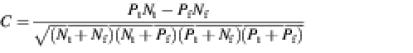Abstract
Given a set of homologous or functionally related RNA sequences, the consensus motifs may represent the binding sites of RNA regulatory proteins. Unlike DNA motifs, RNA motifs are more conserved in structures than in sequences. Knowing the structural motifs can help us gain a deeper insight of the regulation activities. There have been various studies of RNA secondary structure prediction, but most of them are not focused on finding motifs from sets of functionally related sequences. Although recent research shows some new approaches to RNA motif finding, they are limited to finding relatively simple structures, e.g. stem–loops. In this paper, we propose a novel genetic programming approach to RNA secondary structure prediction. It is capable of finding more complex structures than stem–loops. To demonstrate the performance of our new approach as well as to keep the consistency of our comparative study, we first tested it on the same data sets previously used to verify the current prediction systems. To show the flexibility of our new approach, we also tested it on a data set that contains pseudoknot motifs which most current systems cannot identify. A web-based user interface of the prediction system is set up at http://bioinfo. cis.nctu.edu.tw/service/gprm/.
INTRODUCTION
Transcriptional regulation is an important topic in bioinformatics. Much effort has been made to develop useful analysis tools to accelerate the progress in this research. An equally important but much less studied topic is post-transcriptional regulation. Similar to transcriptional regulation, post-transcriptional regulation is often accomplished by the binding of proteins to specific motifs in mRNA molecules (1–3). Unlike DNA binding proteins, which recognize motifs composed of conserved sequences, RNA protein binding sites are more conserved in structures than in sequences. The motif prediction algorithms that only consider conserved sequence profiles (4–8) may fail to identify RNA motifs. A set of post-transcriptionally coregulated RNAs can be characterized by base-pair interactions that organize the molecules into domains and provide a framework for functional interactions. If a new sequence is found to contain the common motifs, it may have the same characteristics as those coregulated RNAs. We are interested in finding the consensus motifs in a family of coregulated RNA sequences.
There has been much work on RNA secondary structure prediction. The current main approaches include free-energy minimization (9–12) and comparative sequence analysis (13–15). Although they show positive results of predicting secondary structures of a single sequence, it is questionable to use these methods to find common motifs in a set of sequences. Other approaches such as stochastic context-free grammar, e.g. COVE (16), and genetic algorithms (GAs) (17) have been applied to multiple sequences, but they are aimed at finding a global alignment instead of consensus motifs.
A dynamic programming approach called FOLDALIGN, which takes into account both sequence similarity and structure constraints, was first developed to discover RNA motifs in a set of sequences (18). However, its time complexity is too high for practical use. Recently, a new system called SLASH (19) has been developed. By combining FOLDALIGN and COVE, the time complexity of SLASH is acceptable for real applications, but it is currently limited to finding stem–loop motifs.
In this paper we introduce a new approach called genetic programming for RNA motifs (GPRM), which is capable of discovering structural motifs more complicated than stem– loop structures. To prove GPRM is comparable to the latest approaches, we tested it on the same data sets as used in the experiments of SLASH. Furthermore, we tested GPRM on a published pseudoknot data set to demonstrate its capability that most current prediction methods lack.
MATERIALS AND METHODS
Motif prediction can be seen as a concept learning problem, that is, learning a target concept (i.e. motifs) from a set of training examples (i.e. biosequences) (20). According to its objective and the training examples given, concept learning can be regarded as supervised or unsupervised. From pre-classified training examples, supervised learning is to learn a discriminative concept to distinguish between examples of different classes. On the other hand, unsupervised learning is to learn a characteristic concept to describe a set of unlabeled training examples.
We consider motif prediction a supervised learning problem. Unlike most current approaches, we use both positive and negative examples. Positive examples are a family of coregulated RNA sequences; negative examples are the same number of sequences randomly generated based on the observed frequencies of a sequence alphabet in positive examples. We learn the motifs that can be used to distinguish the given coregulated sequences from the random sequences.
