Abstract
We succeeded in cloning the gene, termed d-ddb1, for a Drosophila homolog of the p127 subunit of the human damage-specific DNA-binding protein, thought to recognize (6–4) photoproducts and related structures. In Drosophila, the gene product (D-DDB1) also appeared to play a role as a repair factor, d-ddb1 knockout Kc cells generated with a RNAi method being sensitive to UV. In addition, UV or methyl methanesulfonate treatment increased d-ddb1 transcripts. However, we found that the gene is controlled by the DRE/DREF system, which is generally responsible for activating the promoters of proliferation-related genes. Moreover, during Drosophila development, the transcription of d-ddb1 changed greatly, with the highest levels in unfertilized eggs, indicating that external injury to DNA is not essential to D-DDB1 function. Interestingly, as with UV irradiation-induced transfer of D-DDB1 to the nucleus from the cytoplasm, during spermatogenesis the protein transiently shifted from one cell compartment to the other. The results indicate that D-DDB1 not only contributes to the DNA repair system, but also has a role in cell proliferation and development.
INTRODUCTION
The purpose of the present study was to cast light on DNA damage-specific DNA-binding proteins in Drosophila melanogaster. In man a number of such gene products have been identified, including DDB (or XPE), a heterodimeric protein composed of 127 and 48 kDa subunits, which has been shown to be essential for DNA excision repair (1). DDB reportedly recognizes many types of DNA lesions (2–7) and is inducible by treatment with DNA-damaging agents (3,8,9). However, DDB was found not to be required in nucleotide excision repair (NER) reconstitution studies in vitro (10–12). The xeroderma pigmentosum group E (XPE) gene product, homologous to DDB, is involved in NER, and the damaged DNA-binding activity of DDB is absent from cells of a subset of XPE patients (13–16). Recently, functions of DDB other than directly in DNA repair have been suggested (17). DNA repair is complex, with multiple overlapping and intersecting pathways with contributions of replication, transcription, meiotic recombination and gene silencing. A large fraction of mutations isolated on the basis of meiotic recombination defects or embryo developmental anomalies are conferred by and predispose to mutagen hypersensitivity.
To generate a deeper comprehension of the nature of DDB, we have chosen D.melanogaster as a suitable model animal. In Drosophila many genes associated with DNA metabolism and morphogenesis are well characterized (18) and mutants can be easily prepared. This factor, coupled with a refined system for genetic analysis, provides a valuable research resource.
In this report, we document characterization of Drosophila DDB1, the larger subunit of DDB, and details of its relation to repair, cell proliferation and development, including spermatogenesis. Interestingly, the gene was found to be controlled by the DRE/DREF system, which is responsible for activating the promoters of nucleus encoded genes for proliferating cell nuclear antigen (PCNA), the 180 and 73 kDa subunits of DNA polymerase α and cyclin A, among others. Our results provide evidence that Drosophila DDB1 acts as a cell proliferation- or development-associated factor as well as a repair factor.
MATERIALS AND METHODS
Separation of Drosophila UV-damaged site-binding proteins
Aliquots of 10 g of Drosophila Kc cells were homogenized in 4 vol of buffer containing 10 mM Tris–HCl, pH 8, 1 mM EDTA and 5 mM dithiothreitol, and then 40 ml of ice-cold buffer (containing 50 mM Tris–HCl, pH 8, 10 mM MgCl2, 2 mM dithiothreitol, 25% sucrose and 50% glycerol) and thereafter neutralized, saturated ammonium sulfate solution (10 ml) was added slowly to the homogenized suspension. After gentle stirring for another 30 min on ice, the homogenate was centrifuged at 50 000 r.p.m. (optima L-70k/70Ti, Beckman) for 3 h at 4°C. The supernatant was dialyzed against the same buffer (TNMD) as that used for UV cross-linking analysis (containing 20 mM Tris–HCl, pH 7.5, 200 mM NaCl, 5 mM MgCl2, 0.5 mM dithiothreitol and three protein inhibitors, 1 µg/ml pepstatin A, 1 µg/ml leupeptin and 1 mM phenylmethylsulfonyl fluoride). The dialysate was loaded onto a UV-irradiated single-strand DNA–cellulose column (2.5 × 5.0 cm φ) equilibrated with TNMD buffer. After washing the column with 150 ml of 200 mM NaCl in TNMD buffer, elution was performed with a three-step gradient, using 50 ml each of 0.5, 1 and 2 M NaCl in the same buffer. Aliquots of 4 ml of each fraction were tested for UV-damaged site-binding activity.
Determination of UV-damaged site-binding polypeptide size by UV cross-linking analysis
UV cross-linking analysis was carried out as described earlier (19) with modifications. Aliquots of 30 ng of oligonucleotide TC31-3′ (5′-AAGCTTTATGCCTGCATCATC) and 15 ng of oligonucleotide TC31 (5′-AATTCGAGCTCGTACGATGA CGATGATGCATCATCGGTCATCGTACGATGACGATGATGCAGGCATAAAGCTT) were mixed in 41.5 µl of a solution containing 33 mM Tris acetate, pH 7.9, 10 mM magnesium acetate, 66 mM potassium acetate and 0.5 mM dithiothreitol and incubated for 3 min at 95°C, followed by 10 min at 25°C. Then the solution was mixed with 8.5 µl of reaction mixture containing 118 µM each dATP, dGTP and dCTP, 1850 KBq [α-32P]dATP and 4 U of Escherichia coli DNA polymerase I large fragment. DNA was uniformly labeled with 32P by incubation at 37°C for 1 h and then chased for 15 min with 10 µM unlabeled dCTP. The 32P-labeled probe (1.75 ng) was incubated with Kc cell nuclear extract (10 µg protein) or 5 µl of each fraction separated on a UV-irradiated single-strand DNA–cellulose column for 15 min on ice, in 17 µl of buffer containing 20 mM Tris–HCl, pH 7.5, 200 mM NaCl, 5 mM MgCl2, 0.5 mM dithiothreitol, 0.2 µg poly(dI–dC). Uncapped Eppendorf tubes containing reaction mixtures were placed at 8 cm distance from a 254 nm UV transilluminator (model UVGL-58; UVP Inc.) and irradiated on ice for 20 min. UV dose under these conditions was 4.19 kJ/m2. Solutions of CaCl2 and MgCl2 were added to final concentrations of 10 and 100 mM, respectively. Digestion by 2 U DNase I and 9 U exonuclease III was carried out at 30°C for 30 min. The reaction was terminated by adding 20 µl of loading buffer containing 100 mM Tris–HCl, pH 6.8, 4% SDS, 0.2% bromophenol blue, 20% (v/v) glycerol and 0.2 M dithiothreitol. The samples were heated and applied to 10% polyacrylamide gels containing 0.1% SDS. After electrophoresis, the gels were stained with Coomassie Brilliant Blue, photographed, dried and autoradiographed. The molecular weights of the protein bands were estimated by comparing their mobilities with those of marker proteins (Bio-Rad).
Determination of UV-damaged site-binding polypeptides by electrophoretic mobility shift assay using native gels
The methods used were basically the same as described in an earlier study by Todo et al. (20). The binding reaction mixture was the same as for the UV cross-linking analysis. The mixture was incubated for 20 min on ice. It was then electrophoresed in a 6% polyacrylamide gel in a buffer containing 6.7 mM Tris–borate pH 7.5, and 1 mM EDTA (TBE buffer) at 50 V for 3–4 h, after which it was dried and then used to expose Fuji RX medical X-ray film. The gels were also exposed to X-ray films for 6 h.
Production of recombinant D-DDB1
The D-DDB1 fragment (amino acids 592–1140) was expressed as an N-terminal His-tagged fusion protein using the pET-28b (Invitrogen) vector in E.coli BLR21 (DE3). The recombinant fragments were purified by chromatography through His-bind resin. Purified recombinant D-DDB1 (10 µg) fragment was separated on a 12.5% SDS–polyacrylamide gel and the recombinant fragment was visualized using Coomassie Brilliant Blue staining.
Analysis of DNA sequences
DNA sequencing was performed by the dideoxy chain termination method with a sequencing kit and DNA sequencer from Applied Biosystems. DNA sequence analysis was performed using Gentetix Mac v.10 (Software Development Co. Ltd).
Production of polyclonal antibody and western blot analysis
The purified recombinant D-DDB1 fragment was used for immunization of a mouse. Polyclonal antibodies reacting with D-DDB1 were affinity purified by D-DDB1 protein- conjugated Sepharose column chromatography. Western blotting analysis was carried out by the method of Towbin et al. (21). Anti-mouse IgG conjugated with alkaline phosphatase (Promega) was used as the secondary antibody. Color was developed with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate as the substrates of alkaline phosphatase.
