids are resistant to phenol extraction and urea denaturation (38), the total RNA extracted from the PNA-treated cells will probably contain b2a2 mRNA bound to PNA. It follows that the RNA which is bound to PNA will not be amplified by RT–PCR. In this case, the observed reduction in b2a2 mRNA level would provide indirect evidence that antisense PNA added to the culture medium enters the cells and binds to the corresponding intracellular target. The fact that PNA–RNA heteroduplexes do not induce cleavage of the mRNA target by RNase H suggests that any non-specific effects may be limited, as the possible interaction of asPNA with other mRNA molecules will only result in a temporary arrest of translation. In contrast, non-specific binding of phosphate and phosphorothioate antisense oligonucleotides that induce RNase H may result in irreversible degradation of critical cellular mRNAs with consequent serious side effects.
We were surprised to observe that NLS-asPNA appeared to be a weaker antisense agent than free asPNA. In using NLS-asPNA we reasoned that this molecule, by virtue of its ability to penetrate the nucleus, would be able to hybridise with the b2a2 RNA target at both the nuclear and cytoplasmic levels and efficiently block translation. Since this is not supported by the experimental data, it is possible that NLS-asPNA, due to the positive charges of Lys residues, interacts non-specifically with cellular proteins.
The reduction in p210BCR/ABL appears stronger at 24 and 48 than at 72 h. This is an obvious consequence of the fact that the antisense effect that we observed follows a single PNA administration. This behaviour is in keeping with the results of a previous study in which the amount of p210BCR/ABL was reduced by circular antisense oligonucleotides directed against the b2a2 junction (38). Moreover, a significant inhibition of p210BCR/ABL was also reported after 36 h incubation in the presence of antisense oligonucleotides (39). As p210BCR/ABL has a half-life of ∼48 h (40), after 24 h exposure to asPNA one would have expected a reduction in protein level of 25–30% and not of 60%, as experimentally observed by us and other researchers. How can this apparent discrepancy be rationalised? Possible explanations are: (i) as the reported half-life for p210BCR/ABL was measured by inhibiting total protein synthesis with cycloheximide (proteases included), a value of 48 h is probably an overestimate; (ii) asPNA may bind non-specifically to proteins and reduce their half-life; (iii) p210BCR/ABL, being involved in signal transduction pathways, is usually complexed with adaptor proteins so that its half-life refers to the complexed state. When the level of p210BCR/ABL is reduced by asPNA, its interaction with adaptor proteins is not favoured and p210BCR/ABL dissociates. It is possible that the free protein has a shorter half-life than the complexed protein.
Finally, there is evidence that p210BCR/ABL activates mitogenic pathways and confers a growth advantage to Ph-positive cells over normal haematopoietic cells (8–10). In particular, p210BCR/ABL activates, through an interaction with adaptor proteins, the ras pathway, which promotes cell proliferation. The observation that KYO-1 cells treated with asPNA exhibit an incomplete cell cycle and slower cell proliferation is in keeping with down-regulation of the bcr/abl gene by the antisense PNA.
Acknowledgments
ACKNOWLEDGEMENTS
This research was supported by grants from the National Research Council of Italy (CNR), from the Friuli Venezia Giulia Region (FVG, 3887/2Univ.20) and the Ministry of University and Scientific and Technological Research (MIUR 2001)
REFERENCES
- 1.Nielsen P.E. (1999) Peptide nucleic acids as therapeutic agents. Curr. Opin. Struct. Biol., 9, 353–357. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen P.E., Egholm,M., Berg,R.H. and Buchardt,O. (1991) Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science, 254, 1497–1500. [DOI] [PubMed] [Google Scholar]
- 3.Uhlmann E., Peyman,A., Breipohl,G. and Will,D.W. (1998) PNA: synthetic polyamide nucleic acids with unusual binding properties. Angew. Chem. Int. Ed., 27, 2796–2823. [DOI] [PubMed] [Google Scholar]
- 4.Egholm M., Buchardt,O., Christensen,L., Berhens,C., Freier,S.M., Driver,D.A., Berg,R.H. and Nielsen,P.E. (1993) PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature, 365, 5466–5468. [DOI] [PubMed] [Google Scholar]
- 5.Uhlmann E. (1998) Peptide nucleic acids (PNA) and PNA-DNA chimeras: from high binding affinity towards biological function. Biol. Chem., 379, 1045–1052. [PubMed] [Google Scholar]
- 6.Jensen K.K., Ørum,H., Nielsen,P.E. and Norden,B. (1997) Kinetics for hybridization of peptide nucleic acids (PNA) with DNA and RNA studied with the BIAcore technique. Biochemistry, 36, 5072–5077. [DOI] [PubMed] [Google Scholar]
