Abstract
The pyrrole–imidazole (Py–Im) triamide–cyclopropa pyrroloindole (CPI) conjugates ImPyImLDu86 (7) and ImImPyLDu86 (14) were synthesized and their alkylating activities and inhibitory effects on DNA hydrolysis by restriction endonucleases were examined. Sequencing gel analysis demonstrated that conjugates 7 and 14 specifically alkylated DNA at 5′-CGCGCG-3′ and 5′-PyGGCCPu-3′, respectively. Agarose gel electrophoresis indicated that incubation of a supercoiled plasmid, pSPORT I (4109 bp), with conjugate 7 effectively inhibited its hydrolysis by BssHII (5′-G_CGCGC-3′), whereas conjugate 14 had no effect on this hydrolysis. These results suggest that conjugate 7 sequence-specifically inhibits the hydrolysis of DNA by BssHII. Sequence-specific alkylation by the Py–Im triamide–CPI conjugates was further confirmed by inhibition of the Eco52I (5′-C_GGCCG-3′) hydrolysis of conjugate 14-treated pQBI PGK (5387 bp). In clear contrast, hydrolysis of pQB1 PGK by DraI (3′-TTT_AAA-3′) was not inhibited by 5 µM conjugate 14. That ImImPy did not inhibit the hydrolysis of pQB1 PGK indicates that covalent bond formation is necessary for inhibition. A similar experiment, using linear pQBI PGK, achieved the same extent of protection of the DNA with approximately half the concentration of conjugate 14 as was required to protect supercoiled DNA from hydrolysis.
INTRODUCTION
Small sequence-specific DNA-binding molecules, exemplified by N-methylpyrrole (Py) and N-methylimidazole (Im) polyamides, uniquely recognize each of the four Watson–Crick DNA base pairs in the minor groove according to a simple set of pairing rules dictated by the side-by-side binding of the aromatic amino acids. They have attracted intense interest as powerful tools in molecular biology and human medicine (1–6). Trauger et al. (7) have demonstrated elegantly that such polyamides bind to predetermined DNA sequences with affinities and specificities similar to those of DNA-binding proteins. Py–Im polyamides that uniquely recognize each of the four Watson–Crick base pairs can be used as novel recognition components of sequence-specific DNA alkylating agents (8–13). We have demonstrated that hybrid molecules constructed from segment A of duocarmycin A and Py–Im diamides or from hairpin polyamides specifically alkylate at predetermined nucleotide sequences within a 450-bp DNA fragment (9,10). Recently, we found that insertion of a vinyl linker (L) between the polyamide and cyclopropapyrroloindole (CPI) groups alters the location of the reactive cyclopropane ring of the alkylating moiety and allows highly sequence-specific cooperative double-strand alkylation of DNA (11). More recently, we developed a novel DNA interstrand cross-linking agent that cross-links double strands only in the presence of ImImPy, at the 9-bp sequence, 5′-PyGGC(T/A)GCCPu-3′ (14).
Targeting specific sequences in the human genome with such Py–Im polyamides should provide a powerful gene-regulating tool. In fact, Gottesfeld et al. (15) and Dickinson et al. (16) demonstrated that hairpin polyamides inhibit basal and activated transcription from the promoters of RNA polymerases II and III by disrupting specific transcription factor–DNA interactions. The inhibition of transcription by hairpin polyamides is achieved by competitive binding to the binding sites of regulatory proteins. Therefore, the binding of Py–Im polyamides to the coding region cannot effectively inhibit transcription by RNA polymerase (12). Alkylating Py–Im polyamides that form covalent bonds with DNA should have more potential to regulate transcription and can target, not only the regulatory region, but also the coding region. Inhibition of the hydrolysis of plasmid DNA catalyzed by restriction endonucleases has been used effectively to examine sequence-specific binding of peptide nucleic acid (17). As a first step in evaluating the potential of alkylating Py–Im polyamides in the regulation of specific genes, we examine here the alkylation of plasmid DNA and its inhibitory effects on the hydrolysis of DNA by restriction endonucleases.