Northern hybridization
Aliquots of 30 µg of total RNA extracted from D.melanogaster bodies at several different stages and from Kc cells, either incubated for 0–10 h after irradiation with 70 J/m2 UV or for 0–7 h after addition of 0.1% methyl methanesulfonate (MMS), were resolved on 1.2% formaldehyde–agarose gels and transferred to nylon membranes (Hybond-N+; Amersham). After prehybridization, the filters were probed with 32P-labeled d-ddb1 cDNA (nucleotides 1774–3423) or with 32P-labeled full-length ribosomal protein 49 (Rp-49) cDNA as a control at 42°C for 16 h, followed by washing twice with 2× SSPE + 1% SDS at room temperature for 15 min and twice with 1× SSPE + 0.1% SDS at 50°C for 20 min. Blots were exposed to Kodak X-Omat XAR films and quantified with a NIH imaging analyzer.
Double-stranded RNA production
DNA fragments 600 bp in length (nucleotides 1–600), containing the coding sequences for D-DDB1 to be ‘knocked out’ were amplified using PCR. Each primer contained a T7 RNA polymerase-binding site (GAATTAATACGACTC ACTATAGGGAGA) followed by sequences specific for the targeted genes. The PCR products were purified using a High Pure PCR purification kit (Roche Molecular Biochemicals) and employed as templates with a Megascript T7 transcription kit (Ambion, Austin, TX) to produce double-stranded (ds)RNA. The dsRNA products were ethanol precipitated and resuspended in water and annealed by incubation at 65°C for 30 min followed by slow cooling to room temperature. Aliquots of 6 µg were analyzed by 1% agarose gel electrophoresis to ensure that the majority of the dsRNA existed as a single band of ∼600 bp. The remainder was stored at –20°C.
Conditions for RNA interference (RNAi) in Drosophila cell culture
RNAi was carried out by the method of Clemens et al. (22). Drosophila Kc cells were diluted to a final concentration of 1 × 106 cells/ml in serum-free M3 medium (Sigma) and 1 ml aliquots were plated per well of 6-well cell culture dishes (Corning). dsRNA was added directly to the medium to a final concentration of 2.5 or 5 µg/ml. For dsRNAs of ∼600 nt, this corresponds to 1 mg dsRNA. This was followed immediately by vigorous agitation. The cells were incubated for 30 min at room temperature followed by addition of 2 ml of M3 medium containing fetal calf serum. The cells were incubated for an additional 3 days to allow for turnover of the target protein.
Immunostaining of Kc cells
Kc cultured cells derived from D.melanogaster embryos were grown at 25°C in M3 medium supplemented with 2% fetal calf serum. They were incubated for 1 h with anti-D-DDB1 IgG, diluted 1:50 in phosphate-buffered saline (PBS) containing 1% BSA, then treated for 1 h with Alexa594 anti-mouse IgG (Sigma), diluted to 1:100, as secondary antibody. The cells were also stained with a solution of 20 µg/l DAPI for 5 min, examined under a fluorescence microscope (Olympus BX-50) (×1600) and photographed using Tri X Pan 400 film (Kodak).
Monoclonal antibodies
Monoclonal antibodies to DREF, Mab-1 and Mab-4, were raised as described previously (23).
Electrophoretic mobility shift assays and preparation of nuclear extracts
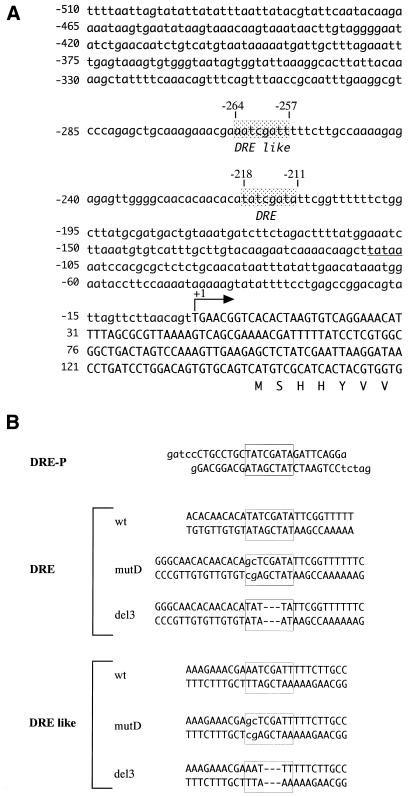
The sequences of double-stranded oligonucleotides containing DREF-binding sites of the PCNA gene promoter (DRE-P) and of their derivatives in the d-ddb1 promoter were as follows.
DRE-P,
gatccCTGCCTGCTATCGATAGATTCAGGa
GGACGGACGATAGCTATCTAAGTCCtctag
DRE wt,
ACACAACACATATCGATATTCGGTTTTT
TGTGTTGTGTATAGCTATAAGCCAAAAA
DRE mutD,
GGGCAACACAACACAgcTCGATATTCGGTTTTTTC
CCCGTTGTGTTGTGTcgAGCTATAAGCCAAAAAAG
DRE del3,
GGGCAACACAACACATAT---TATTCGGTTTTTTC
CCCGTTGTGTTGTGTATA---ATAAGCCAAAAAAG
DRElike-wt,
AAAGAAACGAAATCGATTTTTCTTGCC
TTTCTTTGCTTTAGCTAAAAAGAACGG
DRElike-mutD,
AAAGAAACGAgcTCGATTTTTCTTGCC
TTTCTTTGCTcgAGCTAAAAAGAACGG
DRElike-del3,
AAAGAAACGAAAT---TTTTTCTTGCC
TTTCTTTGCTTTA---AAAAAGAACGG
Nucleotides deleted from or inserted into the wild-type sequence are shown by lower-case letters with underlining. 32P-labeled probes (10 000 c.p.m.) were incubated in 16 µl of reaction mixture containing 25 mM HEPES pH 7.9, 100 mM NaCl, 1 mM EDTA, 10% (v/v) glycerol, 0.1% Tween-80, 0.02% 2-mercaptoethanol, 0.5 µg poly(dI–dC) and 0.5 µg salmon sperm DNA on ice for 5 min. When necessary, double-stranded oligonucleotides were added as competitors at this step. Then, Kc cell nuclear extract was introduced and the reaction mixture was incubated for 20 min on ice. In experiments with antibodies, Kc cell nuclear extracts were preincubated with these for 1 h on ice. DNA–protein complexes were electrophoretically resolved on 4% polyacrylamide gels in 100 mM Tris–borate buffer, pH 8.3, 2 mM EDTA containing 2.5% (v/v) glycerol at 25°C. The gels were dried and then autoradiographed.
DNA transfection and luciferase assay
Drosophila S2 cells (2 × 106 per dish) were grown in 60-mm plastic dishes for 24 h and transfected with 500 ng reporter plasmid DNA and 1 ng pRL-actin5C DNA using CellFectin reagent (Gibco BRL). Cells were harvested 48 h thereafter. The luciferase assay was carried out by means of a Dual-Luciferase Reporter Assay System (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity. To construct the plasmids p5′-510DDBluc(wt), p5′-510DDBwt-mutDluc(wt-mutD), p5′-510DDBwt-del3luc(wt-del3), p5′-510DDBmutD-wtluc(mutD-wt), p5′-510DDBdel3-wtluc(del3-wt), p5′-510DDBmutD-mutDluc (mutD-mutD), p5′-510DDBmutD-del3luc(mutD-del3), p5′-510DDBdel3-mutDluc(del3-mutD), p5′-510DDBdel3-del3luc(del3-del3) and p5′-210DDBluc(-DRE) for the luciferase transient expression assay, a DNA fragment containing the d-ddb1 gene fragment was isolated and inserted into plasmid PGVB. The mutation was introduced using Mutan(R)-Super Express™ (Takara). Plasmid pRL-actin5C containing the actin 5C gene promoter was used as an internal control in the luciferase transient expression assay.
Fixation and immunocytochemical staining of larval tissues
The imaginal discs were dissected from larvae, then fixed with Tris-buffered saline (TBS) solution (50 mM Tris–HCl, pH 8.3, and 150 mM NaCl) containing 3.5% formaldehyde. Tissues were blocked with TBS solution containing 10% goat serum and 0.15% Triton X-100 (TBT). Incubation with primary antibodies was carried out in the same solution for 16 h at 4°C. Samples were washed extensively in TBS containing 0.3% Triton X-100, reblocked with TBT and incubated with alkaline phosphatase-conjugated secondary antibody for western immunoblotting analysis. Stained samples were mounted in a solution containing glycerol. Samples were examined under a microscope (Olympus BX-50) and photographed using Tri X Pan 100 film (Kodak).
