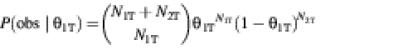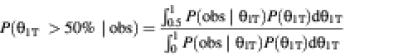Abstract
We have developed an automated method for discovering tissue-specific regulation of alternative splicing through a genome-wide analysis of expressed sequence tags (ESTs). Using this approach, we have identified 667 tissue-specific alternative splice forms of human genes. We validated our muscle-specific and brain-specific splice forms for known genes. A high fraction (8/10) were reported to have a matching tissue specificity by independent studies in the published literature. The number of tissue-specific alternative splice forms is highest in brain, while eye_retina, muscle, skin, testis and lymph have the greatest enrichment of tissue-specific splicing. Overall, 10–30% of human alternatively spliced genes in our data show evidence of tissue-specific splice forms. Seventy-eight percent of our tissue-specific alternative splices appear to be novel discoveries. We present bioinformatics analysis of several tissue-specific splice forms, including automated protein isoform sequence and domain prediction, showing how our data can provide valuable insights into gene function in different tissues. For example, we have discovered a novel kidney-specific alternative splice form of the WNK1 gene, which appears to specifically disrupt its N-terminal kinase domain and may play a role in PHAII hypertension. Our database greatly expands knowledge of tissue-specific alternative splicing and provides a comprehensive dataset for investigating its functional roles and regulation in different human tissues.
INTRODUCTION
Recently, there has been growing interest in alternative splicing as a mechanism for expanding the repertoire of gene functions. Different combinations of exons can be spliced together to produce different mRNA isoforms of a gene, encoding structurally and functionally different protein products (1). The discovery from large-scale genomics studies that alternative splicing may occur in a very large fraction of human genes (35–59%) suggests a major role for alternative splicing in the production of functional complexity in the human genome (2–7).
This hypothesis implies extensive regulation of alternative splicing. Alternative splicing can display strong specificity to a particular tissue or developmental stage (8,9), modulating the functional characteristics of protein isoforms in specific tissues (10). It has also been estimated that ∼15% of disease-causing mutations in human genes involve misregulation of alternative splicing (11) and errors in mRNA processing have been associated with cancer and other human diseases (12–16).
Despite growing interest in how alternative splicing is regulated (2,17–25), relatively little is known about tissue-specific alternative splicing and its regulation, especially when compared with the sheer volume of information known about other mechanisms of functional control such as transcriptional regulation. For example, tissue specificities for only a small number of alternatively spliced genes (about 50) are listed in current alternative splicing databases (25,26).
If alternative splicing plays as large and important a role in gene function regulation as recent genomics studies suggest, much more information is needed. There are many questions that need to be answered (27). What fraction of alternative splicing is tissue-specific? What proportion of tissue-specific splicing is associated with gross subdivisions of tissues (such as an entire organ like the brain) versus very specific cell types and developmental states? How can we efficiently identify the complete regulatory machinery controlling tissue specificity? What are the regulatory factors that mediate this process and what are the control sites that they recognize? What are the functional consequences of these alternative splicing events?
To answer any of these questions, one essential prerequisite is large-scale discovery and characterization of tissue-specific alternative splicing, for example by microarray analysis (28–30). This is needed both to provide biologists with information on whether ‘their gene’ is alternatively spliced in a tissue-specific manner (enabling them to study its functional consequences) and to give splice regulation researchers a big enough dataset to study mechanisms of splice regulation in many genes and tissues.
One possible approach is to use genomics datasets such as expressed sequence tags (ESTs) for large-scale analysis of tissue specificity. This poses two challenges. Although the EST database provides some information about tissue source, these data are incomplete and inconsistent. Thus, a consistent tissue classification of this large dataset is needed to enable detection of tissue specificity. Much more importantly, interpretation of these data requires filtering for statistical significance. EST data often have poor coverage (i.e. only a small number of ESTs from a given tissue for a region of interest in a gene) and many sampling artifacts. For example, there can be dramatically different numbers of ESTs from different libraries or tissues, creating sample bias. This could give the erroneous impression that a given splice form is specific to one tissue (simply because many ESTs for this gene have been sequenced from that tissue and few from other tissues).