MATERIALS AND METHODS
Materials
Polyacrylamide gel electrophoresis was performed on a Hitachi 5500-S DNA Sequencer. Ex Taq DNA polymerase and Suprec-02 purification cartridges were purchased from Takara Co.; the Thermo Sequenase core sequencing kit and loading dye (DMF with fuchsin red) were from Amersham Co. Ltd; 5′-Texas Red-modified DNA oligomer (18mer) was synthesized by Kurabo Co. Ltd; and 50% Long Ranger™ gel solution was obtained from FMC Bioproducts. Plasmids pSPORT I and QBI PGK were obtained from Gibco BRL and Takara Shuzo Co. Ltd, respectively. P1 nuclease and calf intestinal alkaline phosphatase (AP, 1000 U/ml) were purchased from Roche Diagnostics. Restriction endonucleases, BssHII, Eco52I, DraI and HindIII were purchased from Takara Shuzo Co. Ltd. Syntheses of Py–Im–CPI conjugates ImPyImLDu86 (7) and ImImPyLDu86 (14) are described in the Supplementary Material. 7: 1H NMR (DMSO-d6) δ 12.39 (s, 1H), 10.57 (s, 1H), 10.29 (s, 1H), 9.76 (s, 1H), 7.60 (s, 1H), 7.56 (d, J = 14.5 Hz, 1H), 7.43 (s, 1H), 7.40 (d, J = 2.0 Hz, 1H), 7.26 (brs, 1H), 7.20 (d, J = 2.0 Hz, 1H), 7.00 (d, J = 14.5 Hz, 1H), 4.20 (m, 2H), 3.95 (s, 3H), 3.87 (s, 3H), 3.81 (s, 3H), 3.74 (s, 3H), 3.48 (m, 1H), 2.47 (s, 3H), 2.11 (dd, J = 4.0 and 4.5 Hz, 1H), 2.02 (s, 3H), 1.36 (t, J = 4.0 Hz, 1H). ESIMS m/e calc. for C34H33N10O4 (M-H) 693.7, found 693.6. 14: 1H NMR (DMSO-d6) δ 12.35 (s, 1H), 10.34 (s, 1H), 10.29 (s, 1H), 9.33 (s, 1H), 7.57 (d, J = 15.5 Hz, 1H), 7.57 (s, 1H), 7.50 (s, 1H), 7.41 (d, J = 2.0 Hz, 1H), 7.01 (d, J = 2.0 Hz, 1H), 6.83 (brs, 1H), 6.56 (d, J = 14.5 Hz, 1H), 4.29 (d, J = 10.0 Hz, 1H), 4.19 (dd, J = 4.5 and 10.0 Hz, 1H), 4.00 (s, 3H), 3.97 (s, 3H), 3.72 (s, 3H), 3.71 (s, 3H), 3.45 (m, 1H), 2.46 (s, 3H), 2.04 (s, 3H), 2.02 (dd, J = 4.0 and 4.5 Hz, 1H), 1.29 (t, J = 4.0 Hz, 1H). ESMS m/e calc. for C34H34N10O7 (M + H) 695.3, found 695.3.
Preparation of 5′-Texas Red-end-modified 450-bp DNA fragments
The 5′-Texas Red-modified 450-bp DNA fragments pUC18 F378*-827 and pUC18 R1861*-2310 (they are complementary) were prepared using a PCR method and the 5′-Texas Red-modified 18mers 5′-TGTAAAACGACGGCCAGT-3′ (pUC18 forward 378–396), and 5′-TGCTGGCCTTTTGCTCAC-3′ (pUC18 reverse 1861–1879) as primers, and purified by filtration using Suprec-02. Product concentrations were determined by ethidium bromide staining. The asterisks (above) indicate Texas-Red modifications, and the nucleotide numbering starts at the replication site.
High resolution gel eletrophoresis
The 5′-Texas Red-labeled DNA fragment (50 µg) was alkylated with various concentrations of conjugates 7 or 14 in 10 mM Na phosphate buffer (10 µl, pH 7.0) containing 10% DMF at 25°C overnight. The reaction was quenched by the addition of calf thymus DNA (5 mM, 1 µl) with heating for 5 min at 90°C. DNA was precipitated with ethanol. The pellet was dissolved in 8 µl of loading dye (formamide with fuchsin red), heated at 94°C for 30 min, and then immediately cooled to 0°C. A 2-µl aliquot was separated electrophoretically on a 6% denaturing polyacrylamide gel using a Hitachi 5500-S DNA sequencer.