In situ immunostaining of Drosophila adult bodies
Paraffin-embedded, paraformaldehyde-fixed tissue specimens were used for in situ immunostaining to detect D-DDB1 proteins as described (24). Samples were examined under a microscope (Olympus BX-50) and photographed using Tri X Pan 100 film (Kodak).
Fixation and staining of testes
Adult testes were dissected and fixed according to Pisano et al. (25). Fixed preparations were washed twice (10 min each) in PBS, 0.1% Tween-20, and once for 5 min in PBS, before incubation with the antibody. The purified recombinant D-RPA30 protein expressed as an N-terminal His-tagged fusion protein using the pET-28b vector (Invitrogen) in E.coli BLR21 (DE3) was employed for immunization of a rabbit. Polyclonal antibodies reacting with D-RPA30 were affinity purified on a D-RPA30 protein-conjugated Sepharose column. Slides were incubated for 1 h with 40 µl of anti D-DDB1 and anti-RPA30 antibodies diluted 1:100 in PBS in a humid chamber at room temperature. They were then washed twice in PBS (5 min each) and incubated for 1 h with the secondary antibody (Alexa488 anti-mouse IgG or Alexa594 anti-rabbit IgG) diluted 1:100 in PBS. After two washes in PBS (5 min each) the immunostained slides were stained with a solution of 20 µg/l DAPI for 5 min, examined under a microscope (Olympus BX-50) and photographed using Tri X Pan 400 film (Kodak).
RESULTS
Characterization of UV-damaged DNA-binding proteins in Kc cells in D.melanogaster
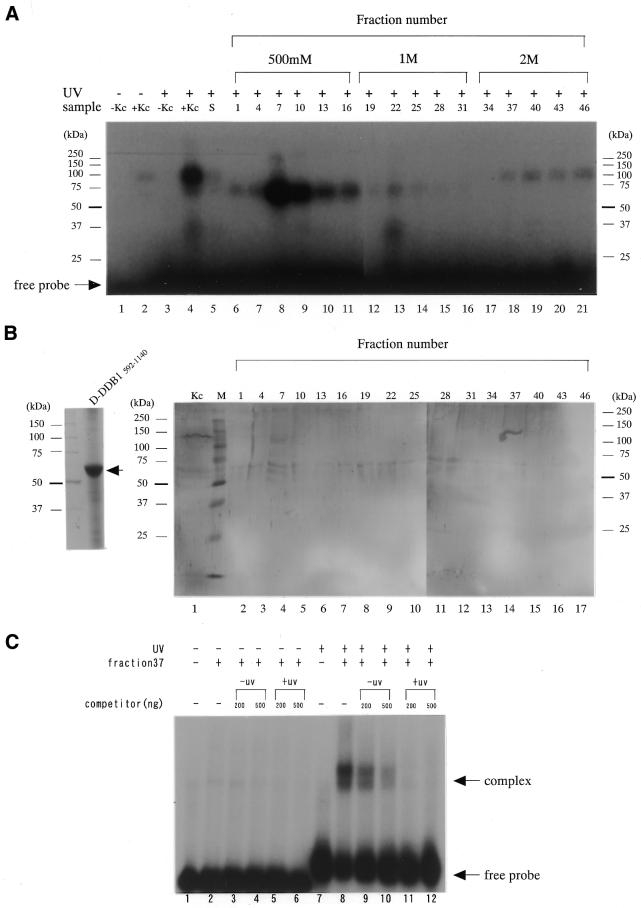
To screen for UV-damaged DNA-binding proteins that might play a role in DNA repair-specific events, crude extracts were generated from cultured D.melanogaster Kc cells and first characterized by UV-damaged DNA–cellulose column chromatography. Then they were subjected to a DNA cross-linking assay for UV-damaged DNA-binding protein activity. Figure 1A shows UV-damaged DNA–cellulose elution patterns of proteins in D.melanogaster Kc cells. The components were separated into at least three peaks in the column at 0.5, 1 and 2 M NaCl. The major fraction eluted at 0.5 M NaCl appeared to be a CPD or (6–4) photolyase with a mol. wt of ∼60 kDa, as reported by Todo et al. (20) (lanes 8–11), and the 30 kDa protein eluted at 1 M NaCl might be a UV-damage-specific endonuclease 1 (D-DDB P1), as we reported previously (26,27) (lane 13). Here, we have concentrated our attention on the 126 kDa protein fraction eluted at 2 M NaCl, because it demonstrated the highest affinity for UV-damaged DNA in D.melanogaster. The 126 kDa protein in fractions 37–46 of the column was collected, purified by SDS–PAGE and used to raise rabbit antiserum against the polypeptide. The antiserum recognized multiple polypeptides, including the 126 kDa protein, in Kc cell extracts.
Figure 1.
(A) UV cross-linking analysis of UV-damaged site-binding polypeptides. Manley’s whole cell extract from Kc cells (10 µg protein) was incubated for 15 min with a UV-unirradiated 32P-labeled probe (lanes 1 and 2) or UV-irradiated 32P-labeled probe (lanes 3 and 4) in a solution containing 20 mM Tris, pH 7.5, 200 mM NaCl, 5 mM MgCl2, 0.5 mM dithiothreitol, 5% (v/v) glycerol, 2 µg poly (dI–dC). Similarly, ∼930 ng protein from Kc cells, separated on a UV-irradiated single-strand DNA–cellulose column, was incubated for 15 min with a UV-irradiated 32P-labeled probe (lanes 5–21). After UV irradiation, followed by digestion with DNase I and exonuclease III, the reaction products were electrophoresed on a 12.5% polyacrylamide gel containing 0.1% SDS. Migration positions of marker ptoteins are indicated. (B) SDS–PAGE analysis of the purified recombinant D-DDB1 fragment. A part of D-DDB1 protein (amino acids 592–1140) was expressed in E.coli BLR21 (DE3) as an N-terminal His-tagged fusion protein obtained using pET-28b (Invitrogen) vector and purified for preparation of an antibody for use in western blotting and immunostaining (left). Western blot analysis revealed that the anti-D-DDB1 antibody specifically recognized the 126 kDa D-DDB1 protein from Kc cells (right panel, lane 1). Lanes 2–17 contained the same fraction separated with the UV-irradiated single-strand DNA–cellulose column indicated in (A). Fraction 37 contained D-DDB1. The results indicated the UV cross-linking band detected in the 2 M eluted fraction [(A) lanes 18–21] to be D-DDB1. (C) UV-irradiated DNA-specific binding activity was detected by EMSA. UV-irradiated or unirradiated TC31 oligonucleotides were radiolabeled and used as a probe in EMSA using fraction 37, containing the D-DDB. Fraction 37 was added to the reaction mixture (lanes 2–6 and 8–12) with increasing amounts of unlabeled unirradiated TC31 (lanes 3, 4, 9 and 10) or inducing amounts of unlabeled irradiated TC31 (lanes 5, 6, 11 and 12). The higher order band corresponds to two independent binding events on the same probe.
Expression screening of a Kc cell cDNA library using the antiserum resulted in isolation of six clones containing partial DNA sequences with high homology to human DDB1 (p127) (28). We then carried out a detailed analysis of the 126 kDa protein homologous to human DDB1, provisionally designated Drosophila damage-specific DNA-binding protein 1 (D-DDB1). The nucleotide sequence data have been lodged in the DDBJ/EMBL/GenBank nucleotide sequence databases with accession no. AF132145. The cDNA of d-ddb1, obtained by RT–PCR using specific primers, was characterized as follows. The open reading frame predicted a protein of 1140 amino acid residues, with a molecular weight of 126 038. Overall similarity of the sequence to the human and rat DDB1 sequences was 49.8 and 49.4%, respectively (28). Figure 1B shows a western blot of the gel pattern of Figure 1A using the anti-DDB1 antibody generated against D-DDB1 recombinant fragment (see left panel). The 126 kDa protein indicated to be D-DDB1, and the molecular mass was 126 kDa (Fig. 1B, lane 1). Fraction 37 in Figure 1A was tested by SDS–PAGE and confirmed to have another small subunit of ∼50 kDa (Drosophila DDB2) (data not shown). Figure 1C shows the results of a native gel electrophoretic mobility shift assay using fraction 37 (containing D-DDB1) in Figure 1A. After UV-irradiation, shifted protein bands occurred in the native gel (Fig. 1C, lane 8). When 32P-labeled unirradiated TC31 was used, the shifted protein bands did not occur (Fig. 1C, lanes 1–6). Shifted bands were detected when 32P-labeled irradiated TC31 was used, which were greatly reduced by addition of increasing amounts of unlabeled irradiated TC31 (Fig. 1C, lanes 11 and 12). Although two bands were observed, the same result was found for human DDB (29).
