Abstract
Computational methods seeking to automatically determine the properties (functional, structural, physicochemical, etc.) of a protein directly from the sequence have long been the focus of numerous research groups. With the advent of advanced sequencing methods and systems, the number of amino acid sequences that are being deposited in the public databases has been increasing steadily. This has in turn generated a renewed demand for automated approaches that can annotate individual sequences and complete genomes quickly, exhaustively and objectively. In this paper, we present one such approach that is centered around and exploits the Bio-Dictionary, a collection of amino acid patterns that completely covers the natural sequence space and can capture functional and structural signals that have been reused during evolution, within and across protein families. Our annotation approach also makes use of a weighted, position-specific scoring scheme that is unaffected by the over-representation of well-conserved proteins and protein fragments in the databases used. For a given query sequence, the method permits one to determine, in a single pass, the following: local and global similarities between the query and any protein already present in a public database; the likeness of the query to all available archaeal/bacterial/eukaryotic/viral sequences in the database as a function of amino acid position within the query; the character of secondary structure of the query as a function of amino acid position within the query; the cytoplasmic, transmembrane or extracellular behavior of the query; the nature and position of binding domains, active sites, post-translationally modified sites, signal peptides, etc. In terms of performance, the proposed method is exhaustive, objective and allows for the rapid annotation of individual sequences and full genomes. Annotation examples are presented and discussed in Results, including individual queries and complete genomes that were released publicly after we built the Bio-Dictionary that is used in our experiments. Finally, we have computed the annotations of more than 70 complete genomes and made them available on the World Wide Web at http://cbcsrv.watson.ibm.com/Annotations/.
INTRODUCTION
The automatic elucidation of protein function directly from sequence has been the focus of research activity for many years. Such an elucidation has an obvious appeal for it tries to minimize the amount of associated manual labor by reducing a large number of possibilities to one or a handful of choices. This is typically achieved by tapping into repositories of previously accumulated knowledge and by trading computation (i.e. in silico approaches) for typically tedious manual analysis. The discovery of protein function directly from sequence, in an automated or semi-automated manner, has become a fundamental question as thousands of unknown proteins and increasing numbers of complete genomes are made available daily in the public domain. Of course, one should not lose sight of the fact that protein annotation is the first step in the attempt to fully describe a particular organism through characterization of its metabolic pathways and transcription regulation networks.
During the last three decades, numerous methods have been proposed for determining protein function from sequence, all of which are essentially instances of a ‘guilty by association’ approach to solving this problem. Depending on the nature of the information exploited and the manner in which the information is used, these methods can essentially be divided into a handful of well-differentiated categories.
The chronologically earliest examples of protein annotation methods rely on the determination of a local or global similarity between a query protein and proteins with known annotation that are contained in a database (1–4). If two sequences of comparable length share a large portion of their extent, the previously uncharaterized sequence will inherit the function of the annotated one. The validity of this scheme relies on the implicit assumption that two sequences that ‘look the same’ at the sequence level also have the same function and structure. This is a reasonable assumption, but counter-examples such as the dehydrogenase/z-crystallin case have also been documented in the literature (for a discussion of this particular case see for example 5). The methods in this category are known as ‘similarity-based’ or ‘homology-based’ and are numerous. The approach we present in this paper belongs to this category as well.
A recurrent situation within the similarity-method category that is pertinent to our discussion relates to the inclination of annotators to use either the first or the best ‘hit’ from the output of a database search that is carried out by one of the similarity search algorithms such as FASTA (3), BLAST (4), Smith-Waterman (2), etc. In the presence of domains that are shared by numerous proteins (6), choosing the first or the best hit may not be optimal. As a matter of fact, the multi-domain organization of proteins can lead to incorrectly annotated database entries (Fig. 1 shows a characteristic example of such a misannotation). Use of a domain scan and the exploitation/analysis of the generated output when annotating a query can substantially improve the results. Such a domain scan can be implemented, for example, with the help of the PROSITE, PRINTS, PFAM, BLOCKS or PRODOM databases (7–11). For a description of other sources of potential concern in protein annotation and some recommended solutions the reader is referred to previous publications (12–16).
Figure 1.
An example of incorrect protein annotation as a result of multiple domain sharing. The sequences have been aligned in a manner that shows their common domains. The ‘d-arabino 3-hexulose 6-phosphate formaldehyde lyase’ function has been propagated from the HUMS_BACSU sequence to the rest of the sequences in this group. Instances of the same domain have been shown in the sequences that contain it using the same color and shading scheme.
A second category of methods has become known as the Rosetta stone approach (also known as the domain fusion method) (17–19). Here, one seeks to determine groups of proteins that are distinct in a given organism but appear as a single product in another organism, the result of an assumed fusion event. The distinct proteins in the original group are assumed to be physically interacting and, depending on the specifics of each case, this information can be helpful in determining protein function.
The methods in the third category seek to determine groups of proteins that repeatedly appear as neighbors of one another in the chromosomes of different organisms. The proteins involved are assumed to have a functional relationship (this methodology is similar in flavor to the Rosetta stone approach but distinct with respect to the type of information that it uses). Exploitation of this idea has found a best fit in the case of prokaryotic genomes where proximal gene organization is manifested in the form of operons, and it has been used successfully to guide functional annotation (20). It is not evident, however, whether this idea carries over to eukaryotic organisms due to the fact that, in general, the latter lack operons. A closely related variation, which does extend to eukaryotic organisms, operates on the assumption that if an organism is in need of a specific pathway then the organism will carry all or most of the genes comprising the pathway, and vice versa. For example, the work of Marcotte (17) and similar work done by others attempt to define function in terms of the pathways and complexes in which the protein participates, rather than suggest a specific biochemical activity: in this framework a protein is associated with a function via its linkages to other proteins.
Finally, in recent years, a fourth category has emerged. Here, one tackles the problem of protein function elucidation through the analysis of correlated mRNA expression of the type that is encountered in the context of DNA- and microarray-chip experiments. The underlying assumption is that functionally related proteins will exhibit correlated mRNA expression levels under multiple experimental settings. Consistent participation of a previously uncharacterized protein in clusters comprising proteins with a well-understood function imposes constraints on the unknown protein’s possible behavior by restricting its candidate memberships within the context of a metabolic pathway (21). In principle, this method can help resolve a protein’s function. A more recent variation of this general approach measures levels of protein expression (instead of mRNA) with the help of mass spectrometry or 2D gel electrophoresis in an attempt to determine clusters of strongly co-expressed proteins: these clusters can be used to determine the function of uncharacterized proteins.
We next present and discuss a new approach to the problem of protein annotation. At the center of our approach is the Bio-Dictionary, an exhaustive collection of amino acid patterns, heretofore referred to as seqlets, that completely covers the natural sequence space of proteins to the extent that the latter is sampled by the currently available biological sequences. In earlier studies, we showed that the seqlets can capture both functional and structural signals that have been reused during evolution within and across families of related proteins. Our approach relies on the seqlets contained in the Bio-Dictionary and the associated information that is available in a well-maintained database such as SwissProt/TrEMBL (22), derives from an earlier prototype system we built to carry out similarity searching (23,24) and employs a weighted, position-specific scoring scheme that is not affected by the over-representation of well-conserved proteins and protein fragments that are present in the public databases. Although similar in intent to systems like GeneQuiz (25), our method goes beyond simply stating the presence of local and global similarities between a query and one or more database sequences: in fact, we also report information about the secondary structure characteristics of the query, the presence of known domains, signal peptides, active sites, post-translationally modified sites, cytoplasmic/extracellular behavior, the similarity of the query to each of the three phylogenetic domains as a function of amino acid position, etc.
MATERIALS AND METHODS
Background
The Bio-Dictionary idea was introduced and discussed in earlier works (26–28); therein we described how one can use the Teiresias pattern discovery algorithm (29,30) to process a very large public database of amino acid sequences and fragments and derive a collection of amino acid patterns that, by design, appear within as well as across family boundaries. These patterns are referred to as seqlets and have been shown to capture functional and structural signals; moreover, they have the very important property of completely describing the processed input at the amino acid level. Following are some seqlet examples that include the name of the represented feature or of the represented protein family, taken from Rigoutsos et al. (26): GDG[IVAMTD]ND[AILV][PEAS] [AMV][LMIF]..A (cation-transporting ATPases), V.I.G. G..G...A (NAD/FAD-binding flavoproteins), G..G.GK[ST]TL (ATP/GTP-binding P-loop), KMSKS[LKDIR][GNDFQ]N (class I aminoacyl-tRNA synthetases), H.....HRD.K..N (serine/threonine protein kinases), etc. In terms of the notation used, [LKDIR] means a choice of exactly one among L, K, D, I and R, whereas ‘.’ denotes a single position wild-card character that can replace any of the symbols in the alphabet.