Preparation of linear DNA
Linear DNA was generated from plasmid DNA (pQBI PGK, 20 µg) by treatment with HindIII (30 U) in 10 mM Tris–HCl buffer (200 µl, pH 7.5) containing 7 mM MgCl2 and 60 mM NaCl at 37°C for 1 h. After phenol/CHCl3 treatment, linear DNA was isolated by ethanol precipitation. DNA concentrations were determined by the absorbance of double-stranded DNA (dsDNA) at 260 nm (1 A260 unit = 50 µg/ml for dsDNA).
Agarose gel electrophoresis
Reaction mixtures (2.5 µl) containing plasmid DNA (75 ng), and the indicated amounts of conjugates 7 or 14, 10 mM Tris–HCl buffer (pH 8.9), 3 mM MgCl2, 100 mM NaCl and 10% DMF were incubated at 25°C for 2 days. After incubation, restriction endonuclease (4 U) was added to the reaction mixture (total volume 5 µl including 0.01% bovine serum albumin), and the solution was incubated at 37°C for 2 h. The reaction mixture was directly loaded onto a 1.2% E-gel™ (Invitrogen®) containing 0.5 mg/ml ethidium bromide with 2 µl of loading buffer (1% sodium dodecyl sulfate, 50% glycerol and 0.05% bromophenol blue) and 13 µl of H2O. Electrophoresis was carried out under a constant electric field of 70 V for 30 min at 25°C. DNA bands were detected and photographed under UV light (302 nm).
RESULTS AND DISCUSSION
Sequence-specific DNA alkylation by Py–Im triamide–CPI conjugate
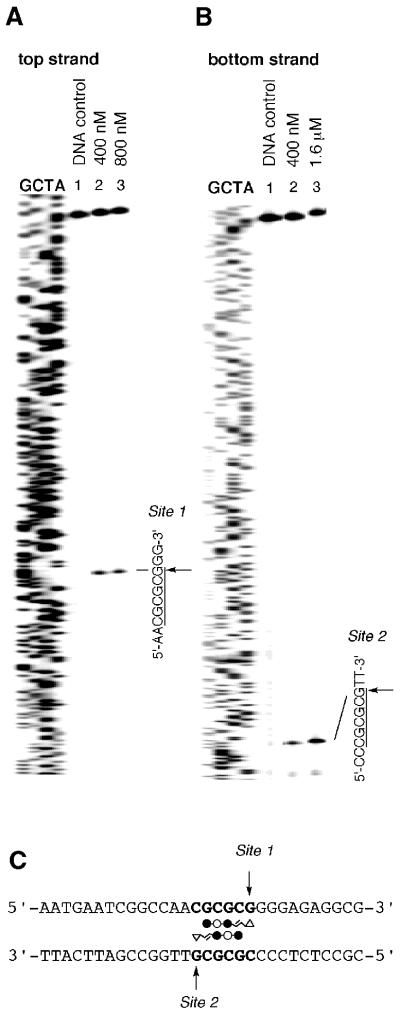
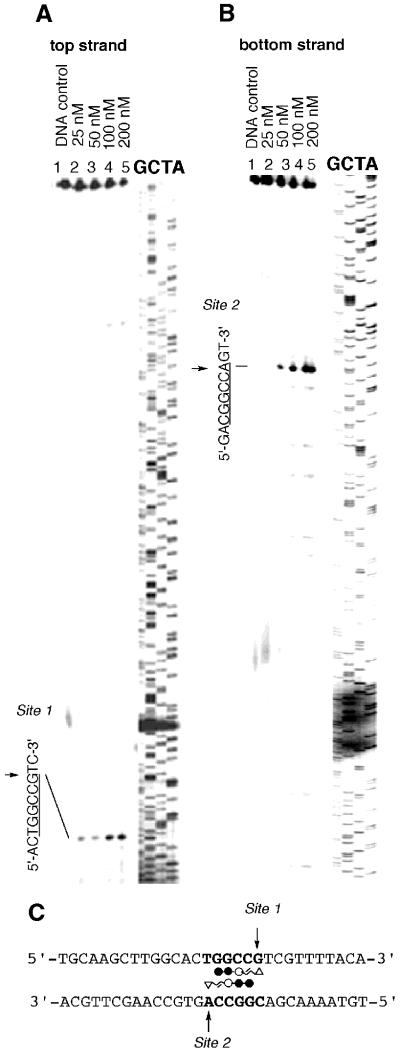
The detailed synthesis procedures of Py–Im triamide–CPI conjugates ImPyImLDu86 (7) and ImImPyLDu86 (14) are described in the Supplementary Material. Sequence-specific DNA alkylation by conjugate 7 was investigated on both strands of a 5′-Texas Red-labeled 450-bp DNA fragment prepared by PCR using pUC18 as the template. As shown in Figure 1, conjugate 7 at a concentration of 400 nM produced highly selective alkylation of the G in the sequence 5′-CGCGCG-3′/3′-GCGCGC-5′ on both strands (Sites 1 and 2) within a 450-bp DNA fragment, with high efficiency. The results are fully consistent with Dervan’s pairing rule and clearly indicate that the imidazolyl acrylic part of the