As RNA motifs may vary in both sequences and structures, we need an expressive representation to describe a wide variety of motifs, and an effective strategy to search a large problem space for the right motifs. Genetic programming (GP) operates on a population of concept hypotheses. Individuals in the population can be described by linear structures, trees or graphs (21). Unlike conventional GAs, GP does not require an encoding scheme to encode putative solutions into bit strings before the evolutionary process, or a decoding scheme to decode finally converged bit strings back to an interpretable representation. By GP, the hypotheses can converge to the comprehensible target concept through evolution. Because of its generality and effectiveness, we adapt GP to develop GPRM. Since RNA secondary structures are typically formed by base-pairing interactions, GPRM is focused on finding Watson–Crick complementary base pairs. There are three components in GPRM. The first is a population of putative structural motifs. The second is a fitness function that measures the quality of each motif. The third is the genetic operators that simulate the natural evolution process. The details are described in the following sections.
Representing individuals in a population
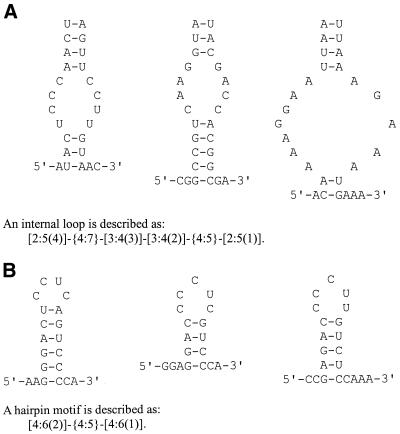
Each individual in a population is a putative motif. We use two kinds of segments to describe structural motifs. A segment is either a Watson–Crick complementary segment or a non-pairing segment. A Watson–Crick complementary segment is a helix, and it may also contain GU wobble pairs. A non-pairing segment, on the other hand, is single-stranded. With different combinations of segments, a wide variety of RNA motifs can be easily represented. For example, in Figure 1A, we show an interior loop in AUACUCCCAACUAGUUCCUUGUAAC, CGGCCGUCAAGGUAUACGACCACGGCGA and ACAAAAGGAAUUAAUUAAAGAAAUGAAA. The common motif is composed of four complementary segments and two non-pairing segments. It is described as [2:5(4)]-{4:6}-[3:4(3)]-[3:4(2)]-{4:5}-[2:5(1)]. In this representation, we use brackets to indicate a complementary segment; braces, a non-pairing segment. The range of segment length is kept inside the brackets and braces, e.g. {4:6} means the length of the non-pairing segment is between 4 and 6 nt. The number within parentheses in a complementary segment is the index of its corresponding pairing segment, e.g. (4) in complementary segment [2:5(4)] means that this segment is paired with the fourth complementary segment in the motif. Similarly, in Figure 1B, we show a hairpin motif in AAGGGACUCCUCAGUCCCCA, GGAGGACCCCUCGUCCCA and CCGAGACCCCUUGUCUCCAAA.
Figure 1.
(A) A common internal loop motif of three sequences, AUACUCCCAACUAGUUCCUUGUAAC, CGGCCGUCAAGGUAUACGACCACGGCGA and ACAAAAGGAAUUAAUUAAAGAAAUGAAA. The internal loop is composed of four complementary segments and two non-pairing segments. (B) A common hairpin motif of three sequences, AAGGGACUCCUCAGUCCCCA, GGAGGACCCCUCGUCCCA and CCGAGACCCCUUGUCUCCAAA. The hairpin is composed of two complementary segments and one non-pairing segment.
To find the motifs from a family of RNA sequences, the user of GPRM is required to specify the maximum number of segments and the range of segment length allowed in a motif. According to the specification, GPRM generates the initial population of putative motifs. The number of segments and the segment length in each motif are randomly assigned but conform to the user’s specification. The pairing relation between complementary segments is determined at random. After the initial population is created, GPRM applies genetic operators to the population to generate a better population of motifs. This evolution process is repeated until no improvement can be found.