The derived collection of seqlets can thus be treated as a (redundant) vocabulary for the natural sequence space of proteins to the extent that the latter is uniformly sampled by the currently available biological sequences. Associating the entries of this vocabulary-like collection with the annotation information contained in a typical entry of the SwissProt database allows us to convert the collection into a dictionary. We have been using the term Bio-Dictionary to refer to the collection of seqlets that has been augmented so as to include the ‘annotation meaning’ for each of the entries. The key elements behind the Bio-Dictionary, and details of its construction, can be found in Rigoutsos et al. (26); analysis of the 3D structural properties associated with the seqlets of a dictionary built out of 17 complete archaeal and bacterial genomes are given in Rigoutsos et al. (27); finally, a discussion and description of potential uses for it appears in Rigoutsos et al. (28). In more recent work, we applied the Bio-Dictionary to in silico prokaryotic gene finding and built a system with exceptional performance (31): unlike approaches that are based on Markov models where each distinct genome requires that a different model be built, our gene finding system is universal in that a single instance of it is used across all archaeal and bacterial genomes.
The earlier work
By carrying out pattern discovery on a given sequence database D, we can use the generated pattern collection C to carry out similarity searches for instances of a query or its fragments in D as follows: a pattern from the derived collection C of patterns that matches a region of the query under consideration pinpoints a potential local similarity between the matched region of the query and all of the sequence fragments from the input database that the pattern represents (recall that by the definition of pattern discovery, patterns appear k or more times in the processed input, with k ≥ 2).
In earlier work, we used the Teiresias algorithm to process Release 34 of the SwissProt database and built a prototype system for similarity searching using only a subset C′ from the derived collection C of patterns. A given query sequence was examined for matches of patterns contained in C′ and the query and database regions corresponding to the matches were aligned, scored, and finally sorted according to the computed score. Following the sorting, one could proceed in one of two distinct ways: (i) the user was presented with the collection of patterns that matched the query and was asked to identify those of biological importance, then alignments were generated using those patterns alone; (ii) those alignments that resulted from patterns whose database instances carried an associated annotation (namely the ‘FT’ line) were reported to the user for further study (23,24).
This early system was meant to be a proof of concept. Consequently, complete coverage of the input database by the patterns in the collection C′ was neither achieved nor pursued. As a matter of fact, this early system used a mere 565 432 patterns which covered ∼50% of the processed SwissProt database at the amino acid level. Neither the existing over-representation of various protein families in the database nor the system’s real-time performance were design considerations at that time. However, this early excursion provided an invaluable learning experience that helped guide us toward the system which we present next.
The method: description
The first and foremost consideration of the new approach is the achievement of a complete coverage of the natural sequence space as it is currently known. To that end we used as our domain of knowledge the 14 May 2001 release of SwissProt/TrEMBL, a large, publicly available and curated database (22). This particular release comprised 532 621 amino acid sequences and fragments with a grand total of 170 762 058 amino acids.
We processed this input database in two phases. First, we ran Teiresias using L = 8, W = 8 and K = 2 and generated variable length seqlets that contained no wild-cards. For each one of these seqlets, we located and masked all of its instances in the input database except for the one that appeared in the longest among the sequences containing instances of the seqlet. We then reran Teiresias on the masked input, but this time using L = 6 and W = 15. For more information about this scheme and other methodological details the reader is referred to Rigoutsos et al. (26). The processing required ∼45 CPU days worth of computation using IBM RS64III processors with a clock speed of 450 MHz. With the help of a parallel implementation of Teiresias that we have developed for shared memory architectures, we completed this computation in 2 days on a 24 processor IBM S-80 supercomputer.
The two pattern discovery phases generated the Bio-Dictionary that we used in our analysis and which contained a combined total of 42 996 454 seqlets [compare the size of the current collection with the 565 432 patterns used in Floratos et al. (23,24)] that accounted for 98.2% of the processed input at the amino acid level (this degree of coverage essentially implies that, on average, a mere five amino acids per protein sequence cannot be accounted for by this seqlet collection). The length and density distributions of these seqlets match closely the ones shown in Rigoutsos et al. (26), whereas the average length of a seqlet is ∼12–13 amino acids. It should be noted that this Bio-Dictionary contains redundant seqlets, i.e. a given amino acid position in the processed input will typically participate in and is covered by multiple seqlets; this redundancy of representation is a desired property which we exploit during annotation.
Each seqlet in the collection carries along the meaning(s) associated with the regions of the proteins that contained an instance of and gave rise to the seqlet. Instead of a static description of each seqlet’s meaning(s) in the manner that we described in Rigoutsos et al. (28), we opted for composing the full entry of each seqlet during the run time as required. We currently derive labels for meanings from only the DE, OC and FT lines of the respective SwissProt/TrEMBL entry; obviously, we can tap into any database containing complementary information and attach additional meanings to each seqlet. One obvious choice is the PDB (32,33): in previous publications (27,28) we described how 3D structure can be associated with seqlets and are currently in the process of extending the approach presented herein in order to reconstruct local 3D structure using the structural hypotheses generated by partially overlapping seqlets (D.Platt, I.Rigoutsos, Y.Gao and L.Parida, submitted for publication).
Recall that the DE or ‘description’ line of SwissProt contains general descriptive information about the respective entry. Similarly, the OC or ‘organism classification’ line contains the taxonomic classification of the source organism. And the FT or ‘feature table’ line contains a simple and precise means for the annotation of the sequence data: a fixed abbreviation with a defined meaning relating to the feature being reported is followed by the residue numbers indicating the end points (extent) of the named feature; the line ends with a description containing additional information about the feature. Of the available SwissProt/TrEMBL abbreviations contained in an FT line we only make use of the ones listed in Table 1.
Table 1. FT line labels used in our work (see also text).
| mod_res | lipid | disulfid | thioeth | thiolest |
| carbohyd | metal | binding | transit | signal |
| propep | chain | peptide | ca_bind | domain |
| dna_bind | np_bind | transmem | zn_fing | similar |
| act_site | site | init_met | non_cons | non_ter |
| helix | strand | turn | se_cys |
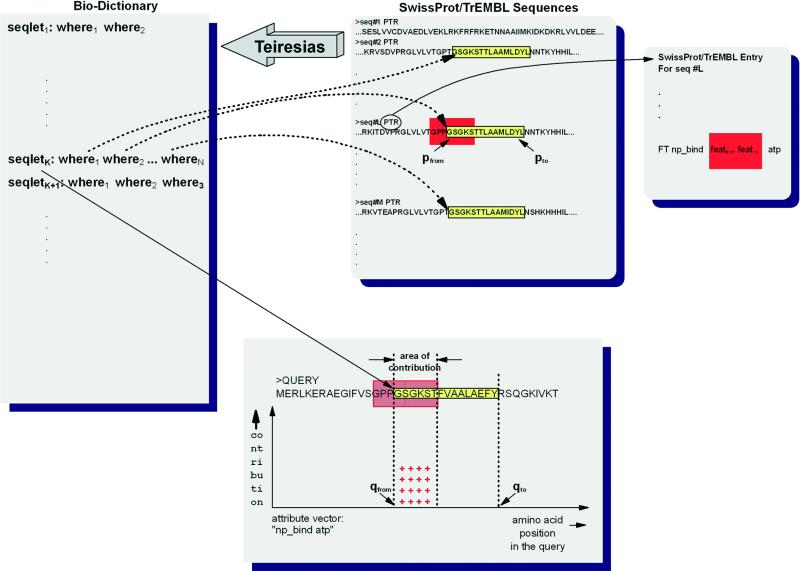
When presented with a query Q to annotate, we carry out the steps outlined in Figure 2, a markedly different approach than the one used in our early prototype. First, we generate the ‘intersection’ of the Bio-Dictionary with the query sequence to find those seqlets that match somewhere in the query. For each of the seqlets in this intersection, we examine the corresponding SwissProt/TrEMBL entries for all of the sequences that gave rise to the seqlet during the Bio-Dictonary formation, thus building the corresponding dictionary entry ‘on-the-fly’ by dynamically attaching to the seqlet all the meaning(s) extracted from those entries. The extracted meanings essentially ‘color’ each seqlet and by extension the region of the query where the seqlet matches. Note that a given seqlet can carry multiple ‘colors’, i.e. attributes. Con sequently, a region of the query can be associated with multiple attributes. If the seqlet under consideration is attached to an attribute that has not been encountered before, then a new attribute vector is introduced: the attribute vector has the same length as the query and initially contains zeroes everywhere; the current seqlet assigns its contribution CONTRIB(.,.) to this new attribute vector at precisely the region corresponding to the seqlet’s match in the query. If the seqlet carries an attribute that has been encountered before, the seqlet adds its contribution CONTRIB(.,.) to the appropriate region of the already existing attribute vector. Multiple seqlets that carry the same attribute will add their individual contributions to the attribute’s vector: the regions to which the seqlets contribute may or may not be overlapping. The manner in which we decide what amount a seqlet will contribute to an attribute vector is described in detail below.