molecule plays the same key role as its pyrrolyl counterpart in the dialkylation system (11). The simultaneous double-strand dialkylation was further confirmed at the oligonucleotide level by HPLC product analysis. Importantly, this agent can dialkylate double DNA strands without the slipped binding mode, which was observed with ImPyLDu86 as a minor alkylation site (11). Similarly, sequencing gel electrophoresis of conjugate 14-treated labeled DNA fragments demonstrated that conjugate 14 specifically alkylated DNA at 5′-TGGCCG-3′/3′-ACCGGC-5′ (Sites 1 and 2) on both strands, by forming a homodimer according to the pairing rule of Py–Im polyamides as shown in Figure 2. The double-strand alkylation by conjugate 14 was observed in a self-complementary sequence, 5′-CAACGGCCGTTG-3′, indicating that alkylation by conjugate 14 occurs at either G or A (data not shown).
Figure 1.
Thermally induced strand cleavage of the alkylated 5′-Texas Red-labeled 450-bp DNA duplex by ImPyImLDu86 (7). Results using 5′-labeled top strand (pUC18 F378–827) (A) and 5′-labeled bottom strand (pUC18 R1861–2310) (B) DNA fragments are shown. These two DNA fragments are complementary sequences containing Site 1 and Site 2. Lane 1 contains the DNA control; G, C, T, A are the Sanger sequencing standards. Concentrations of conjugate 7 are indicated in lanes 2 and 3. Sequences containing dialkylation sites (Site 1 and Site 2) are represented (C). The arrows indicate the sites of alkylation.
Figure 2.
Thermally induced strand cleavage of the alkylated 5′-Texas Red-labeled 426-bp DNA duplex by ImImPyLDu86 (14). Results using 5′- labeled top strand (pUC18 R2207–2632) (A) and 5′-labeled bottom strand (pUC18 F56–481) (B) DNA fragments are shown. These two DNA fragments are complementary sequences containing Site 1 and Site 2. Lane 1 contains the DNA control; G, C, T, A are the Sanger sequencing standards. Concentrations of conjugate 14 are indicated in lanes 2–5. Sequences containing dialkylation sites (Site 1 and Site 2) are represented (C). The arrows indicate the sites of alkylation.