Influence of UV-irradiation and MMS on expression of the d-ddb1 gene and results of knocking out the gene by RNAi
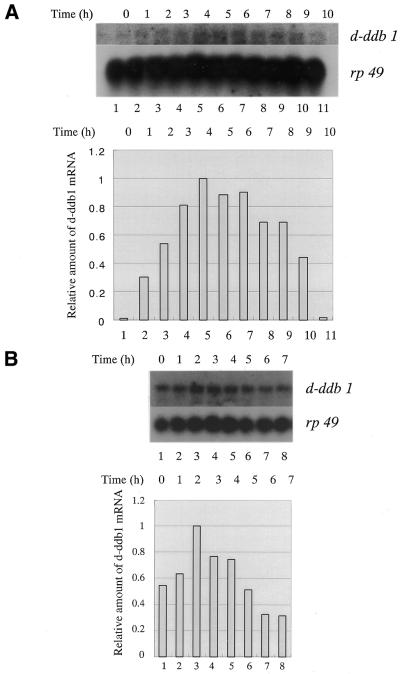
We measured levels of mRNA for the d-ddb1 gene in Kc cells after UV-irradiation. Since >90% of (6–4) photoproducts in UV-damaged nuclear DNA could be repaired within 3 h (30), and since DDB binds to (6–4) photoproducts, we checked the amount of mRNA every hour after UV-irradiation at 70 J/m2. The amount of mRNA increased after UV-irradiation, reached a maximum within 4 h, and then gradually reduced, indicating the transcripts to be UV-inducible (Fig. 2A). Transcripts of d-ddb1 were also induced by 0.1% MMS treatment, although they reached a maximum more quickly (2 h) (Fig. 2B).
Figure 2.
(A) Northern hybridization analysis of total RNA from Kc cells. Cells were incubated for 1–10 h after irradiation at 70 J/m2, separated on a 1.2% agarose–formaldehyde gel, transferred to a nylon membrane and probed with random primed 32P-labeled d-ddb1 cDNA (1774–3420 bp). The blot was reprobed for the rp49 message as a loading control. (Lower panel) Amounts of d-ddb1 mRNA as determined using an imaging analyzer and expressed relative to the highest amount of transcripts 4 h after irradiation. (B) Northern hybridization analysis of total RNA from Kc cells. Cells were incubated for 0–10 h after addition of 0.1% MMS, separated on a 1.2% agarose–formaldehyde gel, transferred to a nylon membrane and probed with a random primed 32P-labeled d-ddb1 cDNA (1774–3420 bp). The blot was reprobed for the rp49 message as a loading control. (Lower panel) Amounts of d-ddb1 mRNA as determined using an imaging analyzer and expressed relative to the highest amount of transcripts 2 h after addition of MMS.
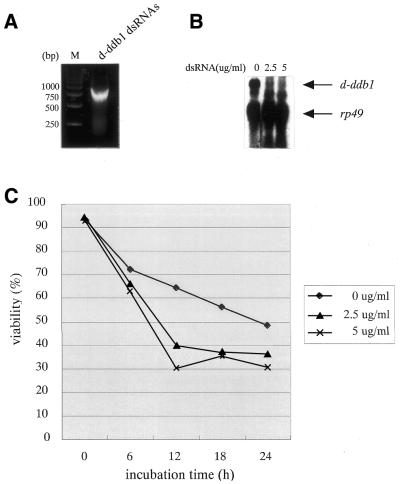
Figure 3 illustrates the effects of UV-irradiation on Kc cells in which the d-ddb1 gene was knocked out by the RNAi method using 600 bp dsRNA (Fig. 3A). As shown in Figure 3B, the transcripts of d-ddb1 were significantly reduced by the RNAi treatment at 2.5 and 5.0 µg/ml compared with same amount of rp49 transcript. The signals between d-ddb1 and rp49 are non-specific bands, because the subjacent band was only detected in Kc cells (see Fig. 5, lane 14). In Figure 3C, data for viability of cells every 6 h after UV-irradiation at 70 J/m2 are presented. The cells were clearly more sensitive to UV after gene knockout. To our knowledge, these results represent the first such description, although there is a report that a mutation in human DDB2 makes the cells more UV-sensitive (17).
Figure 3.
RNAi in Drosophila Kc cells. (A) The 600 bp d-ddb1 dsRNAs are indicated. (B) Northern hybridization analysis of total RNA from Kc cells prepared 3 days after addition of 0, 2.5 or 5.0 µg d-ddb1 dsRNAs is illustrated on the right. Total RNA was separated on a 1.2% agarose– formaldehyde gel, transferred to a nylon membrane and probed with a random primed 32P-labeled d-ddb1 cDNA (1774–3420 bp). (C) Kc cells were incubated for 0–24 h with the indicated concentrations of d-ddb1 dsRNAs (1–600 nt). Cells were prepared after 3 days and tested for survival after UV-irradiation at 70 J/m2.
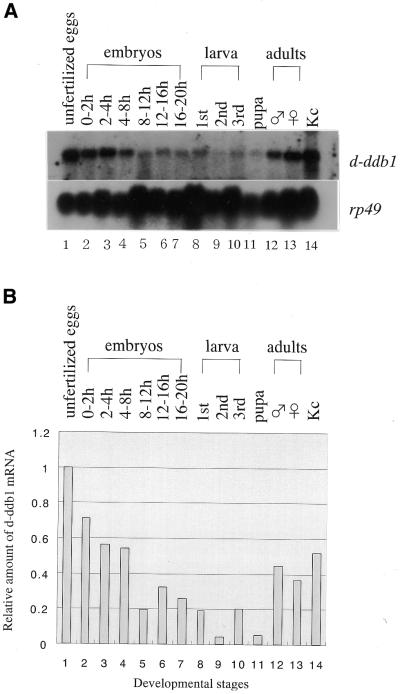
Figure 5.
Northern hybridization analysis of total RNA from D.melanogaster at various developmental stages. (A) An aliquot of 30 µg total RNA was applied to each lane. Radiolabeled probes were used successively on the same membrane. rp-49 mRNA served as a loading control. (B) Relative amounts of d-ddb1 mRNA (1774–3420 bp), normalized to rp-49 mRNA and expressed relative to the value for transcripts in unfertilized eggs.
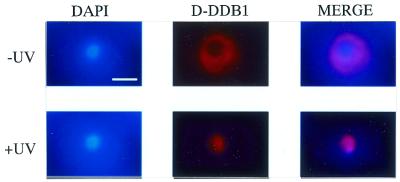
We also tested the effects of UV-irradiation on the cellular localization of D-DDB1 protein as a function of time by immunostaining with the anti-D-DDB1 antibody. As shown in Figure 4, after UV irradiation at 70 J/m2, nuclear accumulation of D-DDB1 protein in Kc cells was found in 8% of all the cells examined after 12 h, although no accumulation was observed in unirradiated cells. The protein seemed to be moved into the nucleus from the cytoplasm.
Figure 4.
Subcellular localization of D-DDB1 in Drosophila Kc cells after UV-irradiation (×1600). Bar indicates 5 µm. Kc cells were stained with DAPI (left) and antibodies against D-DDB1 (middle). Merged images are shown on the right. (Upper panels) Without UV irradiation; (lower panels) with incubation for 12 h after 70 J/m2 UV-irradiation. Note the nuclear localization of D-DDB1 after UV-irradiation.
Changes in expression of the d-ddb1 gene during Drosophila development and its regulation by the DRE/DREF system
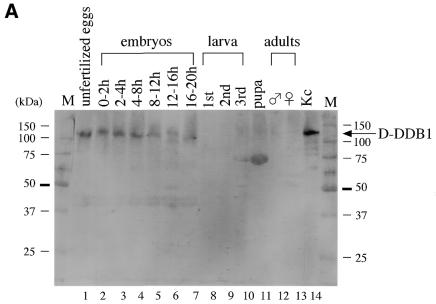
As described briefly in the Introduction, DDB is not only reported to recognize many types of DNA lesions, but might also have a role in other processes. To investigate its relationship to Drosophila development, northern hybridization experiments were performed using the d-ddb1 cDNA as a probe and total RNA extracted from Drosophila at various developmental stages: unfertilized eggs, 0–2, 2–4, 4–8, 8–12, 12–16 and 16–20 h embryos, first, second and third instar larvae, pupae, adult males and females, and Kc cells (Fig. 5). A single hybridization band of ∼4 kb was detected throughout all stages, the size corresponding to that of the full-length cDNA. The amount of d-ddb1 mRNA changed considerably during development. Although UV-irradiation was not performed, the highest level of mRNA was detected in unfertilized eggs, then early embryonic stages from 0 to 8 h, adults and Kc cells (Fig. 5). Low levels were also detected in the 8–20 h embryos, larvae and pupae (Fig. 5). The expres sion pattern was similar to those for the genes for DNA polymerases α and ε and PCNA (31,32).