We have sought to address both these problems. In this paper we present an automatic method to detect tissue-specific alternative splicing events using EST and genomic sequences. After constructing a tissue list of 46 human tissues with 2 million human ESTs, we generated a database of novel human alternative splices that is four times larger than our previous report (7) and used Bayesian statistics to compare the relative abundance of every pair of alternative splices in these tissues. Using several statistical criteria for tissue specificity, we have identified 667 tissue-specific alternative splicing relationships and analyzed their distribution in human tissues. We have validated our results by comparison with independent studies. This genome-wide analysis of tissue specificity of alternative splicing will be made available as a part of the Human Alternative Splicing Database (http://www.bioinformatics.ucla.edu/HASDB) (7), to provide a useful resource to study the tissue-specific functions of transcripts and the association of tissue-specific variants with human diseases.
MATERIALS AND METHODS
Data sources
Our analysis is based on three sources of data: human genomic sequence assemblies (5), human ESTs from the UniGene database (31) and human EST library information. Human genomic assembly sequences (accession no. NT_XXXX) and ‘draft’ BAC clone sequences (accession nos ACXXXX, ALXXXXX) were downloaded from NCBI (ftp://ftp. ncbi.nih.gov/genomes/H_sapiens and ftp://ftp.ncbi.nih.gov/ genbank/gbhtgXX.seq.gz). Human ESTs and library information were downloaded from UniGene (ftp://ftp.ncbi.nih.gov/repository/UniGene). Additional EST library information about human tissue sources was obtained from the NCBI Library Browser, downloaded from www.ncbi.nlm.nih.gov/UniGene/lbrowse.cgi?ORG=Hs. The work described in this paper is based on the January 2002 release of the human genome and UniGene data.
Library classification
Tissue source information for approximately 6900 human EST libraries was exhaustively examined to produce a consistent classification of human tissues suitable for tissue specificity calculations. We checked and refined the NCBI Library Browser classification (200 categories) to produce a considerably reduced classification (46 categories). For many libraries with unclear or incomplete tissue information in UniGene, we checked their dbEST entries for extra information about tissue source. Libraries recorded as having the same tissue source (e.g. ‘brain’) were combined into a single category, including both tumor and normal samples from that tissue. We sought to avoid mixing of multiple tissues during this procedure. If a library could not be clearly assigned to a single tissue (for example, if it was a pool of multiple samples from different tissues, or lacked clear information designating it as a sample from a single tissue), it was excluded from the final set. We excluded a total of 2652 EST libraries by these criteria (see Table 1). Our final classification consisted of 4271 EST libraries in just 46 single tissue categories (see Table 2).
Table 1. Construction of our EST library tissue classification.
| Analysis stage | No. of classes | No. of libraries | No. of ESTs |
|---|---|---|---|
| UniGene January 2002 | 6964 | 2 971 844 | |
| NCBI Library Browser categories | 200 | 6923 | 2 971 150 |
| Combined groups from same tissue | 73 | 6923 | 2 971 150 |
| Excluded non-specific groups | 27 | 2652 | 739 923 |
| Final tissue classes | 46 | 4271 | 2 231 227 |
Table 2. Our EST library classification database of 46 human tissues.