Sequence-specific protection of plasmid DNA by Py–Im triamide–CPI conjugates
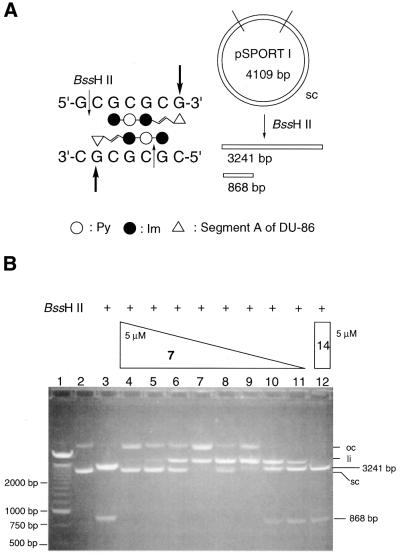
We next investigated the alkylation of supercoiled plasmid DNA (4000–5500 bp) by Py–Im triamide–CPI conjugates 7 and 14. To examine the sequence specificity of conjugate 7 in supercoiled plasmid DNA, the inhibitory effects of conjugate 7 were examined on the hydrolysis of DNA by the restriction endonuclease BssHII, which hydrolyzes dsDNA at 5′-G_CGCGC-3′. If conjugate 7 selectively alkylates supercoiled plasmid DNA at the 3′ end of the 5′-CGCGCG-3′ sequence, which overlaps the recognition sequence for BssHII, an inhibition of hydrolysis would be anticipated. Figure 3 shows the separation by agarose gel electrophoresis of the hydrolysis products of pSPORT I (4109 bp), which contains two BssHII cleavage sites and gives rise to 3241- and 868-bp fragments (Fig. 3B, lane 3). Interestingly, incubation of pSPORT I with conjugate 7 for 2 days (pH 8.9, 25°C) effectively inhibited the hydrolysis by BssHII in a concentration-dependent manner (5 µM to 1.0 nM, Fig. 3B, lanes 4–11). Almost complete inhibition of hydrolysis was observed in the presence of 5 µM conjugate 7 (Fig. 3B, lane 4), whereas conjugate 14, which alkylates DNA at the 3′ end of the 5′-CGGCCG-3′ sequence, had no effect on the hydrolysis of pSPORT I by BssHII (Fig. 3B, lane 12). The results clearly demonstrate that conjugate 7 sequence-specifically alkylates supercoiled plasmid DNA and that this alkylation effectively inhibits the hydrolysis of DNA by BssHII.
Figure 3.
Sequence-specific protection of pSPORT I plasmid DNA from restriction endonuclease BssHII hydrolysis by incubation with ImPyImLDu86 (7). (A) A BssHII restriction map of pSPORT I DNA and schematic representation of the binding mode of conjugate 7 at BssHII hydrolysis sites. The Py and Im rings are represented by open and closed circles, respectively; the triangle represents segment A of DU-86. (B) Agarose gel (1.2%) electrophoresis of triamide–CPI conjugate-treated pSPORT I DNA after BssHII (2.5 U) digestion (lanes 3–12). Lane 1, 250-bp DNA marker; lane 2, pSPORT I DNA control; lane 3, no drug treatment; lanes 4–11, 5 µM, 1 µM, 500 nM, 250 nM, 100 nM, 50 nM, 10 nM, 1 nM concentrations of conjugate 7; lane 12, 5 µM conjugate 14. sc, supercoiled DNA; li, linear DNA; oc, open circle DNA.
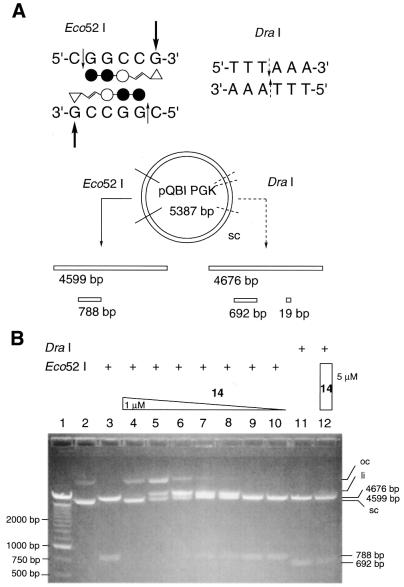
The sequence-specific alkylation of supercoiled plasmid DNA by the alkylating Py–Im triamide 14 was further confirmed by the selective inhibition of the hydrolysis of pQBI PGK (5387 bp) by Eco52I (5′-C_GGCCG-3′). Figure 4 shows the separation by agarose gel electrophoresis of the hydrolysis products of pQBI PGK: 4599- and 788-bp fragments generated from the two Eco52I cleavage sites (Fig. 4B, lane 3). Incubation of DNA with conjugate 14 inhibited the hydrolysis of DNA by Eco52I in a concentration-dependent manner (1 µM to 1 nM, Fig. 4B, lanes 4–10). However, hydrolysis by DraI, which hydrolyzes dsDNA at 5′-TTT_AAA-3′ and cleaves pQBI PGK into 4676-, 692- and 19-bp fragments, was not inhibited by incubation with 5 µM conjugate 14 (lane 12). The results clearly indicate that conjugate 14 selectively alkylates the target sequence in supercoiled plasmid DNA and sequence-specifically inhibits the hydrolysis of DNA by Eco52I.
Figure 4.