Figure 2.
Pseudo-code showing the computational steps of the method.
After all seqlets in the intersection have been exhausted, and separately for each attribute category (e.g. DE, FT, etc.), the attribute vectors are sorted and ranked based on the accumulated support and the top T ranking vectors of the category are reported (T is a user-specified parameter). Each of these vectors will contain non-zero values at precisely those regions that were matched by possibly overlapping, distinct seqlets that carried the same attribute.
Before we describe the scoring scheme it is important to stress some points that are particular to our work. In general, the Bio-Dictionary should not be seen as a collection of seqlets each of which necessarily captures a specific feature such as a kinase domain, a metal binding site, etc. Seqlets that can act as predicates for a feature or protein family do exist in the Bio-Dictionary, but, by design, seqlets may also carry multiple meanings. This is different from the one-to-one correspondence that the reader may be accustomed to and which is typical of predicate-containing databases such as PROSITE, PRINTS and INTERPRO (7,8,34).
As we showcased previously (26), a seqlet can cross functional and structural boundaries and can thus be associated with multiple meanings. Clearly, those of the seqlets that are associated with a unique meaning can function as predicates, but a significant number of them will capture and correspond to multiple meanings.
Similarly, the Bio-Dictionary also contains multiple seqlets all of which capture the same meaning. These seqlets can also have instances that overlap with one another, as indicated by the fact that the product of the number of seqlets contained in the Bio-Dictionary times the seqlets’ average length is a multiple of the actual length of the processed input (26).
Thus, by design, a given position of a processed query will in general be covered by multiple seqlets. Each of the seqlets covering a position within the query will in general carry one or more meanings that are used to ‘color’ the corresponding region of the query. Let a given query position be covered by M distinct seqlets. In order for an attribute, e.g. ‘metal-binding site’, to rank high in the reported results, a large portion of those M seqlets must carry this attribute.
Recall that, by definition, each of the seqlets of the Bio-Dictionary appears in at least two places in the processed database (SwissProt/TrEMBL in our case): thus, if M seqlets cover a given position in the query to annotate, then the following two properties will simultaneously hold:
• there exists a total of F sequence fragments corresponding to all of the instances of the M seqlets in the processed input database; clearly, these fragments will be similar to the amino acid neighborhood surrounding this query position;
• the F sequence fragments in the database will agree on the amino acid identity of the literals (recall that the seqlets contain both literals and wild-cards, i.e. ‘dots’) contained in each of the M seqlets.
These database sequence fragments may or may not agree on the annotation of the query position under consideration. If the annotations for N of these F database sequence fragments state that this site is a metal-binding site then through application of the ‘guilty by association’ approach, our belief that this query position is also a metal-binding site will be proportional to N/F. This very idea is applied to every attribute and attribute category that is attached to a seqlet. The direct implication of this is that a seqlet can be useful and able to contribute to a specific annotation without having to span an attribute (e.g. protein kinase domain) in its entirety. Moreover, the seqlet does not have to be a predicate for an attribute either.
Figure 3 graphically depicts this situation. For discussion purposes, let us assume that when we searched the query Q with the Bio-Dictionary, we determined that seqletK is present in Q in the region [qfrom, qto]. Let us also assume that during the Bio-Dictionary formation, seqletK was determined to have three instances in the processed SwissProt/TrEMBL database. After following these three backpointers to the full entries of the sequences that contain the three instances of seqletK, we determine that in one of the sequences the seqlet instance spans an interval [pfrom, pto] that has a non-empty intersection with a specific region [featfrom, featto] of that sequence that is annotated as np_bind atp, i.e. as an ATP-binding site. Let [ifrom, ito] denote the intersection of the intervals [pfrom, pto] and [featfrom, featto]. In this example, seqletK will corroborate the presence of a partial ATP-binding domain in the query that is being annotated by incrementing the support at the locations [qfrom + (ifrom – pfrom), qfrom + (ito – pfrom)] of the np_bind atp attribute vector.
Figure 3.
Accumulating seqlet contributions. Seqlets do not have to span a feature in its entirety in order to corroborate an attribute. Nor do they have to be specific enough to act as predicates for the attribute. See also text for more details.
It should now be clear why any given seqlet does not have to serve as a predicate for the attribute(s) that it corroborates. The term ‘attribute’ is overloaded in our discussion and should be interpreted rather loosely: it can mean a local similarity, a global similarity, an active site, a phylogenetic domain, a post-translationally modified location, etc.
If the query being annotated contains a true instance of a given attribute, then each one of the numerous seqlets that will cover the region spanned by the attribute more than once will cumulatively and independently provide support for the attribute at the respective positions: as the accumulated support for the attribute increases so does the likelihood of its presence in the query.
If the query is a true member of a known protein family, then we expect the attribute vector for this family to obtain support along its entire length from practically every single one of the seqlets that match in this query. If a query contains a known domain, then the attribute vector for the domain will have non-zero support over the region of the query that corresponds to the domain’s instance. In an analogous manner, if the processed query shares only a local region with a well-characterized family or an individual protein, then the corresponding attribute vector will have non-zero support only over the shared region.
The manner in which we use the seqlets and accumulate scores has proven particularly useful in situations that, among others, include the following: the query is a fragment of a known sequence; the query contains one or more known domains in a novel order; the query has been assembled using an incorrect exon collection (e.g. one or more true exons are missing, introns have been mislabeled as exons and included in the assembly, exons that correspond to distinct genes have been assembled together, etc.).
Moreover, the fact that our seqlets have lengths that typically span between 6 and 18 amino acids (for a detailed discussion see 26) permits us to easily and correctly process very short input queries, e.g. 8–10 amino acids, without the thresholding constraints and limitations that one typically encounters when using heuristics-based similarity search algorithms (3,4).
In real-world applications, situations arise where the query represents or contains only a fragment from a known domain, for example a query involving the first few tens of amino acids from say a ‘protein kinase domain’. In order to alert the user to this situation, we also include, wherever applicable, the ‘minimum’, ‘average’ and ‘standard deviation’ values for the span of each of the T top ranking reported attribute vectors. This permits easy determination of whether the query represents a complete instance of the stated attribute or only a fragment.
The method: scoring
How much to contribute. Above we described how we determine the extent of an attribute vector region to which a seqlet matching the query will contribute. We now discuss how we determine the amount that the seqlet will contribute.
Let seqletK be present in query Q and let qi1qi2qi3…qil and pj1pj2pj3…pjl be its instances in the query and in some database sequence d, respectively; let {i1, … il} and {j1, … jl} denote the indices of the positions spanned by the seqlet in the query Q and sequence d, respectively. For simplicity, we will assume that the instance of seqletK completely spans an annotated region of d that corresponds to an attribute A.
SeqletK brings together two sequence fragments with lengths equal to the span of the seqlet; one fragment comes from the query that is being analyzed while the other is from the sequence d of the database. Obviously, the more similar these two fragments are the more likely it is that upon completion of the annotation process the attribute A that is associated with the database region pj1pj2pj3…pjl will be carried over to the query region qi1qi2qi3…qil through the ‘guilty by association’ approach. There is a rather straightforward manner in which seqletK can contribute to the vector for attribute A; we simply use one of the available scoring matrices and generate contributions in a position- and content-dependent manner as follows:
for m = 1 to l {attribute_vector[i1 + m – 1] += f(scoring_matrix[qi1 + m – 1][pj1 + m – 1])}
(the symbol += is shorthand notation for ‘increment by amount shown on the right of the = sign’). In other words, the seqlet will contribute to the (i1 + m – 1)th position of the attribute vector an amount that relates to the degree of similarity between the amino acids occupying the positions qi1 + m – 1 and pj1 + m – 1, respectively. A good choice for the function f(.) above is f(x) = 2x + constant; with regard to the scoring matrix to use one can employ any of the standard PAM or BLOSUM scoring matrices (35,36).
In order to avoid the over-counting that would be the consequence of a given protein family or fragment being over-represented in the SwissProt/TrEMBL database, we impose the additional constraint that a given seqlet cannot contribute to the same attribute vector and vector position(s) more than once. In other words, if seqletK captures a very well-conserved region appearing in a large number of SwissProt/TrEMBL sequences, only one of the seqlet’s numerous annotated database instances will contribute to the respective attribute vector.
How to normalize. As mentioned already, a given seqlet with distinct possible meanings will contribute in turn to each of the attribute vectors that correspond to those meanings. And these contributions will depend on how well a known database instance of the attribute matches its alleged instance in the query. Different attribute vectors will accumulate different amounts of contribution and these contributions will also depend on the position within the attribute vector.