Since this suggested a link with cell proliferation, we searched for transcription factor-binding sequences in the d-ddb1 gene and, interestingly, found a DRE site (5′-TATCGATA) and a DRE-like site (5′-AATCGATT, which matched 6 of the 8 bp DRE consensus sequence) in the upper cascade, respectively located at positions –218 and –264 with respect to the transcription initiation site of the d-ddb1 gene (Fig. 6A). Therefore, we next tested whether gene expression is regulated by the DRE/DREF system.
Figure 6.
(A) Upstream genomic sequence of the d-ddb1 gene. The bent arrow indicates the transcription initiation site. The boxes enclose the DRE sequence (5′-TATCGATA), located at position –218, and DRE-like sequence (5′-AATCGATT), located at position –264 with respect to the transcription initiation site. The putative TATA box is underlined. (B) Sequences of double-stranded oligonucleotides containing a DRE (DRE-P) in the PCNA promotor and double-stranded oligonucleotides containing DREF-binding sites or their derivatives in the d-ddb1 promoter.
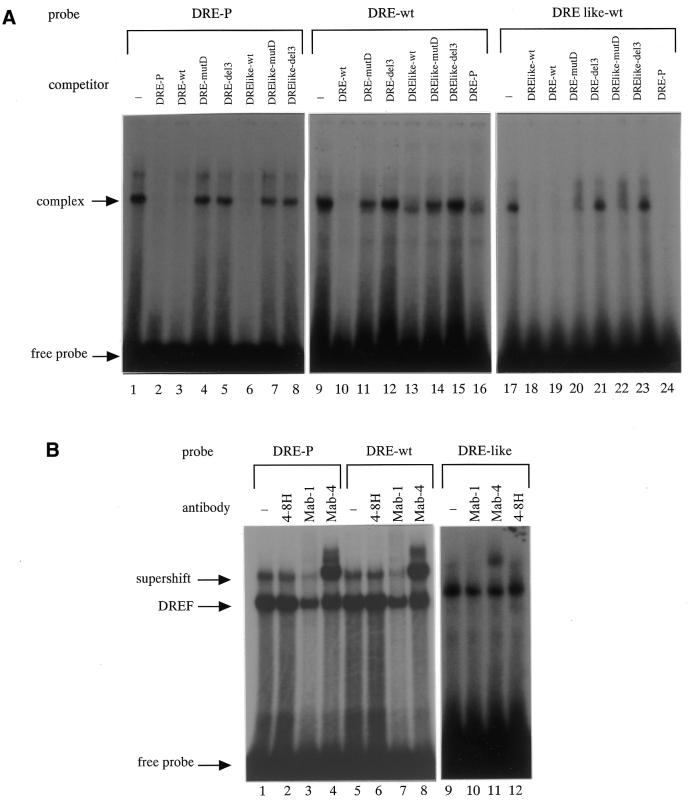
As shown in Figure 7, the 32P-labeled DRE-P, DRE-wt or DRE like-wt oligonucleotides shown in Figure 6B were incubated with or without Kc cell nuclear extract and several competitor complex bands were detected. The 32P-labeled DRE-P probe was used as a positive control and when non-labeled DRE-P, DRE-wt and DRE like-wt were added as competitors the complex band was reduced (Fig. 7A, lanes 2, 3 and 6). Similarly, 32P-labeled DRE-wt or DRE like-wt probes competed with unlabeled DRE-P, DRE-wt and DRE like-wt (Fig. 7A, lanes 10, 13, 16, 18, 19 and 24). A probe mutated in the DRE site did not compete or only very weakly with DRE-wt, DRE like-wt and DRE-P (Fig. 7A, lanes 4, 5, 7, 8, 11, 12, 14, 15 and 20–23). Next, to confirm that the complex band was the DRE/DREF complex, we examined the effects of anti-DREF monoclonal antibodies (Mab-1 and Mab-4) on the binding reaction with Kc cell nuclear extract. Mab-1 inhibited binding of DREF to DRE-P, and Mab-4 supershifted the band with DRE-P (23,33) (Fig. 7B, lanes 1–4). As shown in Figure 7B, one of the shifted bands with the DRE-wt and DRE like-wt probes was diminished by addition of Mab-1 (lanes 7 and 10) and was supershifted by Mab-4 (lanes 8 and 11).
Figure 7.
(A) Complex formation between the probe (DRE-P, DRE-wt or DRE like-wt) and the Kc cell nuclear extract. A radiolabeled double-stranded oligonucleotide of DRE-P (lanes 1–8), DRE-wt (lanes 9–16) or DRE like-wt (lanes 17–24) was incubated with the Kc cell nuclear extract (4 µg protein) in the presence or absence of competitor oligonucleotides (DRE-wt, DRE-mutD, DRE-del3, DRElike-wt, DRElike-mutD, DRElike-del3 or DRE-P, 100 ng/lane). (B) DREF bound to oligonucleotides containing the DRE-wt and DRE like-wt sequences. 32P-labeled DRE-P (lanes 1–4), DRE-wt (lanes 5–8) or DRE like-wt (lanes 9–12) were incubated with Kc cell nuclear extract in the presence of anti-DREF monoclonal antibody (Mab-1) (lanes 3, 7 and 10), anti-DREF monoclonal antibody (Mab-4) (lanes 4, 8 and 11), anti-DNA polymerase α monoclonal antibody (4-8H) as a negative control (lanes 2, 6 and 12) or in the absence of antibody (lanes 1, 5 and 9).
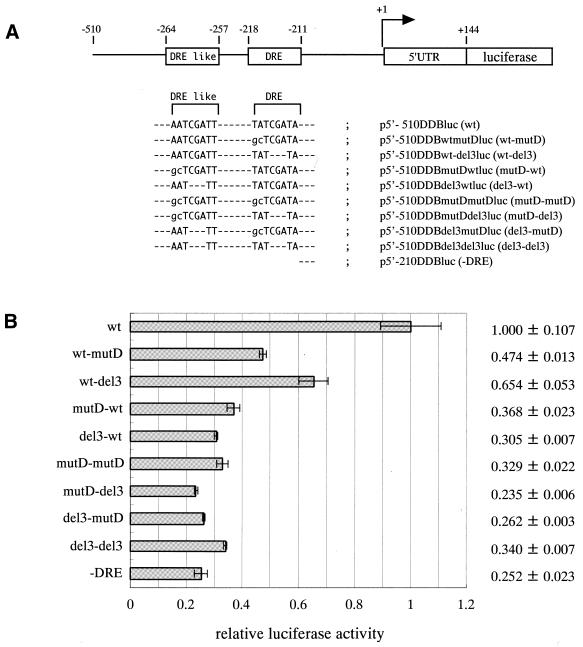
For further confirmation of the stimulatory effects of the two DREF recognition sequences on the promoter, we prepared luciferase expression constructs having the wild-type DRE-wt and DRE like-wt with several mutations in the DRE and DRE-like sequences ligated with the the luciferase gene in a PicaGene basic vector (Fig. 8A). The plasmids carrying these constructs were then transfected into Drosophila S2 cells and luciferase expression levels were determined. As shown in Figure 8B, mutation in any of the two DREF-binding sequences caused a reduction in luciferase expression levels. The two DREF-binding sequences appeared to cooperate to provide high promoter activity.
Figure 8.
(A) Constructs of d-ddb1–luciferase fusion genes [p5′-510DDBluc (wt), p5′-510DDBwt-mutDluc (wt-mutD), p5′-510DDBwt-del3luc (wt-del3), p5′-510DDBmutD-wtluc (mutD-wt), p5′-510DDBdel3-wtluc (del3-wt), p5′-510DDBmutD-mutDluc (mutD-mutD), p5′-510DDBmutDdel3luc (mutD-del3), p5′-510DDBdel3-mutDluc (del3-mutD), p5′-510DDBdel3-del3luc (del3-del3) and p5′-210DDBluc (-DRE)] are shown. (B) Effects of mutations in DRE and DRE-like sites on d-ddb1 gene promotor activety in Drosophila S2 cells, assessed with 500 ng aliquots of luciferase plasmids harboring wild-type or mutant d-ddb1 promotors (indicated on the left). p5′-510DDBluc (wt), p5′-510DDBmutDluc (mutD), p5′-510DDBdel3luc (del3), p5′-510DDBmutDmutDluc (mutD-mutD), p5′-510DDBmutDdel3luc (mutD-del3) and p5′-210DDBluc (-DRE) were transfected into S2 cells. Cell extracts were prepared to determine the luciferase expression levels. Averaged values obtained from two independent dishes with standard deviations are shown by closed bars as luciferase activity relative to those of p5′-510DDBluc (wt).