Fitness function
In GP, the fitness function is used to measure the quality of individuals in a population. The higher the fitness of an individual, the better its chances of survival to the next generation. We are interested in the motifs that can reflect the characteristics conserved in a family of coregulated RNA sequences, e.g. the RNA protein binding sites. We design a fitness function that assigns higher values to those motifs commonly shared by the given set of RNAs, and rarely contained in random sequences.
Our fitness function is derived from the F-score (22) used in the field of information retrieval with the aim to balance the importance of two measures, recall (i.e. sensitivity) and precision (i.e. positive predictive value). Given a positive example set and a negative example set, we define the fitness function as follows:
The motif that will be chosen to participate in the genetic operation, e.g. mutation, is dependent on fitness. Motifs with higher fitness have better chances of being selected. We adapt the tournament selection mechanism. It parallels the competition in nature among individuals for the right to take part in evolution. Unlike fitness-proportionate selection, tournament selection does not need a centralized calculation of the average fitness of the population, and it is somewhat faster than rank selection (23,24). It first randomly picks two individuals from the population. Then the one with better fitness gets selected for the genetic operation.
Genetic operators
Reproduction models the self-replication process in nature. Instead of selecting one motif at a time, and passing it to the next generation, GPRM accelerates the reproduction process by passing the better half of the population sorted by fitness from generation to generation.
Similar to the mutation operation in nature that causes sporadic and random alterations in the genetic materials, GPRM’s mutation operator changes the segment configuration of a motif selected from the population. It first randomly picks a segment of the motif for alteration. If a complementary segment is selected, its corresponding pairing segment and its length range are then randomly changed. For example, a segment of 5–7 nt in length previously paired with the third complementary segment may be changed to a new segment of 4–6 nt in length now paired with the fourth complementary segment. On the other hand, if a non-pairing segment is chosen, only its length range is changed. Note that the segment length can only be randomly altered within the range specified by the user.
Unlike mutation, the crossover operation is performed on two individuals. Its purpose is to exchange the segment configuration between two tentative motifs to generate two offspring. After two motifs are selected from the population, either a pair of complementary segments or a non-pairing segment is chosen at random for exchange.
Implementation
GPRM is an optimization procedure that iteratively applies genetic operators to improve the fitness of tentative solutions. After the creation of the initial population, GPRM goes through three basic steps in each optimization cycle. The three steps are fitness evaluation, individual selection and population generation. The process is repeated until no improvement of fitness can be found, or it reaches the limit of generations. The pseudocode of GPRM is shown in Figure 2. By masking out the motifs already found in sequences, we can repeatedly apply GPRM to find multiple motifs if they exist in a set of RNA sequences.
Figure 2.
Pseudocode of GPRM.
The first step of GPRM is to find all possible pairs of complementary segments in the training examples, and put them in POOL. Suppose the lower and the upper bound of complementary segment length are l and u, and let f = u – l + 1. The time complexity of finding all possible pairs of complementary segments is O(f·L2·N) = O(L3·N) if f ≈ L, where L is the maximum sequence length and N is the number of total sequences. GPRM computes the fitness of each putative motif in the population by iteratively comparing each complementary segment of the motif with the entire POOL. Let the maximum number of complementary segments in a motif be m, which is specified by the user. The time complexity of fitness computation is O(m2·L3·N·P) where P is the constant population size. Compared with the fitness evaluation, the time complexity of crossover and mutation operations is negligible. The total time complexity of GPRM is thus O(L3·N) + O(m2·L3·N·P·G) = O(m2·L3·N), where P is the constant population size, and G is the constant generation limit. In the current version of GPRM, P and G are set to be 1000 and 50, respectively. Note that m is the maximum number of complementary segments specified by the user. Given a family of coregulated RNAs, we are interested in the common motifs for the RNA regulatory protein binding sites instead of a global alignment. Therefore, the number of complementary segments in a motif is relatively small. If m << L and m << N, the total time complexity can be reduced to O(L3·N).
RESULTS
There are two purposes of our experiments. The first is to demonstrate that GPRM is competitive with current RNA motif prediction systems. The second is to show that GPRM can identify complicated motifs that most current systems cannot find.