During the annotation of the query we maintain a book-keeping array, total_contrib, with length equal to that of the query; for every seqlet with an instance qi1qi2qi3…qil in the query, we update total as follows:
for m = 1 to l {total_contrib[i1 + m – 1] += f(scoring_matrix[qi1 + m – 1][pj1 + m – 1])}
In other words, the ith position of total_contrib is a measure of the number of seqlets that contribute to it, with each contribution weighted by the degree of similarity between the amino acids in the query and their input database counterparts. The function f(.) is the same as in the previous section. Note that at all times during processing, the value of total_contrib[i] is greater than or equal to the maximum value one will encounter in the ith position of any of the active attribute vectors for this query.
Once all of the seqlets matching the query have been examined, we normalize the contents of the ith position of each attribute vector by dividing by the value of total_ contrib[i]. Multiplying this normalized value by 100 gives us, for each attribute vector, a measure of the fraction of the total contribution that this attribute has received, as a function of position within the query. Well-conserved attributes will have values close to 100% whereas less conserved attributes will have fewer seqlets contributing to them and thus will have smaller values. Note that this particular way of normalizing has the additional property of alleviating the situation where equal length regions of the query receive disproportionately different contributions due to differences in the number of contributing seqlets: this normalization will permit all regions in the query to have equally ‘strong voices’.
How to rank. Once the contents of the attribute vectors have been normalized, we sort them based on their received contributions and report the top T of them. We have implemented a scheme that will rank a narrow, well-conserved region higher than a wider region which is not as well conserved. This permits us to report attributes such as well-conserved active sites or post-translationally modified sites among the top ranking positions of the results. Finally, when we report local similarities, we further require of the attribute vectors pertaining to similarities that any set of consecutive non-zero positions be at least X positions wide; the value of X is user-defined and typical values range in the interval [10, 20].
The method: how to find matches in the query
In order to efficiently implement the above method we need to be able to quickly determine which of the Bio-Dictionary seqlets match where within the query. A simplistic approach would require that, for the ∼43 000 000 seqlets and every single query position, we check whether there is a match; this would of course be very slow. The problem of identifying such matches is complicated by the presence of wild-cards (‘don’t care characters’) in the seqlets that we use.
To deal with this situation, we have designed and implemented a novel and very efficient method for solving precisely this problem: our method makes use of a very efficient hashing scheme that subselects among the Bio-Dictionary seqlets prior to using the ones that survive in conjunction with a modified version of the Aho-Corasick algorithm (37). The resulting scheme permits us to fully annotate a 300 amino acid protein in ∼10 s on a single IBM RS64III processor running at a clock speed of 450 MHz. The description of this matching algorithm extends beyond the scope of this presentation and will be given elsewhere (M.Lewenstein, T.Huynh and I.Rigoutsos, in preparation).
RESULTS
We next showcase the capabilities of our approach by annotating a carefully selected collection of example queries and discussing the results obtained. All of the results we report in this section can be reproduced using the Web-based implementation of our method, available at http://cbcsrv.watson.ibm.com/Tpa.html. The underlined text in the figures generated by the Web-based tool is in fact hyperlinks which permit the user to issue a search request to SwissProt/TrEMBL and retrieve all of the database entries with the property stated by the text. This capability is meant to facilitate cross-comparisons and verification of the reported results. Moreover, upon completion of an annotation, the user can view the Bio-Dictionary patterns that matched within the query, as well as each pattern’s estimated log probability and the actual position within the query where the match begins.
Example 1. UBIQ_HUMAN
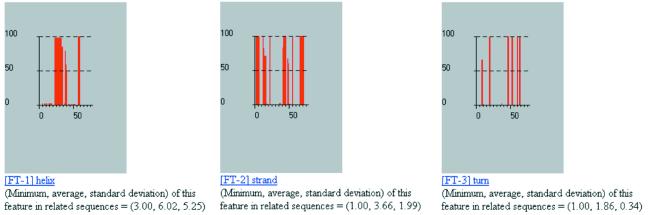
As our first example, we examine the annotation of the 76 amino acid human ubiquitin, UBIQ_HUMAN. Some of the results of the analysis are shown in Figures 4–56. As can be seen from these figures, the SwissProt/TrEMBL database contains enough information for our method to correctly determine the secondary structure of the fragment: notice the localization of the helices, strands and turns and their interweaving in Figure 4.
Figure 4.
Some results from processing the human ubiquitin UBIQ_HUMAN by our method.
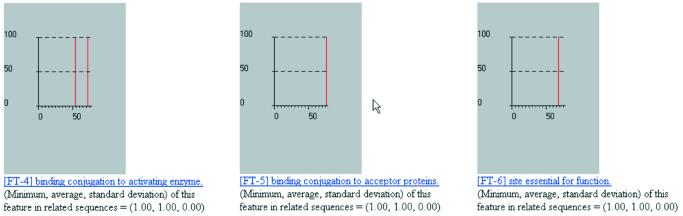
Figure 5.
Additional results from processing the human ubiquitin UBIQ_HUMAN by our method.
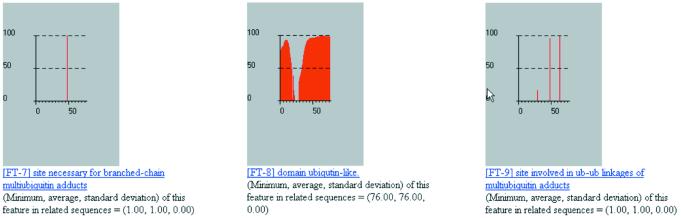
Figure 6.
More results from processing the human ubiquitin UBIQ_HUMAN by our method.
It is not always the case that there will be enough information in SwissProt/TrEMBL for us to be able to make statements about the local secondary structure of a query. This limitation can be alleviated in one of two ways: (i) we can rely on SwissProt/TrEMBL’s continuing augmentation and updates—as the database becomes bigger and more enriched, our capability to annotate local structure will also improve; (ii) we can make use of the information in the PDB database in the manner that we have described (26,27). The seqlets’ meanings will be enriched by incorporating structural information from the much more comprehensive PDB; we are currently in the process of augmenting our annotation method so that it will include this component. Finally, note how our method correctly determines the nature and position of seven sites that are relevant to the function of ubiquitin as well the presence and extent of the ubiquitin domain.
Example 2. A very short fragment
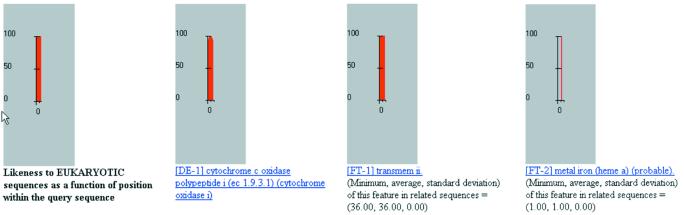
For our second example, we have selected the 8 amino acid fragment VVVTAHAF, a fragment that is too short to be used with heuristics-based similarity search algorithms such as FASTA and BLAST/PSI-BLAST. As shown in Figure 7, when we process this fragment with our method we can correctly determine that: (i) it is an amino acid combination encountered only in the eukaryotic domain; (ii) it belongs to a cytochrome c oxidase; (iii) it is part of a transmembrane domain; and (iv) it has a metal-binding site (iron) at the sixth position from the beginning, i.e. at the position of the histidine.
Figure 7.
Some of the results obtained from processing the fragment VVVTAHAF with our method.
Example 3. ACTR_BOVIN
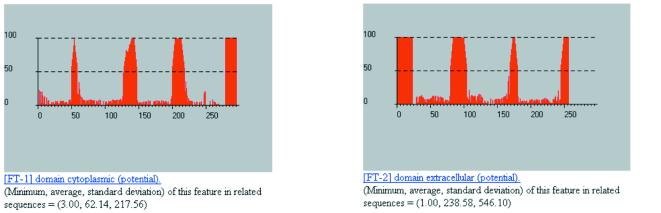
Another capability of our system is the determination of cytoplasmic, transmembrane and extracellular regions in a given query. We showcase this using ACTR_BOVIN, an adrenocorticotropic hormone receptor protein from Bos taurus. Figure 8 shows the plots for the cytoplasmic and extracellular behavior of the query as determined by our method: note that the regions of the query that are not accounted for by these two plots correspond precisely to the seven transmembrane domains of the ACTR_BOVIN (the corresponding transmembrane plots are not included in this figure).
Figure 8.
Partial results from processing the adrenocorticotropic hormone receptor protein ACTR_BOVIN by our method.