| Tissue | No. of libraries | No. of ESTs |
|---|---|---|
| Adipose | 9 | 2389 |
| Adrenal | 16 | 19 838 |
| Aorta_vena | 14 | 13 628 |
| Bladder | 49 | 20 501 |
| Blood | 28 | 42 653 |
| Bone_marrow | 274 | 61 806 |
| Brain | 218 | 347 298 |
| Breast | 1009 | 95 487 |
| Cartilage | 4 | 7843 |
| Connective_tissue | 4 | 2779 |
| Ear | 2 | 12 473 |
| Epididymis | 1 | 105 |
| Esophagus | 8 | 3000 |
| Eye_retina | 30 | 52 539 |
| Foreskin | 3 | 19 803 |
| Gall | 3 | 2489 |
| Genitourinary_tract | 1 | 4737 |
| Greater_omentum | 6 | 322 |
| Heart | 22 | 54 798 |
| Intestine | 6 | 4978 |
| Kidney | 84 | 131 432 |
| Larynx | 6 | 765 |
| Liver | 44 | 60 110 |
| Lung | 299 | 195 760 |
| Lymph | 40 | 88 079 |
| Mammary_gland | 1 | 2321 |
| Mouth_oral | 15 | 2974 |
| Muscle | 21 | 60 913 |
| Nerve | 479 | 39 065 |
| Nose_pharynx | 9 | 2222 |
| Ovary | 142 | 74 592 |
| Pancreas | 33 | 77 221 |
| Parathyroid | 4 | 19 105 |
| Placenta | 339 | 166 277 |
| Prostate | 292 | 117 740 |
| Salivary_gland | 5 | 1349 |
| Skin | 33 | 74 972 |
| Spleen | 7 | 9138 |
| Stomach | 278 | 36 834 |
| Testis | 170 | 96 979 |
| Thymus | 17 | 4958 |
| Thyroid | 19 | 6773 |
| Tonsil | 6 | 41 862 |
| Trachea_bronchus | 3 | 23 |
| Uterus | 218 | 150 297 |
| Sum | 4271 | 2 231 227 |
Tissue specificity scoring
Reliable detection of tissue specificity is complicated by poor EST library coverage (in many cases there are few ESTs from a given tissue for a given exon) and sampling bias problems (different tissues may have dramatically different numbers of ESTs sequenced). We therefore needed a statistical measure of tissue specificity that properly accounts for these sources of uncertainty and bias. Specifically, we cannot assume that the observed numbers of ESTs from one tissue exactly reflect the true proportions of different splice forms in that tissue. A larger number of ESTs gives a more confident estimate of those proportions; a smaller number of ESTs causes increased uncertainty. To take this into account rigorously when attempting to compare the proportions of a given splice form between different tissues we used Bayesian statistics, treating the true proportions as hidden variables and calculating confidence from the available observations.
Suppose gene G has two mutually exclusive (i.e. alternative) splices S1 and S2. By ‘mutually exclusive’ we mean two splices that share one splice site but differ at the other splice site and which thus cannot both be present in a single transcript (7). Throughout this paper we will refer to the set of transcripts containing splice S1 as ‘splice form S1’.
For our hidden variables, let θ1T represent the hidden frequency of S1 in a specific tissue T and let θ1∼ be its frequency in the pool of all other tissues (i.e. all tissues except T, symbolized by ∼). Similarly, let θ2T and θ2∼ represent the hidden frequencies of S2 in tissue T versus in the pool of all other tissues ∼. By definition, these probabilities must be normalized:
θ1T + θ2T = 1
θ1∼ + θ2∼ = 1
For our observations, let N1T and N2T be the total number of ESTs in tissue T observed to have splice S1 or S2, respectively. Similarly, let N1∼ and N2∼ be the total number of ESTs in the pool of all other tissues ∼ observed to have splice S1 or S2, respectively. Since our model assumes two mutually exclusive splice forms, the likelihood of the observations should follow a simple binomial distribution. For example, in tissue T
We first calculated the confidence that splice S1 is preferred in tissue T as a Bayesian posterior probability:
We used P(θ1T) = 1 as an uninformative prior probability. We also computed the posterior probability that splice S1 is preferred in the pool of all other tissues[P(θ1∼ > 50%|obs)] in the same way, from the counts N1∼ and N2∼.
We defined a tissue specificity (TS) score of splice S1 for tissue T as the difference between this posterior probability for tissue T versus the pool of other tissues,
TS = 100[P(θ1T > 50%|obs) – P(θ1∼ > 50%|obs)]
To assess how stable the TS value is to possible error models, we calculated a robustness value rTS, which measures how much the TS value drops when a single EST observation of splice S1 is removed from tissue T. Specifically, we computed the negative log value of the relative change of the TS value caused by removing that EST,
rTS = –log10(ΔTS/TS),
where ΔTS = |TS(N1T) – TS(N1T – 1)|. rTS = 1 means TS drops by 10–1 = 10% and rTS = 0 means TS drops to 0. Because each EST makes an equal contribution to the TS value, only one resampling step was required to calculate rTS. We also calculated rTS∼, the effect of removing one EST observation of splice S2 from the pool of other tissues, defined in the same way as rTS, using ΔTS = |TS(N2∼) – TS(N2∼ – 1)|.