It is important to use the same data sets in experiments to keep the consistency of a comparative study. As SLASH (19) is the latest RNA motif prediction system, we first tested GPRM on the same data sets as used in SLASH’s experiments to show GPRM’s competitive performance. Moreover, we used a published pseudoknot data set to demonstrate GPRM’s flexibility that is lacking in most current systems, including SLASH. These data sets are described in the following sections.
Data sets
The first data set is one of the data sets used to test SLASH. It contains 34 archaea 16S ribosomal sequences (19). This data set was originally derived from a set of 311 sequences extracted from the SSU rRNA database (http://www-rna.uia.ac.be/ssu/) (25). The archaea set of 311 sequences was further reduced to 34, filtering out the sequences that miss base assignments or are >90% identical. The final 34 sequences are the following, where the number in parentheses is its GenBank accession number: Acidianus brierleyi (D26489), Caldococcus noboribetus (D85038), Cenarchaeum symbiosum (U51469), Desulfurococcus mobilis (M36474), Metallosphaera sp. (D85508_D38776), Pyrobaculum aerophilum (L07510), Pyrodictium occultum (M21087), Stygiolobus azoricus 2 (D85520), Sulfolobus metallicus 2 (D85519), Sulfolobus solfataricus 2 (D26490), Sulfurisphaera ohwakuensis (D85507_ D38775), Thermofilum pendens (X14835), Thermoproteus tenax (M35966), Archaeoglobus fulgidus (X05567_Y00275), Bacterial sp. 34 (X92171), Bacterial sp. 36 (X92172), Haloarcula vallismortis (U17593), Halobacteriaceae gen. sp. 2 (AJ002946), Halorubrum sodomense (D13379), Natrono bacterium magadii (X72495), Methanobacterium sp. (AF028690), Methanobacterium thermoautotrophicum 5 (AE000940_AE000666), Methanothermus fervidus (M32222), Methanococcus jannaschii 3 (U67517_L77117), Methano coccus vannielii (M36507), Methanoculleus marisnigri (AF028693), Methanosarcina frisius (X69874), Methano spirillum hungatei (M60880), Methanothrix soehngenii (X16932_X51423), Pyrococcus sp. 2 (Z70247), Thermo coccus mexicalis (Z75218), Thermococcus stetteri (Z75240), Ferromonas metallovorans (AJ224936) and Thermoplasma acidophilium (M38637_M20822). To ensure that the sequences can only be aligned locally, Gorodkin et al. (19) further randomly truncated each sequence at both ends by up to 20 nt.
The second data set is another data set used in the experiments of SLASH. It is the ferritin IRE-like data set (iron response element) constructed by Gorodkin et al. (19). They first obtained 14 sequences from the UTR database (26). Since the selected IRE regions are significantly conserved not only in structure but also in sequence, even sequence motif finding algorithms can identify them within the UTRs. Therefore, they modified the IREs and their UTRs to make the search more difficult. By iteratively shuffling the sequences and randomly adding 1 nt to the IRE conserved region, they obtained a set of 56 IRE-like sequences from the 14 IRE UTRs. The new structure motifs are as shown in schemes 1 and 2 below.
where the parentheses indicate base pairing, N ∈ {A, G, C, T}, W ∈ {A, U}, H ∈ {A, C, U}, and X is a random nucleotide. They are highly variable in sequence, but with conserved structure.
The third data set includes 18 viral 3′-UTRs each of which contains a pseudoknot. Seven of the RNA sequences are the soil-borne rye mosaic viruses; the others are the soil-borne wheat mosaic viruses. We first retrieved the pseudoknot sequences from PseudoBase (27) (http://wwwbio.leidenuniv.nl/∼Batenburg/PKB.html). Their accession numbers in PseudoBase are listed as PKB183–PKB189 and PKB194– PKB204. The pseudoknot sequences and base pairings are presented below.
As the pseudoknots are relatively short, to make the search for the pseudoknots more challenging, we randomly include the flanking of 5–70 nt at both ends of each pseudoknot sequence. All the data sets above are downloadable from http://bioinfo.cis.nctu.edu.tw/service/gprm/.