Example 4. UL78_HCMVA
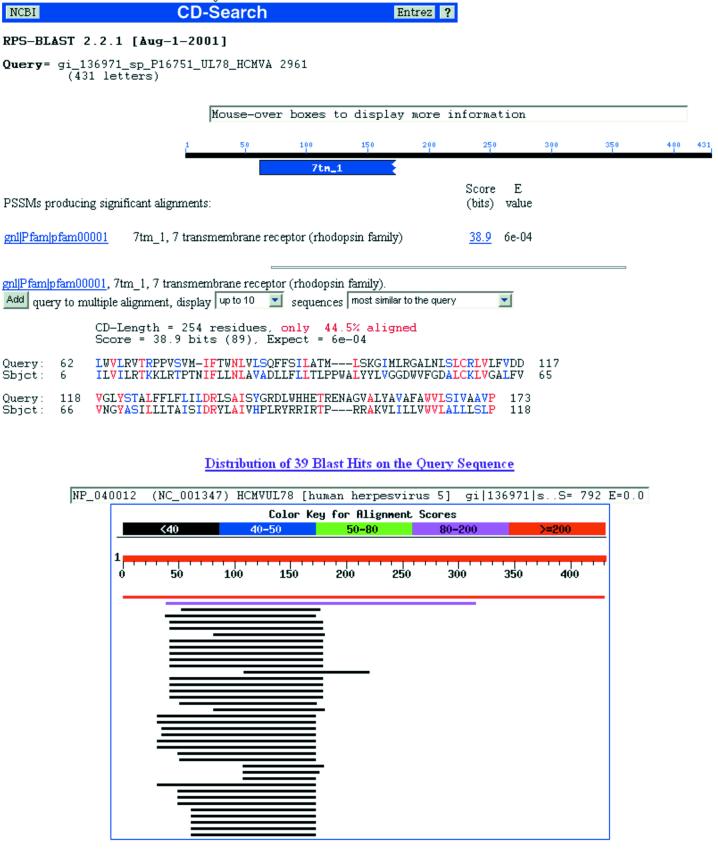
The next example is a sequence that comes from the human herpesvirus 5 (39). In particular, it is the 431 amino acid sequence UL78_HCMVA. In Figure 9 we show the output of both RPS-BLAST and PSI-BLAST (38) on this specific query sequence: as can be seen, the only detectable similarity is with the rhodopsin family and is confined in the region [60, 170]; no other similarities can be determined outside this region (the PSI-BLAST hit at the second position is with an uncharacterized sequence from the Tupaia herpesvirus and thus is not informative). One possible interpretation is that there is no single sequence in the SwissProt/TrEMBL database that resembles UL78_HCMVA (other than the query itself and its Tupaia herpesvirus counterpart), either in terms of the order of any domains that may be present or in terms of its composition.
Figure 9.
Results of RPS-BLAST and PSI-BLAST using the sequence UL78_HCMVA as an input. Default parameter settings were used.
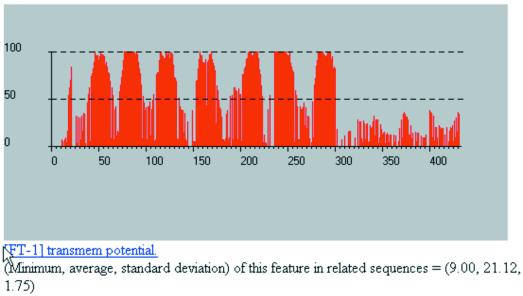
When we process UL78_HCMVA with our method, we discover weak similarities that relate UL78_HCMVA mainly to hypothetical proteins in a manner that is similar to what is shown in Figure 9. However, further inspection of our results provides us with enough information to appropriately categorize the query. In Figure 10 we show the plot for the query’s transmembrane behavior as reported by our method: seven very distinct regions are immediately apparent, thus permitting us to conjecture that this sequence is a G protein-coupled receptor homolog. The seven regions correspond to the intervals 45–61, 74–95, 112–132, 154–174, 202–222, 236–256 and 280–300, respectively, and have well-delineated boundaries. Notably, a similarity search in the GPCRDB database using UL78_HCMVA as the query currently generates no hits to known families.
Figure 10.
Partial results from processing the sequence UL78_HCMVA from human herpesvirus 5. See text for more details.
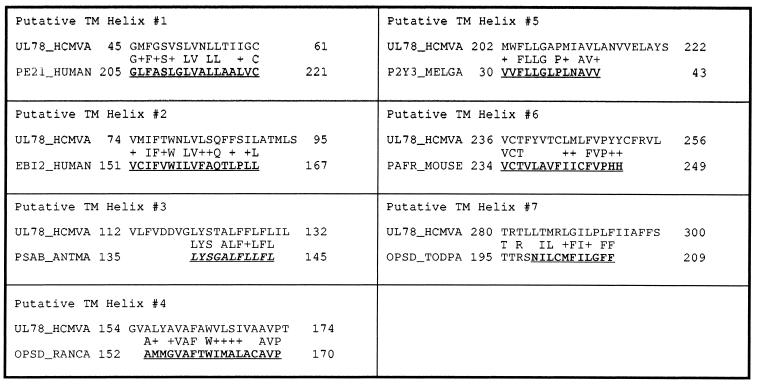
In Table 2 we show the alignment for the best ranking hits obtained when we search the GPCRDB database subset that contains only SwissProt entries (but not TrEMBL) using each of the seven putative transmembrane regions as a query. If the UL78_HCMVA putative transmembrane regions correspond to transmembrane helices of a GPCR homolog one will expect to see them matching known transmembrane regions from sequences in GPCRDB. This is indeed the case, as this table shows: the regions of the GPCRDB/SwissProt hits that are labeled as transmembrane regions of a G protein-coupled receptor are shown underlined bold next to each of the seven queries. The only exception is the query corresponding to the putative TM helix 3, where the best match is to a transmembrane region from PSAB_ANTMA, a photosystem protein. In all seven cases the quality of the conservation is notable. It should also be noted that several sequences other than UL78_HCMVA were reported as GPCR homologs when the analysis of the complete genome of the human herpesvirus 5 was first published (39).
Table 2. Best GPCRDB/SwissProt hits when using UL78_HCMVA regions as the query (see also text).
Example 5. Comparison with the annotations of recently published/updated genomes
We next showcase the capability of our approach by processing the complete genomes of three organisms whose sequences were published after 14 May 2001 and compare our results with the annotations that accompanied the release of the respective genomes. Since the Bio-Dictionary that we used for our experiments was built using the SwissProt/TrEMBL release of 14 May 2001, the results from these comparisons are indicative of our method’s ability to extrapolate and annotate novel sequences. Additionally, we annotated two genomes whose sequences were released into the public domain prior to 14 May 2001. Obviously, the sequences of these organisms are already contained in the input database from which we built the Bio-Dictionary used in these annotations. However, the GenBank database entries for these genomes and the respective annotations were updated several months after 14 May 2001: it is these more recent annotations that we use for our comparative study and not the annotations that accompanied the original genome submission. The purpose behind these comparisons is to determine the extent of agreement between our predictions and the original annotators’ updated predictions when using a sequence database that has been substantially augmented since the genomes under consideration were originally deposited.
It should be stressed that any such comparisons can only provide estimates of what a user can expect when using our method to annotate a genome. Indeed, the very notion of an automated comparison of different annotation collections is, to a certain degree, ill-defined. The following observations will make this last statement clear.
First, the published genomes are sequenced, annotated and released by different research groups which employed different automated tools in conjunction with generally distinct, although overlapping, knowledge bases of annotated biological sequences. Once the automatically obtained gene annotations become available, they are typically curated manually during a ‘genome annotation jamboree’ by a different team of scientists each time and using non-standard nomenclature and abbreviations. As a result of this manual curation, the annotations that accompany a newly published genome contain much more that simply the result of applying a ‘guilty by association’ automated approach. This last observation puts us at a distinct disadvantage when carrying out the comparisons that we report below.
Independently of the annotation approach that is used, there is always the issue of what it means to have ‘annotated a protein’. Even ignoring disagreements in the annotations of individual proteins, several levels of detail are possible when making an annotation. As an example, Table 3 shows valid, non-contradicting annotations for a fictitious protein: the thing to notice here is the different amount of information that is conveyed by each annotation statement. Ideally, one seeks the most detailed description possible for the available knowledge base. The possibility of different levels of annotation detail adds an extra degree of difficulty and can result in annotation disagreements when lists of annotations that have been reported by different groups at different points in time are automatically compared with one another (13,14,40–45).
Table 3. Annotations for a fictitious protein that are non-conflicting with one another but correspond to varying degrees of conveyed annotation detail.
| Non-conflicting annotations for a ficticious protein |
| Cellular process protein |
| Membrane protein |
| Integral membrane protein |
| Protein involved in cellular signaling |
| G protein-coupled receptor |
| Secretin-like protein |
| Corticotropin releasing factor |
Even if one ignores the above difficulties, differences can still arise as a result of using different guidelines and criteria each time, thus leading to substantial variations in the claimed percentage of genes that can be annotated in a newly sequenced genome based on sequence similarity with known proteins. Generally speaking, the current state of the art permits one to report functional hypotheses for ∼70% of the predicted genes in a given prokaryotic genome (43–51). The fraction for eukaryotic genomes is typically much lower, although in the case of specific eukaryotic chromosomes, notable exceptions exist (52).