Criteria for high confidence (HC) tissue specificity were TS > 50, rTS > 0.9, rTS∼ > 0.9; for low confidence (LC) TS > 0, rTS > 0.5, rTS∼ > 0.5. A necessary (but insufficient) condition for the HC group was at least three EST observations of S1 in tissue T; for LC at least two EST observations of S1 in tissue T.
Validation of tissue-specific splices of known genes
To search for alternative splicing information for a given gene, we performed thorough literature searches using PubMed, OMIM, LocusLink and other databases of alternative splicing. We sought information about sequencing of alternative splice forms and their tissue specificity. Isoform data without a complete reported sequence was not considered sufficient validation. To be counted as a match, an alternative splice identified in our database had to match a specific transcript sequence published in the literature. To be counted as a validated tissue specificity the isoform also had to be independently reported to be specific to the same tissue that we identified. For the validation data shown in Table 6 we validated a sample set consisting of all brain-specific and muscle-specific alternative splices identified by our HC criteria on a previous dataset (UniGene and human genomic sequence data February 2001). All procedures and criteria were identical for different runs. We also used the GeneMine software system (32) to visualize and validate versus the literature all aspects of the genomic mapping of our clusters, exons and introns, splices sites, alternative splicing and the impacts on protein structure and function, by examining all the features in the genomic–EST–mRNA multiple sequence alignments.
Table 6. Validation of brain- and muscle-specific isoforms versus independent literature.
| UniGene ID | Gene description | Computational isoform | Published isoform data |
|---|---|---|---|
| Hs.102948 | Enigma (LIM domain protein) | Brain-specific | N/D |
| Hs.105509 | CTL2 gene | Brain-specific | N/D |
| Hs.114034 | Maternal G10 transcript | Brain-specific | N/D |
| Hs.117546 | NNAT neuronatin | Brain-specific | Form found, no tissue data (8) |
| Hs.15098 | Hypothetical protein MGC2652 | Brain-specific | N/D |
| Hs.158947 | Hypothetical protein FLJ20244 | Brain-specific | N/D |
| Hs.159608 | Aldehyde dehydrogenase 3 family, member 3 | Brain-specific | Brain-specific (68,69) |
| Hs.167031 | DKFZp566D133 protein | Brain-specific: 2 | N/D |
| Hs.175941 | B-cell receptor-associated protein BAP29 | Brain-specific | N/D |
| Hs.182503 | ESTs | Brain-specific | N/D |
| Hs.194660 | ESTs | Brain-specific: 2 | N/D |
| Hs.24371 | Uncharacterized hypothalamus protein HT007 | Brain-specific: 4 | N/D |
| Hs.25854 | Crystallin, ζ (quinone reductase)-like 1 | Brain-specific | N/D |
| Hs.26655 | G6PT1 glucose | Brain-specific: 2 | Brain-specific (70,71) |
| Hs.278736 | CDC42 cell division cycle 42 | Brain-specific | Brain-specific (50,54) |
| Hs.279939 | MTCH1 mitochondrial carrier homolog 1 | Brain-specific | N/D |
| Hs.282997 | GBA glucosidase β | Brain-specific: 2 | Form found, no tissue data (72) |
| Hs.28777 | H2AFL H2A histone family, member L | Brain-specific | N/D |
| Hs.57435 | Solute carrirer family 11, member 2 | Brain-specific | Form not found; but another form brain-specific (73,74) |
| Hs.75254 | IRF3 interferon regulatory factor 3 | Brain-specific | Brain-specific (39) |
| Hs.7973 | Hypothetical protein DKFZP434G156 | Brain-specific: 2 | N/D |
| Hs.91747 | PFN2 profilin 2 | Brain-specific | Brain-specific (75,76) |
| Hs.101490 | ESTs | Muscle-specific | N/D |
| Hs.102948 | Enigma (LIM domain protein) | Muscle-specific | N/D |
| Hs.180266 | Tropomyosin 2 (β) | Muscle-specific: 2 | Muscle-specific (9,77) |
| Hs.239069 | Four and a half LIM domains 1 | Muscle-specific | Form not found; but another form muscle-specific (78) |
| Hs.75108 | Ribonuclease/angiogenin inhibitor | Muscle-specific: 2 | Form found, no tissue data (79) |
N/D means no sequencing of isoforms was found in the literature, preventing comparison with our data. A number in the ‘Computational isoform’ column indicates that more than one splice was observed to have that tissue specificity.