Evaluation
We applied the Matthews correlation coefficient (28) to quantify the agreement between the predicted motif and the actual structure assignment. For each sequence in the data set, two secondary structure assignments were compared by counting the number of true positives Pt (base pairs exist in actual assignment and are predicted), true negatives Nt (base pairs do not exist in actual assignment and are not predicted), false positives Pf (base pairs do not exist in actual assignment but are predicted) and false negatives Nf (base pairs exist in actual assignment but are not predicted), respectively. The Matthews correlation coefficient can then be computed as:
Given that the sequence length is sufficiently large, the Matthews correlation coefficient can be approximated in the following way (19):
With the published/curated alignments, we can evaluate the performance of our approach by calculating the Matthews correlation coefficient. Due to its stochastic characteristics, GPRM was repeatedly tested 30 times on each of the data sets. The correlation coefficients averaged over 30 runs are presented in Table 1, and Table 2 is the GP tableau for the RNA secondary structure prediction problem. Owing to limited space, a partial result of the predicted RNA motifs is shown in Figure 3, and a complete GPRM sample output can be found at http://bioinfo.cis.nctu.edu.tw/service/gprm/.
Table 1. The experimental results of GPRM on three data sets.
| Data set | Archaea rRNA | IRE-like | Viral 3′-UTR |
|---|---|---|---|
| Total sequences | 34 | 56 | 18 |
| Minimum sequence length | 90 | 117 | 37 |
| Maximum sequence length | 108 | 330 | 137 |
| Average sequence length | 97.59 | 202.93 | 63.89 |
| Sequence length SD | 3.77 | 59.31 | 25.95 |
| Average coefficient | 0.87 | 0.99 | 0.76 |
| Coefficient SD | 0.02 | 0.02 | 0.05 |
The first row shows the total number of sequences in each data set. Rows 2–4 present the minimum, the maximum and the average sequence length, respectively. The fifth row gives the SD of sequence length. Rows 6 and 7 provide the correlation coefficient averaged over 30 runs, and its SD. In each run, we used a random negative set of the same size as the positive set.
Table 2. Tableau for RNA secondary structure prediction problem.
| Objective | Given a family of functionally related RNA sequences, predict the common structure motifs |
| Terminal set | User-specified pairing and non-pairing segment length ranges, and pairing segment indices |
| Functional set | Watson–Crick complementarity and structure element connections |
| Fitness measure | F-score based on precision and recall |
| Selection method | Tournament selection |
| Parameters | Population size = 1000, maximum number of generations = 50, crossover rate = 50%, mutation rate = 90%, reproduction rate = 50% |
Figure 3.
A partial result of the predicted RNA motifs. The numbers above the sequences are the indices of the nucleotides. The predicted and the published motifs are both shown for reference. A complete sample result can be found at http://bioinfo.cis.nctu.edu.tw/service/gprm/.
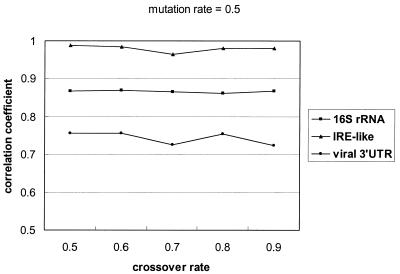
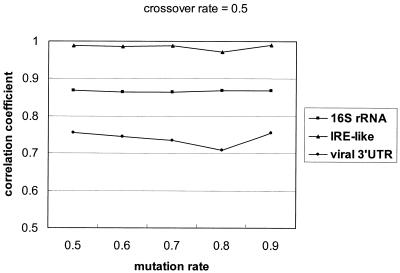
The crossover rate and the mutation rate can affect GPRM’s performance. As the two operators may interact with each other, in order not to complicate the experiments, we fixed one rate at 0.5 and varied the other from 0.5 to 0.9 to measure its effect on the Matthews correlation coefficient. Figure 4 shows the change of correlation coefficients along with varying crossover rates, and Figure 5 presents the results when applying different mutation rates. According to Figure 4, the correlation coefficient for the viral 3′-UTR data set is more sensitive to the change of crossover rate. Its SD is 0.007. Compared with the viral 3′-UTRs, GPRM is more stable when applied to the other two data sets. The SDs of their correlation coefficients are 0.001 and 0.003, respectively. Similarly, Figure 5 shows that the performance of GPRM on the viral 3′-UTRs varied with different mutation rates more noticeably than on the other two data sets, 0.008 compared with 0.0009 and 0.003.