In light of the above observations, we decided to generate our figures by manually comparing, for each and every one of the involved genes, the annotations reported during the release of the genome with those generated by our method. The results are given in Table 4. The first three genomes, namely Rickettsia conorii Malish 7 (53), Staphylococcus aureus Mu50 (54) and Streptococcus pneumoniae TIGR4 (55) were published and made available in the Fall of 2001. The last two genomes, namely Chlamydia pneumoniae J138 (56) and Buchnera sp. APS (57) were published in June and September 2000, respectively, but their GenBank records were updated in the Fall of 2001. For each genome we report the number and percentage of genes that fall into each of the following categories: (i) the latest GenBank annotation and our annotation agree; (ii) the GenBank annotation contains a ‘hypothetical protein’ entry whereas our system proposes a functional hypothesis; (iii) the GenBank annotation lists a functional hypothesis whereas our system reports a ‘hypothetical protein’; and (iv) the GenBank annotation and our annotation disagree.
Table 4. Results from manually comparing our predictions with the annotations that have been reported for several genomes.
| Genome name | Latest GenBank annotation date | No. of predicted genes | Latest GenBank annot. | Latest GenBank annot. | Latest GenBank annot. | Latest GenBank annot. |
|---|---|---|---|---|---|---|
| = B-D annot. (hypothetical proteins included) [% (no. of genes)] | = hypothetical protein && B-D annot. = functional hypothesis [% (no. of genes)] | = functional hypothesis && B-D annot. = hypothetical protein [% (no. of genes)] | ≠ B-D annot. (hypothetical proteins not included) [% (no. of genes)] | |||
| R.conorii Malish 7 | 3 Oct 2001 | 1374 | 88.94% (1222) | 7.06% (97) | 2.04% (28) | 1.96% (27) |
| S.aureus Mu50 | 4 Oct 2001 | 2748 | 91.85% (2524) | 7.28% (200) | 0.18% (5) | 0.69% (19) |
| S.pneumoniae TIGR4 | 3 Oct 2001 | 2094 | 87.87% (1840) | 4.25% (89) | 2.63% (55) | 5.25% (110) |
| C.pneumoniae J138 | 2 Oct 2001 | 1069 | 98.41% (1052) | 0.04% (4) | 0.05% (6) | 0.07% (7) |
| Buchnera sp. APS | 10 Sep 2001 | 564 | 97.69% (551) | 1.24% (7) | 0.01% (1) | 1.06% (6) |
The first three of the genomes listed are novel in that they were published several months after we built the Bio-Dictionary used to generate functional hypotheses. The remaining two genomes were published in 2000, but their GenBank entries were updated in the Fall of 2001. As can be seen, our system’s output matches in quality the annotations that have been made available after manual curation of automated analysis. See main text for details.
As shown in Table 4, for the two genomes that were updated recently, the agreement between our automated predictions and the latest GenBank annotations reaches a level of 98% over the entire genome. It should be noted that this figure also includes those genes for which there is no functional hypothesis (i.e. they are listed as ‘hypothetical proteins’). For the three novel genomes, the agreement between the predictions ranges between 88 and 92%. It is worth reiterating that the annotations that are included in the GenBank entries for the various genomes are the result of manually curating the output of multiple automated tools, whereas our scheme generates annotations in an entirely automated manner using a single unified framework. In recent collaborative work with colleagues from several European laboratories the complete genome of Chlamydia trachomatis serovar D (58) was re-annotated using (i) manual means, (ii) traditional automated tools and (iii) our method. As described in detail (I.Iliopoulos, S.Tsoka, M.A.Andrade, A.J.Enright, M.Caroll, P.Poullet, V.Promponas, T.Liakopoulos, G.Palaios, C.Pasquier, S.Hamodrakas et al., submitted for publication), the annotations that were obtained through manual means and through our Bio-Dictionary-based method achieved the best overall performance reaching an annotation agreement on 862 of the 893 processed sequences, i.e. 96.5% of the entire genome. Of the remaining 31 sequences, 13 could be annotated manually but could not be annotated by our method, whereas the other 18 could be annotated with our method but could not be annotated manually.
Example 6. Annotations on the World Wide Web
Similarly to the previous example, we have annotated the sequences of more than 70 complete genomes across the three phylogenetic domains, including: Methanococcus jannaschii DSM 2661 (59), Halobacterium sp. NRC-1 (60), Sulfolobus solfataricus P2 (61), Mycoplasma genitalium G-37 (62), Synechocystis sp. PCC 6803 (63), Escherichia coli K12-MG165 (64), Helicobacter pylori 26695 (65), Borrelia burgdorferi B31 (66), Aquifex aeolicus VF5 (67), Myco bacterium tuberculosis H37Rv (68), Chlamydia trachomatis serovar D (58), Chlamydophila pneumoniae CWL029 (69), Thermotoga maritima MSB8 (70), Deinococcus radiodurans R1 (71), Yersinia pestis CO92 (72), Saccharomyces cerevisiae S288C (73), Caenorhabditis elegans (74), Drosophila melanogaster (75), Homo sapiens (76,77) and Mus musculus. The annotations of these genomes are available on the World Wide Web and can be viewed and interactively explored by visiting http://cbcsrv.watson.ibm.com/Annotations/.
The system that we make available on the World Wide Web provides the user with several options. Within a specific genome, if the accession number of a gene is known, then it can be used to locate and view the annotation of the gene. Alternatively, one can search the results in the DE and FT attribute categories of the genome using regular expressions that can be entered with the help of a graphical user interface. For example, when run against the DE results, the regular expression
-[1-3]].*calcium.*bind
will locate and report all the sequences in the genome under consideration that ‘share any similarities with calcium binding sequences and are ranked in the top three positions’. Analogously, when run against the FT results, the regular expression
-[1-9]].*domain.*bh[1234].*
will permit the user to search for sequences that ‘contain one or more of the cell apoptosis-associated domains BH1, BH2, BH3 and BH4 and are ranked in the top nine positions’. To list the three top ranking functional hypotheses for each gene in a genome, one can use the regular expression
-[1-3]]
to search through the DE results. At http://cbcsrv.watson. ibm.com/Help/ShowMeHowToSearch.html the user can find information on how to form these regular expressions and the permitted keywords, as well as several specific examples with explanations.
Additionally, we have enabled and made available cross-genomic comparisons/searches: through a graphical user interface, one or more genomes can be selected and their annotations searched for similarities with a specific family (e.g. elongation factor, tRNA-aminoacyl synthetase, etc.) or the presence of a specific feature (e.g. hydrogen bond donor, calcium-binding domain, helix–turn–helix, etc.) with the help of regular expressions similar to those used to analyze individual genomes.
DISCUSSION AND FUTURE DIRECTIONS
In this paper, we have presented and discussed a new method for the automated annotation of amino acid sequences. The method quickly, objectively and exhaustively determines local and global similarities between a given query and any protein already present in a public database, the likeness of the query to all available archaeal/bacterial/eukaryotic/viral sequences in the database as a function of amino acid position within the query, the secondary structure character of the query as a function of amino acid position within the query, the cytoplasmic, transmembrane or extracellular behavior of the query, the nature and position of binding domains, active sites, post-translationally modified sites and signal peptides, etc.
The key concept underlying our method is that of the Bio-Dictionary, which we presented and discussed in earlier work. By design, the presented method is extendable and can make use of any type of attribute that would be of interest to the end user. It can also make use of multiple databases.
Through a carefully selected collection of examples, we have demonstrated the capabilities of our method and the quality of the annotations that it generates. Our system automatically generates results whose quality matches that of publicly available annotations; recall that such annotations are typically the product of a manual curation that has followed the application of automated processes. In terms of actual annotation speed, our system can annotate a 300 amino acid query in ∼10 s on a single IBM RS64III processor running at a clock speed of 450 MHz.
We are currently in the process of enhancing our system with several new components. One extension involves the automatic determination and reporting of all the PubMed references pertaining to the query sequence that is annotated. For each of the reported results in the DE category we will be making available links to all PubMed articles that are relevant for the study of the query sequence and the family described by the caption. This is currently work in progress.
A second extension, which we have already described above, involves the automated generation of local 3D structure through ‘meanings’ that are derived from the contents of the PDB database. This is also work in progress.
Finally, an important topic that we will be studying relates to the fact that the SwissProt/TrEMBL database has up to now used non-standardized nomenclature to label database entries. For example, the following are some of the DE lines that are associated with aldose reductases:
aldose reductase (ec 1.1.1.21) (ar) (aldehyde reductase)
aldose reductase
alcohol dehydrogenase [nadp+] (ec 1.1.1.2) (aldehyde reductase).
When our system is presented with an aldose reductase as a query, e.g. ALDR_HUMAN, then multiple attribute vectors will be reported, one for each of these seemingly distinct (but in reality identical) attributes. A planned future release of our system will alleviate this problem through the use of standardized names.