- 7.Rose D.J. (1993) Characterization of antisense binding properties of peptide nucleic acids by capillary gel electrophoresis. Anal. Chem., 65, 3545–3549. [DOI] [PubMed] [Google Scholar]
- 8.Deininger M.W.N., Goldman,J.M. and Melo,J.V. 2000) The molecular biology of chronic myeloid leukaemia. Blood, 96, 3343–3356. [PubMed] [Google Scholar]
- 9.Faderl S., Talpaz,M., Zeef,E., O’Brien,S., Kurzrock,R. and Kantarjian,H.M. (1999) The biology of chronic myeloid leukemia. N. Engl. J. Med., 241, 164–172. [DOI] [PubMed] [Google Scholar]
- 10.Warmuth M., Danhauser-Riedl,S. and Hallek,M. (1999) Molecular pathogenesis of chronic myeloid leukemia: implications for new therapeutic strategies. Ann. Hematol., 78, 49–64. [DOI] [PubMed] [Google Scholar]
- 11.Daley G.Q., van Etten,R.A. and Baltimore,D. (1990) Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science, 247, 824–830. [DOI] [PubMed] [Google Scholar]
- 12.Elefanty A.G., Hariharan,I.K. and Cory,S. (1990) Bcr-abl, the hallmark of chronic myeloid leukaemia in man, induces multiple haemopoietic neoplasms in mice. EMBO J., 9, 1069–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gewirtz A.M. (1999) Oligonucleotide therapeutics for human leukemia. Antisense Nucleic Acid Drug Dev., 9, 397–401. [Google Scholar]
- 14.Cutrona G., Carpaneto,E.M., Ulivi,M., Roncella,S., Landt,O., Ferrarini,M. and Boffa,L.C. (2000) Effects in live cells of a c-myc anti-gene PNA linked to a nuclear localization signal. Nat. Biotechnol., 18, 300–303. [DOI] [PubMed] [Google Scholar]
- 15.Boffa L.C., Scarfi,S., Mariani,M.R., Damonte,G., Allfrey,V.G., Benatti,U. and Morris,P.L. (2000) Dihydrotestosterone as a selective cellular/nuclear localization vector for anti-gene peptide nucleic acid in prostatic carcinoma cells. Cancer Res., 60, 2258–2262. [PubMed] [Google Scholar]
- 16.Fraser G.L., Holmgren,J., Clarke,P.B.S. and Wahlestedt,C. (2000) Antisense inhibition of delta-opioid receptor gene function in vivo by peptide nucleic acids. Mol. Pharmacol., 57, 725–731. [DOI] [PubMed] [Google Scholar]
- 17.Aldrian-Herrada G., Desarménien,M.G., Orcel,H., Boissin-Agasse,L., Méry,J., Brugidou,J. and Rabié,A. (1998) A peptide nucleic acid (PNA) is more rapidly internalized in cultured neurons when coupled to a retro-inverso delivery peptide. The antisense activity depresses the target mRNA and protein in magnocellular oxytocin neurons. Nucleic Acids Res., 26, 4910–4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tyler B.M., Janse,K., McCormick,D.J., Douglas,C.L., Boules,M., Stewart,J.A., Zhao,L., Lacy,B., Cusack,B., Fauq,A. and Richelson,E. (1999) Peptide nucleic acids targeted to the neurotensin receptor and administered i.p. cross the blood-brain barrier and specifically reduce gene expression. Proc. Natl Acad. Sci. USA, 96, 7053–7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mosmann T. (1983) Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods, 65, 55–63. [DOI] [PubMed] [Google Scholar]
- 20.Diviacco S., Rapozzi,V., Xodo,L., Helene,C., Quadrifoglio,F. and Giovannangeli,C. (2001) Site-directed inhibition of DNA replication by triple helix formation. FASEB J., 16, 2660–2668. [DOI] [PubMed] [Google Scholar]
- 21.Stock R.P., Olvera,A., Sànchez,R., Saralegui,A., Scarfi,S., Sanchez-Lopez,R., Ramos,M.A., Boffa,L.C., Benatti,U. and Alagon,A. (2001) Inhibition of gene expression in Entamoeba histolytica with antisense peptide nucleic acid oligomers. Nat. Biotechnol., 19, 231–234. [DOI] [PubMed] [Google Scholar]
- 22.Buchardt O., Egholm,M., Berg,R.H. and Nielsen,P.E. (1993) Peptide nucleic acids and their potential applications in biotechnology. Trends Biochem., 11, 384–386. [DOI] [PubMed] [Google Scholar]
- 23.Verheijen J.C., Bayly,S.F., Player,M.R., Torrence,P.F., van der Marel,G.A. and van Boom,J.A. (1999) 2-5A-PNA complexes: a novel class of antisense compounds. Nucl. Nucl., 18, 1485–1486. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X., Simmons,C.G. and Corey,D.R. (2001) Liver cell specific targeting of peptide nucleic acid oligomers. Bioorg. Med. Chem. Lett., 11, 1269–1272. [DOI] [PubMed] [Google Scholar]
- 25.Hanvey J.C., Peffer,N.J., Bisi,J.E., Thomson,S.A., Cadilla,R., Josey,J.A., Ricca,D.J., Hassman,C.F., Bonham,M.A., Au,K.G. et al. (1992) Antisense and antigene properties of peptide nucleic acids. Science, 258, 1481–1485. [DOI] [PubMed] [Google Scholar]
- 26.Wittung P., Kajanus,J., Edwards,K., Haaima,G., Nielsen,P., Norden,B. and Malmstrom,B. (1995) Phospholipid membrane permeability of peptide nucleic acid. FEBS Lett., 375, 27–29. [PubMed] [Google Scholar]
- 27.Haughland R.P. (1996) Handbook of fluorescent probes and research chemicals. In Spence,M.T.Z. (ed.), Nucleic Acid Detection. Molecular Probes, Eugene, OR, pp. 143–168.