Figure 4.
The correlation coefficients for different crossover rates. We fixed the mutation rate at 0.5, and varied the crossover rate from 0.5 to 0.9. For all experiments, the size of the negative data set was set the same as the positive data set size.
Figure 5.
The correlation coefficients for different mutation rates. We fixed the crossover rate at 0.5, and varied the mutation rate from 0.5 to 0.9. For all experiments, the size of the negative data set was set the same as the positive data set size.
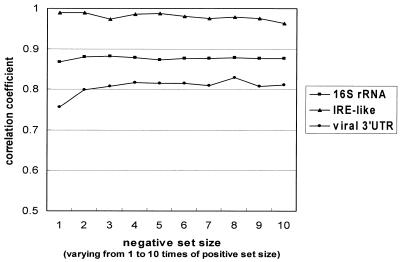
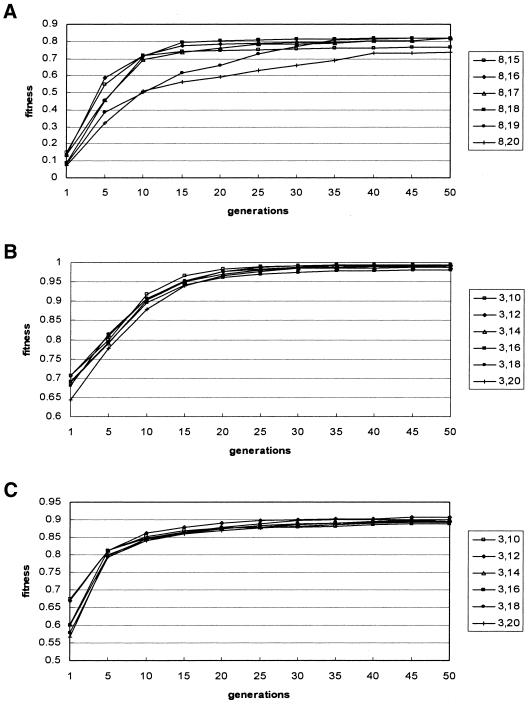
Currently, GPRM uses a random negative set of the same size as the positive set. To investigate the effect of the negative set size on the correlation coefficient, we repeated our experiments with negative sets of different sizes, varying from 1 to 10 times of the positive set size. The correlation coefficients are presented in Figure 6. It shows that the performance for the viral 3′-UTRs data set was affected the most. In addition, we examined the effects of varying complementary segment length ranges on convergence. We fixed the minimum length to 8 bp, and varied the maximum length from 15 to 20 bp for the 16S rRNAs data set. We recorded the fitness values at different generations to see how the segment lengths affect GPRM’s convergence behavior. The result is shown in Figure 7A. Similarly, for the IRE-like and the viral 3′-UTR data sets, we fixed the minimum length to 3 bp, and varied the maximum from 10 to 20 bp. The results are presented in Figure 7B and C, respectively. Figure 7 indicates no significant effects of varying segment lengths. For each test data set, GPRM’s fitness values converged before 50 generations. Similar experiments were also performed on varying non-pairing segment length ranges. The results also showed no significant differences (data not shown).
Figure 6.
The correlation coefficients for negative sets of different sizes. We varied the set size from 1 to 10 times of positive set size. For all experiments, we fixed the mutation rate at 0.9 and the crossover rate at 0.5.
Figure 7.