Acknowledgments
ACKNOWLEDGEMENTS
I.R. would like to thank Terri Attwood for bringing UL78_HCMVA to his attention; this sequence proved to be an invaluable benchmark test. The authors would like to thank Jiri Novotny for commenting on an early draft of the manuscript, and Stephen Chin-Bow for helpful comments and for assistance with the design of the graphical user interfaces. Finally, our thanks also go to the anonymous reviewers for their insightful comments and feedback on the manuscript.
REFERENCES
- 1.Needleman S.B. and Wunsch,C.D. (1970) A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol., 48, 443–453. [DOI] [PubMed] [Google Scholar]
- 2.Smith T.F. and Waterman,M.S. (1981) Identification of common molecular subsequences. J. Mol. Biol., 147, 195–197. [DOI] [PubMed] [Google Scholar]
- 3.Pearson W.R. and Lipman,D.J. (1988) Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 5, 403–410. [DOI] [PubMed] [Google Scholar]
- 5.Baxevanis A. and Ouellette,F. (1998) Bioinformatics: A Practical Guide to the Analysis of Genes and Proteins. John Wiley & Sons, New York, NY.
- 6.Doolittle R. (1995) The multiplicity of domains in proteins. Annu. Rev. Biochem., 64, 287–314. [DOI] [PubMed] [Google Scholar]
- 7.Attwood T.K., Beck,M.E., Flower,D.R., Scordis,P. and Selley,J.N. (1998) The PRINTS protein fingerprint database in its fifth year. Nucleic Acids Res., 26, 304–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofmann K., Bucher,P., Falquet,L. and Bairoch,A. (1999) The PROSITE database, its status in 1999. Nucleic Acids Res., 27, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henikoff S. and Henikoff,J. (1994) Protein family classification based on searching a database of blocks. Genomics, 19, 97–107. [DOI] [PubMed] [Google Scholar]
- 10.Sonnhammer E.L., Eddy,S.R. and Durbin,R. (1997) Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins, 28, 405–420. [DOI] [PubMed] [Google Scholar]
- 11.Sonnhammer E.L. and Kahn,D. (1994) Modular arrangement of proteins as inferred from analysis of homology. Protein Sci., 3, 482–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bork P. and Bairoch,A. (1996) Go hunting in sequence databases but watch out for the traps. Trends Genet., 12, 425–427. [DOI] [PubMed] [Google Scholar]
- 13.Bork P., Dandekar,T., Diaz-Lazcoz,Y., Eisenhaber,F., Huynen,M. and Yuan,Y. (1998) Predicting function: from genes to genomes and back. J. Mol. Biol., 283, 707–725. [DOI] [PubMed] [Google Scholar]
- 14.Galperin M.Y. and Koonin,E.V. (1998) Sources of systematic error in functional annotation of genomes: domain rearrangement, non-orthologous gene displacement and operon disruption. In Silico Biol., 1, 57–67. [PubMed] [Google Scholar]
- 15.Wooton J. and Federhen,S. (1996) Analysis of compositionally biased regions in sequence databases. Methods Enzymol., 266, 554–571. [DOI] [PubMed] [Google Scholar]
- 16.Promponas V., Enright,A., Tsoka,S., Kreil,D., Leroy,C., Hamodrakas,S., Sander,C. and Ouzounis,C. (2000) CAST: an iterative algorithm for the complexity analysis of sequence tracts. Bioinformatics, 16, 915–922. [DOI] [PubMed] [Google Scholar]
- 17.Marcotte E. (2000) Computational genetics: finding protein function by nonhomology methods. Curr. Opin. Struct. Biol., 10, 359–365. [DOI] [PubMed] [Google Scholar]
- 18.Marcotte E., Pellegrini,M., Ng,H.-L., Rice,D., Yeates,T. and Eisenberg,D. (1999) Detecting protein function and protein-protein interactions from genome sequences. Science, 285, 751–753. [DOI] [PubMed] [Google Scholar]
- 19.Enright A., Iliopoulos,I., Kyrpides,N. and Ouzounis,C. (1999) Protein interaction maps for complete genomes based on gene fusion events. Nature, 402, 86–90. [DOI] [PubMed] [Google Scholar]
- 20.Overbeek R., Fonstein,M., D’Souza,M., Pusch,G. and Maltsev,N. (1999) The use of gene clusters to infer functional coupling. Proc. Natl Acad. Sci. USA, 96, 2896–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisen M.B., Spellman,P.T., Brown,P.O. and Botstein,D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA, 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence data bank and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Floratos A., Rigoutsos,I., Parida,L., Stolovitzky,G. and Gao,Y. (1999) Sequence homology detection through large-scale pattern discovery. In Proceedings of the 3rd Annual ACM International Conference on Computational Molecular Biology (RECOMB ’99). Lyon, France. ACM Press, New York, NY.
- 24.Floratos A., Rigoutsos,I., Parida,L. and Gao,Y. (2001) DELPHI: a pattern-based method for detecting sequence similarity. IBM J. Res. Dev., 45, 455–474. [Google Scholar]
- 25.Andrade M.A., Brown,N.P., Leroy,C., Hoersch,S., de Daruvar,A., Reich,C., Franchini,A., Tamames,J., Valencia,A., Ouzounis,C. and Sander,C. (1999) Automated genome sequence analysis and annotation. Bioinformatics, 15, 391–412. [DOI] [PubMed] [Google Scholar]
- 26.Rigoutsos I., Floratos,A., Ouzounis,C., Gao,Y. and Parida,L. (1999) Dictionary building via unsupervised hierarchical motif discovery in the sequence space of natural proteins. Proteins Struct. Funct. Genet., 37, 264–277. [DOI] [PubMed] [Google Scholar]
- 27.Rigoutsos I., Gao,Y., Floratos,A. and Parida,L. (1999) Building dictionaries of 1D and 3D motifs by mining the unaligned 1D sequences of 17 archaeal and bacterial genomes. In Proceedings of the Seventh International Conference on Intelligent Systems for Molecular Biology (ISMB ’99). AAAI Press, Menlo Park, CA. [PubMed]
- 28.Rigoutsos I., Floratos,A., Parida,L., Gao,Y. and Platt,D. (2000) The emergence of pattern discovery techniques in computational biology. Metab. Eng., 2, 159–177. [DOI] [PubMed] [Google Scholar]
- 29.Rigoutsos I. and Floratos,A. (1998) Combinatorial pattern discovery in biological sequences: the TEIRESIAS algorithm. Bioinformatics, 14, 55–67. [DOI] [PubMed] [Google Scholar]
- 30.Rigoutsos I. and Floratos,A. (1998) Motif discovery without alignment or enumeration. In Proceedings of the 2nd Annual ACM International Conference on Computational Molecular Biology (RECOMB). New York, NY. ACM Press, New York, NY.
- 31.Shibuya T. and Rigoutsos,I. (2002) Dictionary-driven prokaryotic gene finding. Nucleic Acids Res., 30, 2710–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abola E.E., Sussman,J.L., Prilusky,J. and Manning,N.O. (1997) Protein data bank archives of three-dimensional macromolecular structures. Methods Enzymol., 277, 556–571. [DOI] [PubMed] [Google Scholar]
- 33.Bernstein F., Koetzle,T., Williams,G., Meyer,E., Brice,M., Rodgers,J., Kennard,O., Shimanouchi,T. and Tasumi,M. (1977) The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol., 112, 535–542. [DOI] [PubMed] [Google Scholar]
- 34.Apweiler R., Attwood,T., Bairoch,A., Bateman,A., Birney,E., Biswas,M., Bucher,P., Cerutti,L., Corpet,F., Croning,M. et al. (2001) The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res., 29, 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dayhoff M.O. (1978) Atlas of Protein Sequence and Structure. National Biomedical Research Foundation, Washington, DC, Vol. 5, pp. 345–352.
- 36.Henikoff S. and Henikoff,J. (1992) Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA, 89, 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gusfield D. (1997) Algorithms on Strings, Trees and Sequences. Cambridge University Press, Cambridge, UK.