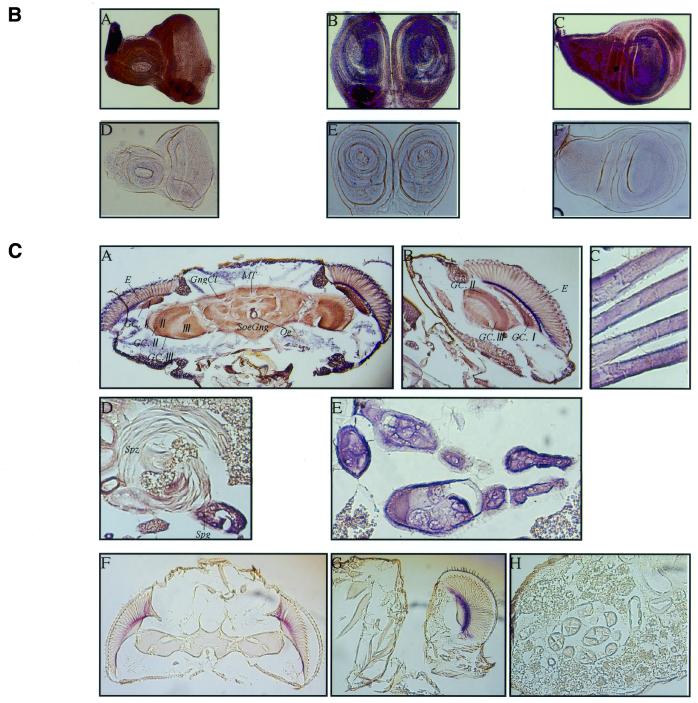
For further investigation, an antibody against D-DDB1 was used for western blot analysis with extracts from Drosophila bodies at various developmental stages. As shown in Figure 9A, the 126 kDa subunit level decreased throughout embryogenesis; D-DDB1 levels were high at early stages when the rate of DNA replication was maximal and decreased as development proceeded (Fig. 9A). Although no or a very slight signal for D-DDB1 protein was detected in extracts from larval, pupal and adult whole bodies, probably due to dilution of D-DDB1 with large amounts of other proteins, a specific signal was detected from the brain lobes and imaginal discs dissected from third instar larvae, testes and ovaries (data not shown). Figure 9B shows imaginal discs dissected from third instar larvae and immunostained with antibody against D-DDB1. Anti-D-DDB1 antibody stained the cells of imaginal discs (A–C). No significant staining signal was observed with normal rabbit IgG as the primary antibody (D–F). In Figure 9C, D-DDB1 protein is evident in ganglion cells in the brain (A, B), flight muscles (C), testis (D) and ovaries (E).
Figure 9.
(Opposite) (A) D-DDB1 protein levels in D.melanogaster at various developmental stages and in Kc cells. Aliquots of 25 µg protein from Drosophila bodies at various developmental stages were separated by 10% SDS–PAGE. The anti-D-DDB1 antibody reacted with one polypeptide band at 126 kDa. (B) Immunochemical localization of D-DDB1 in the imaginal discs of third instar larvae. The imaginal discs were dissected from third instar larvae and fixed. Polyclonal anti-D-DDB1 IgG (A–C) and normal rabbit IgG (D and E) were used as primary antibodies as indicated. Alkaline phosphatase-conjugated goat anti-mouse or anti-rabbit IgG was used as the secondary antibody. (A and D) Eye discs; (B and E) leg discs; (C and F) wing discs. (C) Expression of the D-DDB1 protein was immunohistochemically detected in Drosophila tissues at the adult stage. (A and F) Head frontal sections; (B and G) head median sections; (C) flight muscle sagittal section; (D) testis sagittal section; (E and H) ovary frontal and sagittal sections; (F–H) negative control stained with normal rabbit IgG. E, compound eye; GC.I, GC.II, GC.III, ganglion cells of periopticon, epiopticon and opticon; GngCl, ganglion cells (neurocytes); I, periopticon; II, epiopticon; III, opticon; MT, median fiber bundle; Oe, oesophagus; SoeGng, suboesophageal ganglion; Spg, spermatogonium; Spz, sperm.
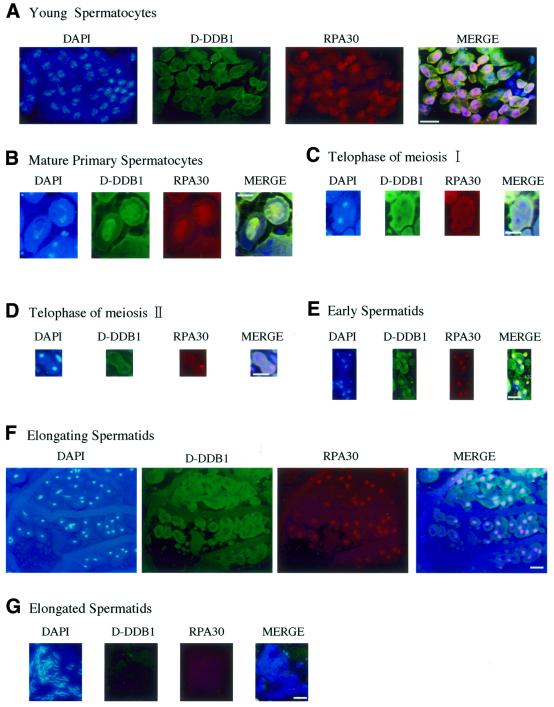
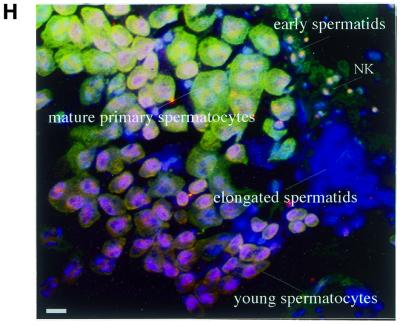
Pursuing the roles of D-DDB1 in relation to development, testis immunostaining was conducted because transcripts of d-ddb1 were found to be abundant in adult male flies (Fig. 5B, lane 12) and D-DDB1 protein was detected in the testis (Fig. 9C). Interestingly, D-DDB1 appeared to be transiently present in meiotic cells during spermatogenesis (Fig. 10). We applied double staining with D-DDB1 and RPA30, because there is a report that DDB mediates NER activity (34) and because RPA30 is known to be an indispensable factor for replication. Triple labeling (D-DDB1 in green, DNA in blue and RPA30 in red) gave, in merged views: green, D-DDB1; blue, DNA; red, RPA30; yellow, D-DDB1 and RPA30. In young spermatocytes D-DDB1 accumulated in the cytoplasm, although RPA30 was present in the nucleus (Fig. 10A). In mature primary spermatocytes both D-DDB1 and RPA30 accumulated in the nucleus (Fig. 10B). At the telophase stage of meioses I and II both D-DDB1 and RPA30 were found in the cytoplasm (Fig. 10C and D). In early spermatids the typical features included a spherical mitochondrial Nebenkern (NK) lacking both D-DDB1 and RPA30 in the NK, but both accumulated in the nucleus (Fig. 10E). In elongating spermatids D-DDB1 disappeared from the nucleus, although RPA30 was present (Fig. 10F). In fully elongated spermatids neither D-DDB1 nor RPA30 was detected. Since at this stage chromatin condensation becomes tighter due to protamine, D-DDB1 and RPA might be removed (Fig. 10G). Thus, during spermatogenesis, D-DDB1 is first located in the cytoplasm, then accumulates in the nucleus of the mature primary spermatocyte, and finally disappears (Fig. 10H). In the nucleus positive stages (6–4) photoproducts were not detected using a specific monoclonal antibody.
Figure 10.
(Opposite) D-DDB1 organization during D.melanogaster spermatogenesis. Microscopic imaging of a testis triple labeled with D-DDB1 in green and DNA in blue and RPA30 in red. The merged view (DNA and D-DDB1 and RPA30) is shown: green, D-DDB1; blue, DNA; red, RPA30; yellow, D-DDB1 and RPA30. (A) In young spermatocytes D-DDB1 accumulates in the cytoplasm, although RPA30 is present in the nucleus. (B) In mature primary spermatocytes D-DDB1 and RPA30 are both in the nucleus. (C and D) At the telophase stage of meioses I and II D-DDB1 and RPA30 are both in the cytoplasm. (E) In early spermatids typical features include a spherical mitochondrial Nebenkern (NK) lacking D-DDB1 and RPA30. (F) In elongating spermatids D-DDB1 is outside the nucleus, although RPA30 is present within. (G) In fully elongated spermatids D-DDB1 and RPA30 are undetectable. (H) The merged overview. Bars, 10 µm.