The fitness values for different base-pairing segment length ranges at different generations. For 16S rRNAs, we fixed the minimum length to 8 bp, and varied the maximum length from 15 to 20 bp. The fitness values at different generations are shown in (A). For both IRE-like data and viral 3′-UTRs, we fixed the minimum length to 3 bp, and varied the maximum length from 10 to 20 bp. The result of the IRE-like data set is presented in (B), and the viral 3′-UTRs result is illustrated in (C). In each experiment, we fixed the mutation rate at 0.9 and the crossover rate at 0.5.
DISCUSSION
We developed a GP approach to finding common structural motifs in a set of coregulated RNA sequences. Those methods designed to identify only consensus sequences are not reliable to find RNA motifs. With flexible GP operators and structural motif representations, our new method, GPRM, is able to identify general RNA secondary motifs.
To show GPRM is comparable to the latest RNA motif prediction systems, we tested it on the same data sets previously used in order to maintain consistency. We first tested GPRM on a set of archaeal rRNA sequences that contain locally aligned stem–loop regions. By comparing them with the curated database alignment, we were able to evaluate our new approach quantitatively by the Matthews correlation coefficient. We obtained a 0.87 correlation coefficient between the predicted structural alignment and the curated database alignment. This is similar to the published experimental results (19). We also tested GPRM on the ferritin IRE-like data set created by Gorodkin et al. (19), and obtained a 0.99 correlation coefficient. GPRM was further tested on a a set of viral 3′-UTR pseudoknot regions extracted from PseudoBase (25). We used this data set to demonstrate its capability that current RNA motif finding algorithms lack. We obtained promising correlation coefficients from 0.75 to 0.83 as shown in Figure 6.
GPRM can be further improved in two directions. First, the current fitness function of GPRM is only based on motif occurrences in training examples. We plan to enhance the fitness function by incorporating background knowledge such as thermodynamic (9,10) or phylogenetic (29) information. Secondly, GPRM is currently limited to find base-pairing structures. We will extend the motif representation and the genetic operators to deal with more complex structures, e.g. multiple compound stem–loops or structures with multi-branch loops.
Acknowledgments
ACKNOWLEDGEMENTS
Thanks to Jan Gorodkin for providing the valuable archaea 16S rRNA sequences and the ferritin IRE-like data set. Thanks also go to the anonymous reviewers for their valuable comments and suggestions. This project is partially supported by the National Science Council, Taiwan (NSC 90-2213-E-009-169).
REFERENCES
- 1.Gygi S.P., Rochon,Y., Franza,B.R. and Aebersold,R. (1999) Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol., 19, 1720–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gray N.K. and Hentze,M.W. (1994) Regulation of protein synthesis by mRNA structure. Mol. Biol. Rep., 19, 195–200. [DOI] [PubMed] [Google Scholar]
- 3.Klaff P., Riesner,D. and Steger,G. (1996) RNA structure and the regulation of gene expression. Plant Mol. Biol., 32, 89–106. [DOI] [PubMed] [Google Scholar]
- 4.Hertz G., Hartzell,G.,III and Stormo,G. (1990) Identification of consensus patterns in unaligned DNA sequences known to be functionally related. Comput. Appl. Biosci., 6, 81–92. [DOI] [PubMed] [Google Scholar]
- 5.Lawrence C., Altschul,S., Boguski,M., Liu,J., Neuwald,A. and Wootton,J. (1993) Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignments. Science, 262, 208–214. [DOI] [PubMed] [Google Scholar]
- 6.Bailey T. and Elkan,C. (1995) Unsupervised learning of multiple motifs in biopolymers using expectation maximization. Machine Learning, 21, 51–80. [Google Scholar]
- 7.Van Helden J., Andre,B. and Collado-Vides,J. (1998) Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol., 281, 827–842. [DOI] [PubMed] [Google Scholar]
- 8.Hu Y., Sandmeyer,S., McLaughlin,C. and Kibler,D. (2000) Combinatorial motif analysis and hypothesis generation on a genomic scale. Bioinformatics, 16, 222–232. [DOI] [PubMed] [Google Scholar]
- 9.Zuker M. and Stiegler,P. (1981) Optimal computer folding of large RNA sequences using thermodynamic and auxiliary information. Nucleic Acids Res., 9, 133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gultyaev A.P., van Batenburg,F.H.D. and Pleij,C.W.A. (1995) The computer simulation of RNA folding pathways using a genetic algorithm. J. Mol. Biol., 250, 37–51. [DOI] [PubMed] [Google Scholar]
- 11.Rivas E. and Eddy,S.R. (1999) A dynamic programming algorithm for RNA structure prediction including pseudoknots. J. Mol. Biol., 285, 2053–2068. [DOI] [PubMed] [Google Scholar]
- 12.van Batenburg F.H.D., Gultyaev,A.P. and Pleij,C.W.A. (1995) An APL-programmed genetic algorithm for the prediction of RNA secondary structure. J. Theor. Biol., 174, 269–280. [DOI] [PubMed] [Google Scholar]
- 13.Westhof E. and Michel,F. (1994) Prediction and experimental investigation of RNA secondary and tertiary foldings. In Nagai,K. and Mattaj,I.W. (eds), RNA–Protein Interactions. IRL Press, Oxford, UK, pp. 26–51.