- 38.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chee M., Bankier,A., Beck,S., Bohni,R., Brown,C., Cerny,R., Horsnell,T., Hutchinson,C., Kouzarides,T., Martignetti,J. et al. (1990) Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol., 154, 125–169. [DOI] [PubMed] [Google Scholar]
- 40.Andrade M., Casari,G., de Daruvar,A., Sander,C., Schneider,R., Tamames,J., Valencia,A. and Ouzounis,C. (1997) Sequence analysis of the Methanococcus jannaschii genome and the prediction of protein function. Comput. Appl. Biosci., 13, 481–483. [DOI] [PubMed] [Google Scholar]
- 41.Tsoka S., Promponas,V. and Ouzounis,C. (1999) Reproducibility in genome sequence annotation: the Plasmodium falciparum chromosome 2 case. FEBS Lett., 451, 354–355. [DOI] [PubMed] [Google Scholar]
- 42.Kyrpides N. and Ouzounis,C. (1999) Whole-genome sequence annotation: going wrong with confidence. Mol. Microbiol., 32, 886–887. [DOI] [PubMed] [Google Scholar]
- 43.Koonin E., Mushegian,A., Galperin,M. and Walker,D. (1997) Comparison of archaeal and bacterial genomes: computer analysis of protein sequences predicts novel functions and suggests a chimeric origin for the archaea. Mol. Microbiol., 25, 619–637. [DOI] [PubMed] [Google Scholar]
- 44.Koonin E., Tatusov,R. and Galperin,M. (1998) Beyond complete genomes: from sequence to structure and function. Curr. Opin. Struct. Biol., 8, 355–363. [DOI] [PubMed] [Google Scholar]
- 45.Ouzounis C., Casari,G., Valencia,A. and Sander,C. (1996) Novelties from the complete genome of Mycoplasma genitalium. Mol. Microbiol., 20, 897–899. [DOI] [PubMed] [Google Scholar]
- 46.Iliopoulos I., Tsoka,S., Andrade,M., Janssen,P., Audit,B., Tramontano,A., Valencia,A., Leroy,C., Sander,C. and Ouzounis,C. (2000) Genome sequences and great expectations. Genome Biol., 2, i0001.1–i0001.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsoka S. and Ouzounis,C. (2000) Recent developments and future directions in computational genomics. FEBS Lett., 480, 42–48. [DOI] [PubMed] [Google Scholar]
- 48.Kyrpides N., Olsen,G., Klenk,H.-P., White,O. and Woese,C. (1996) Methanococcus jannaschii genome: revisited. Microbiol. Comp. Genomics, 1, 329–338. [PubMed] [Google Scholar]
- 49.Kyrpides N. and Ouzounis,C. (1998) Errors in genome reviews. Science, 281, 1457. [DOI] [PubMed] [Google Scholar]
- 50.Bork P., Ouzounis,C., Casari,G., Schneider,R., Sander,C., Dolan,M., Gilbert,W. and Gillevet,P. (1995) Exploring the Mycoplasma capricolum genome: a minimal cell reveals its physiology. Mol. Microbiol., 16, 955–967. [DOI] [PubMed] [Google Scholar]
- 51.Kyrpides N., Ouzounis,C., Iliopoulos,I., Vonstein,V. and Overbeek,R. (2000) Analysis of the Thermotoga maritima genome combining a variety of sequence similarity and genome context tools. Nucleic Acids Res., 28, 4573–4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frishman D. and Mewes,H.-W. (1997) Protein structural classes in five complete genomes. Nature Struct. Biol., 4, 626–628. [DOI] [PubMed] [Google Scholar]
- 53.Ogata H., Audic,S., Renesto-Audiffren,P., Fournier,P.E., Barbe,V., Samson,D., Roux,V., Cossart,P., Weissenbach,J., Claverie,J.M. and Raoult,D. (2001) Protein mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science, 293, 2093–2098. [DOI] [PubMed] [Google Scholar]
- 54.Kuroda M., Ohta,T., Uchiyama,I., Baba,T., Yuzawa,H., Kobayashi,I., Cui,L., Oguchi,A., Aoki,K., Nagai,Y. et al. (2001) Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet, 357, 1225–1240. [DOI] [PubMed] [Google Scholar]
- 55.Tettelin H., Nelson,K.E., Paulsen,I.T., Eisen,J.A., Read,T.D., Peterson,S., Heidelberg,J., DeBoy,R.T., Haft,D.H., Dodson,R.J. et al. (2001) Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science, 293, 498–506. [DOI] [PubMed] [Google Scholar]
- 56.Shirai M., Hirakawa,H., Kimoto,M., Tabuchi,M., Kishi,F., Ouchi,K., Shiba,T., Ishii,K., Hattori,M., Kuhara,S. and Nakazawa,T. (2000) Comparison of whole genome sequences of Chlamydia pneumoniae J138 from Japan and CWL029 from USA. Nucleic Acids Res., 28, 2311–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shigenobu S., Watanabe,H., Hattori,M., Sakaki,Y. and Ishikawa,H. (2000) Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature, 407, 81–86. [DOI] [PubMed] [Google Scholar]
- 58.Stephens R., Kalman,S., Lammel,C., Fan,J., Marathe,R., Aravind,L., Mitchell,W., Olinger,L., Tatusov,R., Zhao,Q., Koonin,E. and Davis,R. (1998) Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science, 282, 754–759. [DOI] [PubMed] [Google Scholar]
- 59.Bult C., White,O., Olsen,G., Zhou,L., Fleischmann,R., Sutton,G., Blake,J., FitzGerald,L., Clayton,R., Gocayne,J. et al. (1996) Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science, 273, 1058–1073. [DOI] [PubMed] [Google Scholar]
- 60.Ng W.V., Kennedy,S.P., Mahairas,G.G., Berquist,B., Pan,M., Shukla,H.D., Lasky,S.R., Baliga,N., Thorsson,V., Sbrogna,J. et al. (2000) Genome sequence of Halobacterium species NRC-1. Proc. Natl Acad. Sci. USA, 97, 12176–12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qunxin S., Singh,R., Confalonieri,F., Zivanovic,Y., Allard,G., Awayez,M.J., Chan-Weiher,C.C.-Y., Clausen,I.G., Curtis,B.A., De Moors,A. et al. (2001) The complete genome of the Crenarchaeote Sulfolobus solfataricus P2. Proc. Natl Acad. Sci. USA, 98, 7835–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fraser C., Gocayne,J., White,O., Adams,M., Clayton,R., Fleischmann,R., Bult,C., Kerlavage,A., Sutton,G., Kelley,M. et al. (1995) The minimal gene complement of Mycoplasma genitalium. Science, 270, 397–403. [DOI] [PubMed] [Google Scholar]
- 63.Kaneko T. and Tabata,S. (1997) Complete genome structure of the unicellular cyanobacterium Synechocystis sp. PCC6803. Plant Cell Physiol., 38, 1171–1176. [DOI] [PubMed] [Google Scholar]
- 64.Blattner F., Plunkett,G., Bloch,C., Perna,N., Burland,V., Riley,M., Collado-Vides,J., Glasner,J., Rode,C., Mayhew,G.F. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- 65.Tomb J.-F., White,O., Kerlavage,A., Clayton,R., Sutton,G., Fleischmann,R., Ketchum,K., Klenk,H., Gill,S., Dougherty,B. et al. (1997) The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature, 388, 539–547. [DOI] [PubMed] [Google Scholar]
- 66.Fraser C., Casjens,S., Huang,W., Sutton,G., Clayton,R., Lathigra,R., White,O., Ketchum,K., Dodson,R., Hickey,E. et al. (1997) Genomic sequence of a Lyme disease spirochete, Borrelia burgdorferi. Nature, 390, 580–586. [DOI] [PubMed] [Google Scholar]
- 67.Deckert G., Warren,P., Gaasterland,T., Young,W., Lenox,A., Graham,D., Overbeek,R., Snead,M., Keller,M., Aujay,M. et al. (1998) The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature, 392, 353–358. [DOI] [PubMed] [Google Scholar]
- 68.Cole S., Brosch,R., Parkhill,J., Garnier,T., Churcher,C., Harris,D., Gordon,S., Eiglmeier,K., Gas,S., Barry,C. et al. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature, 393, 537–544. [DOI] [PubMed] [Google Scholar]
- 69.Kalman S., Mitchell,W., Marathe,R., Lammel,C., Fan,J., Hyman,R., Olinger,L., Grimwood,J., Davis,R. and Stephens,R. (1999) Comparative genomes of C. pneumoniae and C. trachomatis. Nature Genet., 21, 385–389. [DOI] [PubMed] [Google Scholar]
- 70.Nelson K., Clayton,R., Gill,S., Gwinn,M., Dodson,R., Haft,D., Hickey,E., Peterson,J., Nelson,W., Ketchum,K. et al. (1999) Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature, 399, 323–329. [DOI] [PubMed] [Google Scholar]
- 71.White O., Eisen,J., Heidelberg,J., Hickey,E., Peterson,J., Dodson,R., Haft,D., Gwinn,M., Nelson,W., Richardson,D. et al. (1999) Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science, 286, 1571–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Parkhill J., Wren,B., Thomson,N., Titball,R., Holden,M., Prentice,M., Sebaihia,M., James,K., Churcher,C., Mungall,K. et al. (2001) Genome sequence of Yersinia pestis, the causative agent of plague. Nature, 413, 523–527. [DOI] [PubMed] [Google Scholar]
- 73.(1997) The yeast genome directory. Nature (suppl), 387, 5–105. [PubMed] [Google Scholar]
- 74.The C. elegans Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science, 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- 75.Adams M., Celniker,S., Holt,R., Evans,C., Gocayne,J., Amanatides,P., Scherer,S., Li,P., Hoskins,R., Galle,R. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 76.The Genome International Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 77.Venter C., Adams,M., Myers,E., Li,P., Mural,R., Sutton,G., Smith,H., Yandell,M., Evans,C., Holt,R. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]