Sequence-specific protection of pQBI PGK plasmid DNA from restriction endonuclease hydrolysis by ImImPyLDu86 (14). (A) A pQBI PGK DNA restriction map for Eco52I and DraI, and schematic representation of the binding mode of conjugate 14 at the Eco52I hydrolysis sites. (B) Agarose gel (1.2%) electrophoresis of conjugate 14-treated pQBI PGK after Eco52I (lanes 3–10) and DraI (lanes 11 and 12) digestion. Lane 1, 250-bp DNA marker; lane 2, pQBI PGK DNA control; lanes 3 and 11, no drug treatment; lanes 4–10, 1 µM, 500 nM, 250 nM, 100 nM, 50 nM, 10 nM, 1 nM concentrations of conjugate 14; lane 12, 5 µM conjugate 14. sc, supercoiled DNA; li, linear DNA; oc, open circle DNA.
It is important to note that 5 µM ImImPy did not inhibit the hydrolysis of pQBI PGK, even though this concentration is 3-fold higher than the concentration of conjugate 14 that completely inhibited the hydrolysis (Fig. 5). These results clearly indicate that covalent bond formation between DNA and Py–Im polylamides is essential for the inhibition of hydrolysis. Because alkylation in the minor groove influences base pairing and therefore also leads to substantial changes in the major groove, the hydrolytic activity of restriction endonucleases that use major-groove contact might be effectively inhibited. Even though the protection afforded by the alkylating Py–Im triamide relative to that afforded by ImImPy is dramatically different, the concentration required for complete protection was rather high (1–5 µM) relative to the strong binding of the Py–Im polyamide. Part of the reason may be derived from the nature of the protection assay. Since complete inhibition in protection experiments requires complete alkylation of DNA at the target site, unreacted alkylating agent and/or it’s hydrolysis products possibly interfere with the completion of alkylation. Furthermore, the Py–Im–CPI conjugates in the present study do not possess a positively charged terminal end, which is known to enhance the binding affinity to DNA. Incorporation of such a group may further enhance the alkylation activity, and as a result, it may provide more significant protection of the DNA by the alkylating Py–Im polyamide relative to that provided by the non-covalent Py–Im polyamide.
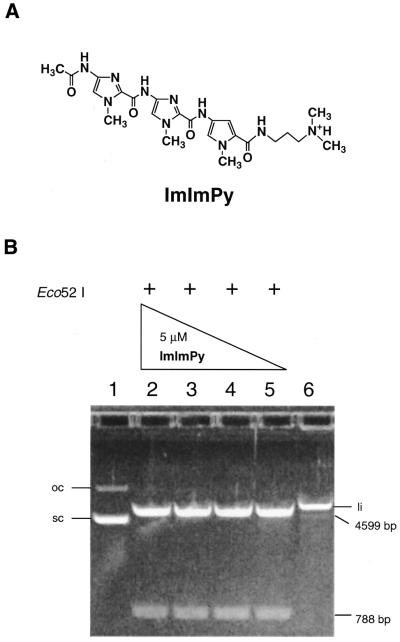
Figure 5.
Hydrolysis of pQBI PGK by Eco52I in the presence of ImImPy. (A) The chemical structure of ImImPy. (B) Agarose gel (1.2%) electrophoresis of the products of Eco52I hydrolysis of pQBI PGK DNA in the presence of various concentrations of ImImPy (lanes 2–5). Lane 1, supercoiled pQBI PGK control (sc); lane 2, 5 µM ImImPy; lane 3, 2 µM; lane 4, 1 µM; lane 5, 500 nM; lane 6, linear pQBI PGK control (li). oc, open circle DNA.