- 14.Gutell R.R., Larsen,N. and Woese,C.R. (1994) Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol. Rev., 58, 10–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laferriere A., Gautheret,D. and Cedergren,R. (1994) An RNA pattern matching program with enhanced performance and portability. Comput. Appl. Biosci., 10, 211–212. [DOI] [PubMed] [Google Scholar]
- 16.Eddy S. and Durbin,R. (1994) RNA sequence analysis using covariance models. Nucleic Acids Res., 22, 2079–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J.-H., Le,S.-Y. and Maizel,J.V. (2000) Prediction of common secondary structures of RNAs: a genetic algorithm approach. Nucleic Acids Res., 28, 991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorodkin J., Heyer,L.J. and Stormo,G.D. (1997) Finding the most significant common sequence and structure motifs in a set of RNA sequences. Nucleic Acids Res., 25, 3724–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorodkin J., Stricklin,S.L. and Stormo,G.D. (2001) Discovering common stem–loop motifs in unaligned RNA sequences. Nucleic Acids Res., 29, 2135–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inge J. (1996) Methods for finding motifs in sets of related biosequences. PhD Thesis, Department of Informatics, University of Bergen, Norway.
- 21.Banzhaf W., Nordin,P., Keller,R.E. and Francone,F.D. (1998) Genetic Programming: An Introduction on the Automatic Evolution of Computer Programs and its Applications. Morgan Kaufmann Publisher, San Francisco, CA.
- 22.Lewis D. and Gale,W.A. (1994) A sequential algorithm for training text classifier. In Proceedings of the 17th Annual International ACM-SIGIR Conference on Research and Development in Information Retrieval. pp. 3–12.
- 23.Koza J. (1992) Genetic Programming: On the Programming of Computers by Means of Natural Selection. The MIT Press, Cambridge, MA.
- 24.Robertson G. (1987) Parallel implementation of genetic algorithms in a classifier system. In Davis,L. (ed.), Genetic Algorithms and Simulated Annealing. Pitman, London, UK.
- 25.Van de Peer Y., De Rijk,P., Wuyts,J., Winkelmans,T. and De Wachter,R. (2000) The European small subunit ribosomal RNA database. Nucleic Acids Res., 28, 175–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pesole G., Liuni,S., Grillo,G., Larizza,A., Malakowski,W. and Saccone,C. (2000) UTRdb and UTRsite: specialized databases of sequences and functional elements of 5′ and 3′ untranslated regions of eukaryotic mRNAs. Nucleic Acids Res., 28, 193–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Batenburg F.H.D., Gultyaev,A.P. and Pleij,C.W.A. (2001) PseudoBase: structural information on RNA pseudoknots. Nucleic Acids Res., 28, 1, 201–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matthews B.W. (1975) Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta, 405, 442–451. [DOI] [PubMed] [Google Scholar]
- 29.Akmaev V.R., Kelley,S.T. and Stormo,G.D. (2000) Phylogenetically enhanced statistical tools for RNA structure prediction. Bioinformatics, 16, 501–512. [DOI] [PubMed] [Google Scholar]