DISCUSSION
Damage-specific DNA-binding protein (DDB) has been implicated in numerous aspects of DNA repair (10–12). To elucidate precise functions we have focused on DDB in an animal model, using D.melanogaster. We thereby succeeded in isolating four UV-irradiated DNA-specific binding proteins from Kc cells and established one of them to be a Drosophila homolog of the p127 subunit of DDB. We then cloned the d-ddb1 gene. D-DDB1 appeared to be a protein that recognizes UV-damaged DNA in vivo.
It has been reported that CREB-binding protein and p300 histone acetyltransferase can interact with the small subunit of DDB (the DDB2 or p48 subunit) (35), while the human STAGA complex (SPT3–TAFII31–GCN5L acetylase) interacts with the large subunit of DDB (the DDB1 or p127 subunit) (36). DDB might have functions as a repair protein factor in the context of chromatin structure for recruitment of NER factors to DNA damage sites, because it physically interacts with histone acetyltransferase subunits, believed to be important in altering chromatin structure. Transcription of ddb2, but not ddb1, is strongly activated by p53 tumor suppressor protein in human cells (1). However, the response of DDB to DNA damage remains to be clarified. We investigated D-DDB1, a homolog of DDB1, and found it to be directly related to the Drosophila DNA repair system. This was confirmed in cells knocked out for the d-ddb1 gene by the RNAi method. To our knowledge there has been no report of ddb1-deficient cells, although lack of ddb2 has been studied (17). D-DDB1 was detected in the cytoplasm of Kc cells at interphase, but not in the nucleus, suggesting that D-DDB1 is ordinarily present in the cytoplasm. Interestingly, after UV-irradiation, D-DDB1 is shifted to the nucleus within 12 h. This phenomenon is in line with recruitment of NER factors to DNA damage sites. It has been reported that DDB2 possesses nuclear localization signals and is required for the nuclear accumulation of DDB1 (37). D-DDB1 was detected in adult brain (Fig. 9C). D-DDB1 protein may play an important role in the central nervous system (CNS) and muscles. DDB reportedly recognizes a class of oxygen free radical-induced base lesions (38). Since O2 consumption in the CNS and muscles is known to be particularly high as compared with other organs, their data suggested that the amount of D-DDB1 protein in the CNS and muscles of Drosophila might correspond to the necessity of NER in the CNS and muscles.
Northern hybridization of developmental stages here revealed a pattern of expression very similar to those of Drosophila enzymes involved in nuclear DNA replication (DNA polymerases α and ε, PCNA, etc.) (31,32). During embryogenesis d-ddb1 transcript and D-DDB1 protein levels were high in the early stages when rates of DNA replication were maximal and decreased markedly as development proceeded. A clear decrease in d-ddb1 mRNA in the embryos from 0 to 20 h is shown in Figure 5. This may indicate that the presence of a AUUUA signal within the 3′-untranslated region of d-ddb1 mRNA is associated with rapid mRNA turnover. D-DDB1 protein level also decreases throughout embryogenesis, as does d-ddb1 mRNA level. This may indicate a short half-life of the D-DDB1 protein. Maternal loading of the d-ddb1 transcript indicates an importance related to blastogenesis. Reportedly, it has been shown that DDB2 interacts with the activation domain of E2F1 and stimulates E2F1-activated transcription in human cells (29). E2F is a transcription factor playing important roles in the regulation of cell proliferation (39). In mouse liver the nuclear levels of both DDB1 and DDB2 increase in late G1 phase and several hours before the peak of DNA replication (40). These phenomena may indicate that DDB has an important role in entering S phase. As shown here, DRE and DRE-like sequences are located upstream of the transcription initiation site of the d-ddb1 gene. The DRE/DREF system regulates the expression of several genes encoding key factors involved in nuclear DNA replication (23) and the present gel mobility shift analyses have demonstrated that the DRE and DRE-like sites of the d-ddb1 gene are essential for formation of stable DNA–protein complexes. Furthermore, mutations in any one of these two DREF-binding sites resulted in an extensive reduction in promoter activity. The data suggest that transcription of the d-ddb1 gene is under control of the DRE/DREF system. The results of western blotting analysis of DREF levels during Drosophila development reported by Yamaguchi et al. (31) are similar to those of northern blotting for d-ddb1. We conclude that D-DDB1 must contribute to both DNA repair and replication.
Like D-DDB1, another DNA repair protein factor, Rad51, is reportedly induced several-fold in murine cells ectopically expressing E2F1 or E2F2 (41). It may thus be required during replication to ensure genomic integrity. The dE2F gene has three tandemly arranged DNA replication-related element (DREs) and is one of the most critical target genes of DREF (42). The d-ddb1 gene has two DRE sites and the fact that its transcription was activated by DREF is clearly of interest in this context. The mechanisms by which DDB stimulates E2F-activated transcription and functions in DNA repair should next be deciphered. Further analysis should provide more precise insights into the molecular roles in different physiological processes.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Drs Masamitsu Yamaguchi, Yoshihiro H. Inoue and Hideki Yoshida of the University of Kyoto Institute of Technology, Dr Takeshi Todo of Radiation Biology Center of Kyoto University, Ms Yoko Ogasawara of Tokyo University of Science, and Ms Kaori Shimanouti and Shizuka Murakami of our laboratory for technical assistance and helpful discussions.
DDBJ/EMBL/GenBank accession no. AF132145
REFERENCES
- 1.Hwang B.J., Ford,J.M., Hanawalt,P.C. and Chu,G. (1999) Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc. Natl Acad. Sci. USA, 96, 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feldberg R.S. (1980) On the substrate specificity of a damage-specific DNA binding protein from human cells. Nucleic Acids Res., 8, 1133–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carew J.A. and Feldberg,R.S. (1985) Recognition of a cytosine base lesion by a human damage-specific DNA binding protein. Nucleic Acids Res., 13, 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reardon J.T., Nichols,A.F., Keeney,S., Smith,C.A., Taylor,J.S., Linn,S. and Sancar,A. (1993) Comparative analysis of binding of human damaged DNA-binding protein (XPE) and Escherichia coli damage recognition protein (UvrA) to the major ultraviolet photoproducts: T[c,s]T, T[t,s]T, T[6-4]T, and T[Dewar]T. J. Biol. Chem., 268, 21301–21308. [PubMed] [Google Scholar]
- 5.Hirschfeld S., Levine,A.S., Ozato,K. and Protic,M. (1990) A constitutive damage-specific DNA-binding protein is synthesized at higher levels in UV-irradiated primate cells. Mol. Cell Biol., 10, 2041–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keeney S., Chang,G.J. and Linn,S. (1993) Characterization of a human DNA damage binding protein implicated in xeroderma pigmentosum E. J. Biol. Chem., 268, 21293–21300. [PubMed] [Google Scholar]
- 7.Payne A. and Chu,G. (1994) Xeroderma pigmentosum group E binding factor recognizes a broad spectrum of DNA damage. Mutat. Res., 310, 89–102. [DOI] [PubMed] [Google Scholar]
- 8.Protic M., Hirschfeld,S., Tsang,A.P., Wagner,M., Dixon,K. and Levine,A.S. (1989) Induction of a novel damage-specific DNA binding protein correlates with enhanced DNA repair in primate cells. Mol. Toxicol., 2, 255–270. [PubMed] [Google Scholar]
- 9.Vaisman A. and Chaney,S.G. (1995) Induction of UV-damage recognition protein by cisplatin treatment. Biochemistry, 34, 105–114. [DOI] [PubMed] [Google Scholar]