Effect of supercoiling on the protection by Py–Im triamide–CPI conjugates
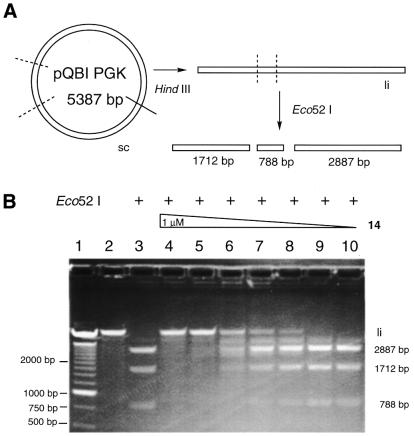
To evaluate the influence of DNA supercoiling on the protection afforded by conjugate 14 against enzyme hydrolysis, a similar experiment was performed using linearized pQBI PGK with HindIII (5′-A_AGCTT-3′) digestion of supercoiled pQBI PGK. The linear DNA (5387 bp) was treated with conjugate 14 and subjected to Eco52I digestion. Figure 6 shows the agarose gel electrophoretic separation of the hydrolysis products of linear pQBI PGK by Eco52I. In the absence of conjugate 14, the linear DNA was digested by Eco52I into three DNA fragments of 2887, 1712 and 788 bp. More efficient inhibition was induced by incubation of linear DNA with conjugate 14 than when supercoiled DNA was incubated with conjugate 14. In the former case, almost complete inhibition of hydrolysis was observed in the presence of 500 nM conjugate 14 (Fig. 6B, lane 5). Even at 50 nM conjugate 14, full-length linear DNA was detected. Densitometric analysis of the unhydrolyzed DNA indicated that approximately half the concentration of conjugate 14 was required to achieve the same extent of inhibition with linear DNA as with supercoiled DNA (Fig. 7). Although supercoiling slightly retarded the polyamide binding to the target sequences in the minor groove, it did not affect the sequence-specificity of conjugate 14. These results might be consistent with the recent observation that hairpin polyamides sequence-specifically bind to the nucleosome core particle (18).
Figure 6.
Sequence-specific protection by ImImPyLDu86 (14) of linear pQBI PGK plasmid DNA from restriction endonuclease hydrolysis. (A) A pQBI PGK DNA restriction map for HindIII and Eco52I. (B) Agarose gel (1.2%) electrophoresis of conjugate 14-treated linear pQBI PGK after Eco52I (lanes 3–10) digestion. Lane 1, 250-bp DNA marker; lane 2, pQBI PGK DNA control (li); lane 3, no drug treatment; lanes 4–10, 1 µM, 500 nM, 250 nM, 100 nM, 50 nM, 10 nM, 1 nM concentrations of conjugate 14.
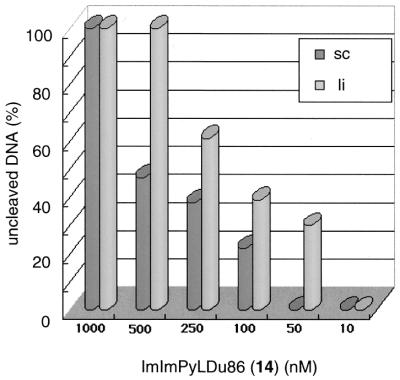
Figure 7.
The amount of pQBI PGK plasmid DNA [supercoiled (sc) and linear (li)] protected from Eco52I hydrolysis by different concentrations of conjugate 14. The amounts of uncleaved supercoiled and linear DNA are given as dark and light shaded bars, respectively: at 1000 nM conjugate 14, 100% sc, 100% li; at 500 nM, 47% sc, 100% li; at 250 nM, 38% sc, 61% li; at 100 nM, 22% sc, 39% li; at 50 nM, 0% sc, 30% li; at 10 nM, 0% sc, 0% li.