- 28.Santa Lucia J., Allawi,H.T. and Seneviratne,P.A. (1996) Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry, 35, 3555–3562. [DOI] [PubMed] [Google Scholar]
- 29.Jensen K.K., Orum,H., Nielsen,P.E. and Norden,B. (1997) Kinetics for hybridization of peptide nucleic acids (PNA) with DNA and RNA studied with the BIAcore technique. Biochemistry, 36, 5072–5077. [DOI] [PubMed] [Google Scholar]
- 30.Bonham M.A., Brown,S., Boyd,A.L., Brown,P.H., Bruckenstein,D.A., Hanvey,J.C., Thomson,S.A., Pipe,A., Hassman,F., Bisi,J.E. et al. (1995) An assessment of the antisense properties of RNase H-competent and steric-blocking oligomers. Nucleic Acids Res., 23, 1197–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campanini F., Santucci,M.A., Pattachini,L., Brusa,G., Piccioli,M., Barbieri,E. and Tura,S. (2001) Competitive polymerase chain reaction as a method to detect the amplification of bcr-abl gene of chronic myeloid leukaemia. Haematologica, 86, 167–173. [PubMed] [Google Scholar]
- 32.Moravcova J., Lukasova,M., Stary,J. and Haskover,C. (1998) Simple competitive two-step RT-PCR assay to monitor minimal residual disease in CML patients after bone marrow transplantation. Leukemia, 12, 1303–1312. [DOI] [PubMed] [Google Scholar]
- 33.McMahon B.M., Stewart,J.A., Jackson,J., Fauq,A., McCormick,D.J. and Richelson,E. (2001) Intraperitoneal injection of antisense peptide nucleic acids targeted to the mu receptor decreases response to morphine and receptor protein levels in rat brain. Brain Res., 904, 345–349. [DOI] [PubMed] [Google Scholar]
- 34.Beletskii A., Hong,Y.-K., Pehrson,J., Egholm,M. and Strauss,W.M. (2001) PNA interference mapping demonstrates functional domains in the noncoding RNA Xist. Proc. Natl Acad. Sci. USA, 98, 9215–9220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Derossi D., Calvet,S., Trembleau,A., Brunissen,A., Chassaing,G. and Prochiantz,A. (1996) Cell internalization of the third helix of the Antennapedia homeodomain is receptor-independent. J. Biol. Chem., 271, 18188–18193. [DOI] [PubMed] [Google Scholar]
- 36.Beck G.F., Irwin,W.J., Nicklin,P.L. and Akhtar,S. (1996) Interactions of phosphodiester and phosphorothioate oligonucleotides with intestinal epithelial Caco-2 cells. Pharm. Res., 13, 1028–1037. [DOI] [PubMed] [Google Scholar]
- 37.Dias N., Dheur,S., Nielsen,P.E., Gryaznov,S., Van Aerschot,A., Herdewijn,P., Helene,C. and Saison-Behmoras,T.E. (1999) Antisense PNA tridecamers targeted to the coding region of Ha-ras mRNA arrest polypeptide chain elongation. J. Mol. Biol., 294, 403–416. [DOI] [PubMed] [Google Scholar]
- 38.Rowley P.T., Kosciolek,B.A. and Kool,E.T. (1999) Circular antisense oligonucleotides inhibit growth of chronic myeloid leukemia cells. Mol. Med., 5, 693–700. [PMC free article] [PubMed] [Google Scholar]
- 39.Bhatia R. and Verfaillie,C.M. (1998) Inhibition of BCR-ABL expression with antisense oligodeoxynucleotides restores beta1 integrin-mediated adhesion and proliferation inhibition in chronic myelogenous leukemia hematopoietic progenitors. Blood, 91, 3414–3422. [PubMed] [Google Scholar]
- 40.Spiller D.G., Giles,R.V., Broughton,C.M., Grzybowski,J., Ruddell,C.J., Tidd,D.M. and Clark,R.E. (1998) The influence of target protein half-life on the effectiveness of antisense oligonucleotide analog-mediated biologic responses. Antisense Nucleic Acid Drug Dev., 8, 281–293. [DOI] [PubMed] [Google Scholar]