- 10.Mu D., Park,C.H., Matsunaga,T., Hsu,D.S., Reardon,J.T. and Sancar,A. (1995) Reconstitution of human DNA repair excision nuclease in a highly defined system. J. Biol. Chem., 270, 2415–2418. [DOI] [PubMed] [Google Scholar]
- 11.Aboussekhra A., Biggerstaff,M., Shivji,M.K., Vilpo,J.A., Moncollin,V., Podust,V.N., Protic,M., Hubscher,U., Egly,J.M. and Wood,R.D. (1995) Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell, 80, 859–868. [DOI] [PubMed] [Google Scholar]
- 12.Kazantsev A., Mu,D., Nichols,A.F., Zhao,X., Linn,S. and Sancar,A. (1996) Functional complementation of xeroderma pigmentosum complementation group E by replication protein A in an in vitro system. Proc. Natl Acad. Sci. USA, 93, 5014–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu G. and Chang,E. (1988) Xeroderma pigmentosum group E cells lack a nuclear factor that binds to damaged DNA. Science, 242, 564–567. [DOI] [PubMed] [Google Scholar]
- 14.Kataoka H. and Fujiwara,Y. (1991) UV damage-specific DNA-binding protein in xeroderma pigmentosum complementation group E. Biochem. Biophys. Res. Commun., 175, 1139–1143. [DOI] [PubMed] [Google Scholar]
- 15.Keeney S., Wein,H. and Linn,S. (1992) Biochemical heterogeneity in xeroderma pigmentosum complementation group E. Mutat. Res., 273, 49–56. [DOI] [PubMed] [Google Scholar]
- 16.Keeney S., Eker,A.P., Brody,T., Vermeulen,W., Bootsma,D., Hoeijmakers,J.H. and Linn,S. (1994) Correction of the DNA repair defect in xeroderma pigmentosum group E by injection of a DNA damage-binding protein. Proc. Natl Acad. Sci. USA, 91, 4053–4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nichols A.F., Itoh,T., Graham,J.A., Liu,W., Yamaizumi,M. and Linn,S. (2000) Human damage-specific DNA-binding protein p48. Characterization of XPE mutations and regulation following UV irradiation. J. Biol. Chem., 28, 21422–21428. [DOI] [PubMed] [Google Scholar]
- 18.Spradling A.C. and Rubin,G.M. (1981) Drosophila genome organization: conserved and dynamic aspects. Annu. Rev. Genet., 15, 219–264. [DOI] [PubMed] [Google Scholar]
- 19.Hirose F., Yamaguchi,M., Handa,H., Inomata,Y. and Matsukage,A. (1993) Novel 8-base pair sequence (Drosophila DNA replication-related element) and specific binding factor involved in the expression of Drosophila genes for DNA polymerase alpha and proliferating cell nuclear antigen. J. Biol. Chem., 268, 2092–2099. [PubMed] [Google Scholar]
- 20.Todo T., Ryo,H., Borden,A., Lawrence,C., Sakaguchi,K., Hirata,H. and Nomura,T. (1997) Non-mutagenic repair of (6–4)photoproducts by (6–4)photolyase purified from Drosophila melanogaster. Mutat. Res., 385, 83–93. [DOI] [PubMed] [Google Scholar]
- 21.Towbin H.S., Staehelin,J. and Gordon,J. (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl Acad. Sci. USA, 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clemens J.C., Worby,C.A., Simonson-Leff,N., Muda,M., Maehama,T., Hemmings,B.A. and Dixon,J.E. (2000) Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl Acad. Sci. USA, 97, 6499–6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirose F., Yamaguchi,M., Kuroda,K., Omori,A., Hachiya,T., Ikeda,M., Nishimoto,Y. and Matsukage,A. (1996) Isolation and characterization of cDNA for DREF, a promoter-activating factor for Drosophila DNA replication-related genes. J. Biol. Chem., 271, 3930–3937. [DOI] [PubMed] [Google Scholar]
- 24.Ingham P. and Howard,K. (1989) In situ hybridization to wax-sectioned material. In Michael,A. (ed.), Drosophila: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 173–180.
- 25.Pisano C., Bonaccorsi,S. and Gatti,M. (1993) The kl-3 loop of the Y chromosome of Drosophila melanogaster binds a tektin-like protein. Genetics, 133, 569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kai M., Takahashi,T., Todo,T. and Sakaguchi,K. (1995) Novel DNA binding proteins highly specific to UV-damaged DNA sequences from embryos of Drosophila melanogaster. Nucleic Acids Res., 23, 2600–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kai M., Todo,T., Wada,M., Ryo,H., Masutani,C., Kobayashi,H., Morioka,H., Ohtsuka,E., Hanaoka,F. and Sakaguchi,K. (1998) A new Drosophila ultraviolet light-damaged DNA recognition endonuclease that selectively nicks a (6-4) photoproduct site. Biochim. Biophys. Acta, 1397, 180–188. [DOI] [PubMed] [Google Scholar]
- 28.Dualan R., Brody,T., Keeney,S., Nichols,A.F., Admon,A. and Linn,S. (1995) Chromosomal localization and cDNA cloning of the genes (DDB1 and DDB2) for the p127 and p48 subunits of a human damage-specific DNA binding protein. Genomics, 29, 62–69. [DOI] [PubMed] [Google Scholar]
- 29.Hayes S., Shiyanov,P., Chen,X. and Raychaudhuri,P. (1998) DDB, a putative DNA repair protein, can function as a transcriptional partner of E2F1. Mol. Cell. Biol., 18, 240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGregor W.G. (1999) DNA repair, DNA replication, and UV mutagenesis. J. Invest. Derm. Symp. Proc., 4, 1–5. [DOI] [PubMed] [Google Scholar]
- 31.Yamaguchi M., Hirose,F., Nishimoto,Y., Naruge,T., Ikeda,M., Hachiya,T., Tamai,K., Kuroda,K. and Matsukage,A. (1995) Expression patterns of DNA replication enzymes and the regulatory factor DREF during Drosophila development analyzed with specific antibodies. Biol. Cell, 85, 147–155. [DOI] [PubMed] [Google Scholar]
- 32.Oshige M., Yoshida,H., Hirose,F., Takata,K., Inoue,Y., Aoyagi,N., Yamaguchi,M., Koiwai,O., Matsukage,A. and Sakaguchi,K. (2000) Molecular cloning and expression during development of the Drosophila gene for the catalytic subunit of DNA polymerase ε. Gene, 256, 93–100. [DOI] [PubMed] [Google Scholar]
- 33.Yamaguchi M., Hayashi,Y., Nishimoto,Y., Hirose,F. and Matsukage,A. (1995) A nucleotide sequence essential for the function of DRE, a common promoter element for Drosophila DNA replication-related genes. J. Biol. Chem., 270, 15808–15814. [DOI] [PubMed] [Google Scholar]
- 34.Wakasugi M., Shimizu,M., Morioka,H., Linn,S., Nikaido,O. and Matsunaga,T. (2001) Damaged DNA-binding protein DDB stimulates the excision of cyclobutane pyrimidine dimers in vitro in concert with XPA and replication protein A. J. Biol. Chem., 276, 15434–15440. [DOI] [PubMed] [Google Scholar]
- 35.Datta A., Bagchi,S., Nag,A., Shiyanov,P., Adami,G.R., Yoon,T. and Raychaudhuri,P. (2001) The p48 subunit of the damaged-DNA binding protein DDB associates with the CBP/p300 family of histone acetyltransferase. Mutat. Res., 486, 89–97. [DOI] [PubMed] [Google Scholar]
- 36.Martinez E., Palhan,V.B., Tjernberg,A., Lymar,E.S., Gamper,A.M., Kundu,T.K., Chait,B.T. and Roeder,R.G. (2001) Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol., 20, 6782–6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu W., Nichols,A.F., Graham,J.A., Dualan,R., Abbas,A. and Linn,S. (2000) Nuclear transport of human DDB protein induced by ultraviolet light. J. Biol. Chem., 275, 21429–21434. [DOI] [PubMed] [Google Scholar]
- 38.Fujiwara Y., Masutani,C., Mizukoshi,T., Kondo,J., Hanaoka,F. and Iwai,S. (1999) Characterization of DNA recognition by the human UV-damaged DNA-binding protein. J. Biol. Chem., 274, 20027–20033. [DOI] [PubMed] [Google Scholar]
- 39.Weinberg R.A. (1996) E2F and cell proliferation: a world turned upside down. Cell, 85, 457–459. [DOI] [PubMed] [Google Scholar]
- 40.Nag A., Datta,A., Yoo,K., Bhattacharyya,D., Chakrabortty,A., Wang,X., Slagle,B.L., Costa,R.H. and Raychaudhuri,P. (2001) DDB2 induces nuclear accumulation of the hepatitis B virus X protein independently of binding to DDB1. J. Virol., 75, 10383–10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishida S., Huang,E., Zuzan,H., Spang,R., Leone,G., West,M. and Nevins,J.R. (2001) Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol., 14, 4684–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sawado T., Hirose,F., Takahashi,Y., Sasaki,T., Shinomiya,T., Sakaguchi,K., Matsukage,A. and Yamaguchi,M. (1998) The DNA replication-related element (DRE)/DRE-binding factor system is a transcriptional regulator of the Drosophila E2F gene. J. Biol. Chem., 273, 26042–26051. [DOI] [PubMed] [Google Scholar]