In conclusion, we have demonstrated that the alkylating Py–Im triamide–CPI conjugates 7 and 14 alkylate supercoiled plasmid DNA, that this sequence-specific alkylation effectively protects DNA from restriction endonuclease hydrolysis, and that supercoiling of DNA has a slightly negative effect on the DNA alkylation. These results indicate that sequence-specific DNA alkylating agents acting in the minor groove strongly affect DNA–protein interactions. Sequence-specific alkylation by Py–Im polyamides may control gene expression, not only in promoter/enhancer-binding regions, but also in the coding regions of genes.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
REFERENCES
- 1.Gottesfeld J.M., Turner,J.M. and Dervan,P.B. (2000) Chemical approaches to control gene expression. Gene Expr., 9, 77–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dervan P.B. and Burli,R.W. (1999) Sequence-specific DNA recognition by polyamides. Curr. Opin. Chem. Biol., 3, 688–693. [DOI] [PubMed] [Google Scholar]
- 3.Wemmer D.E. and Dervan,P.B. (1997) Targeting the minor groove of DNA. Curr. Opin. Struct. Biol., 7, 355–361. [DOI] [PubMed] [Google Scholar]
- 4.Kielkopf C.L., Baird,E.E., Dervan,P.B. and Rees,D.C. (1998) Structural basis for G·C recognition in the DNA minor groove. Nature Struct. Biol., 5, 104–109. [DOI] [PubMed] [Google Scholar]
- 5.Yang X.L., Kaenzig,C., Lee,M. and Wang,A.H.J. (1999) Binding of AR-1–144, a tri-imidazole DNA minor groove binder, to CCGG sequence analyzed by NMR spectroscopy. Eur. J. Biochem., 263, 646–655. [DOI] [PubMed] [Google Scholar]
- 6.White S., Szewczyk,J.W., Turner,J.M., Baird,E.E. and Dervan,P.B. (1998) Recognition of the four Watson–Crick base pairs in the DNA minor groove by synthetic ligands. Nature, 391, 468–471. [DOI] [PubMed] [Google Scholar]
- 7.Trauger J.W., Baird,E.E. and Dervan,P.B. (1996) Recognition of DNA by designed ligands at subnanomolar concentrations. Nature, 382, 559–561. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y., Gupta,R., Huang,L., Luo,W. and Lown,J.W. (1996) Design, synthesis, cytotoxic properties and preliminary DNA sequencing evaluation of CPI-N-methylpyrrole hybrids. Enhancing effect of a trans double bond linker and role of the terminal amide functionality on cytotoxic potency. Anticancer Drug Des., 11, 15–34. [PubMed] [Google Scholar]
- 9.Tao Z.F., Fujiwara,T., Saito,I. and Sugiyama,H. (1999) Sequence-specific DNA alkylation by hybrid molecules between segment A of duocarmycin A and pyrrole/imidazole diamide. Angew. Chem. Int. Ed. Engl., 38, 650–653. [DOI] [PubMed] [Google Scholar]
- 10.Tao Z.F., Fujiwara,T., Saito,I. and Sugiyama,H. (1999) Rational design of sequence-specific DNA alkylating agents based on duocarmycin A and pyrrole-imidazole hairpin polyamides. J. Am. Chem. Soc., 121, 4961–4967. [Google Scholar]
- 11.Tao Z.F., Saito,I. and Sugiyama,H. (2000) Highly cooperative DNA dialkylation by the homodimer of imidazole-pyrrole diamide-CPI conjugate with vinyl linker. J. Am. Chem. Soc., 122, 1602–1608. [Google Scholar]
- 12.Wurtz N.R. and Dervan,P.B. (2000) Sequence specific alkylation of DNA by hairpin pyrrole-imidazole polyamide conjugates. Chem. Biol., 7, 153–161. [DOI] [PubMed] [Google Scholar]
- 13.Chang A.Y. and Dervan,P.B. (2000) Strand selective cleavage of DNA by diastereomers of hairpin polyamide-seco-CBI conjugates. J. Am. Chem. Soc., 122, 4856–4864. [Google Scholar]
- 14.Bando T., Iida,H., Saito,I. and Sugiyama,H. (2001) Sequence-specific DNA interstrand cross-linking by imidazole-pyrrole CPI conjugate. J. Am. Chem. Soc., 123, 5158–5159. [DOI] [PubMed] [Google Scholar]
- 15.Gottesfeld J.M., Neely,L., Trauger,J.W., Baird,E.E. and Dervan,P.B. (1997) Regulation of gene expression by small molecules. Nature, 387, 202–205. [DOI] [PubMed] [Google Scholar]
- 16.Dickinson L.A., Gulizia,R.J., Trauger,J.W., Baird,E.E., Mosier,D.E., Gottesfeld,J.M. and Dervan,P.B. (1998) Inhibition of RNA polymerase II transcription in human cells by synthetic DNA-binding ligands. Proc. Natl Acad. Sci. USA, 95, 12890–12895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Izvolsky K.I., Demidov,V.V., Nielsen,P.E. and Frank-Kamenetskii,M.D. (2000) Sequence-specific protection of duplex DNA against restriction and methylation enzymes by pseudocomplementary PNAs. Biochemistry, 39, 10908–10913. [DOI] [PubMed] [Google Scholar]
- 18.Gottesfeld J.M., Melander,C., Suto,R.K., Raviol,H., Lugar,K. and Dervan,P.B. (2001) Sequence-specific recognition of DNA in the nucleosome by pyrrole-imidazole polyamides. J. Mol. Biol., 309, 615–629. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.