Bioinformatics analysis of tissue-specific protein isoforms
To assess the effects of alternative splicing on the protein product, we predicted protein isoform sequences for each alternative splice, their protein domain composition and motif analysis. These results will be described in detail elsewhere. ORF prediction was performed using standard methods for identifying the longest open reading frame in each transcript. Protein domain prediction was performed using RPS-BLAST (33) on the protein isoform sequences, against a database of protein domain sequences from SMART (34) and PFAM (35), using cut-off thresholds of 10–20 expectation. For all the examples with functional importance shown in this paper we also evaluated the effects of each alternative splice relationship by carefully examining the complete alignment of ESTs to genomic sequence using GeneMine software. Since an alternative splice can change where the coding region starts and ends, we adopted the policy that any alternative splice that alters the protein product will be classified as a ‘coding region’, regardless of its location relative to the GenBank CDS annotation.
RESULTS
Tissue classification of human ESTs
Since the cDNA library source of each public EST sequence is recorded, in principle this dataset could provide large-scale detection of tissue-specific alternative splicing, if each cDNA library could be associated with a specific tissue. Unfortunately, these data are from many different contributors and are not annotated in a uniform way. To provide a reliable basis for analyzing tissue specificity, we have carefully classified the approximately 7000 human EST libraries into distinct tissue classes. We began with the NCBI Library Browser classification, which consists of 200 categories covering 6923 libraries. Manually inspecting public information about cDNA libraries available from Unigene, dbEST and GenBank, we combined different categories that were from the same tissue, excluded many categories (e.g. ‘head and neck’) that did not correspond to a specific tissue and constructed a manually curated library tissue classification database (Table 1). For this study we combined tumor and normal samples from each tissue source (e.g. ‘brain’), although in the future it will be interesting to look for tissue specificities that distinguish these.
Our final classification consisted of 46 tissues containing 4271 cDNA libraries and 2.2 million human ESTs (Table 2). This represents 75% of ESTs in UniGene. This classification is by no means an optimally structured subdivision of the distinct tissues in the human body, but rather is intended to reflect the level of specificity present in the public cDNA library samples themselves. These samples are rarely more specific than an entire organ (e.g. ‘brain’). As an example of our tissue classification, Table 3 lists all the libraries classified as ‘adipose’ tissue.
Table 3. Libraries in the tissue category ‘adipose’.
| Tissue | Library ID | Title | Description |
|---|---|---|---|
| Adipose | 108 | Clontech_adult_human_fat_cell_library_HL1108A | Adipose |
| Adipose | 110 | WATM1 | Adipose, subcutaneous white adipose |
| Adipose | 112 | BATM1 | Adipose, perirenal brown adipose tissue |
| Adipose | 115 | Human_Adipose_tissue | Adipose |
| Adipose | 196 | BATM2 | Adipose, perirenal brown adipose tissue |
| Adipose | 307 | Adipose_tissue,_white_I | Adipose, white adipose |
| Adipose | 341 | Adipose_tissue,_white_II | Adipose, white adipose tissue |
| Adipose | 423 | Adipose_tissue,_brown | Adipose, brown adipose tissue |
| Adipose | 509 | NCI_CGAP_Lip2 | Adipose |
Genome-wide detection of alternative splicing
Using the latest human EST data (UniGene January 2002) and genomic sequence we performed a genome-wide analysis of alternative splicing as previously described (7). This conservative analysis process takes into account many factors, including mapping of EST consensus sequences to unique genomic locations, validation by intronic splice site sequences and very specific match requirements (two mutually exclusive splices that match exactly at one splice site but diverge at the other splice site) to report an alternative splice. This analysis identified 27 790 alternative splices (see Table 5), approximately four times that of our previous analysis (7). These results will be accessible via our online Human Alternative Splicing Database (http://www.bioinformatics.ucla.edu/HASDB).
Table 5. Genome-wide detection of tissue-specific alternative splicing.
| No. of alternatively spliced genes | No. of alternative splice relationships | |
|---|---|---|
| UniGene January 2002 | 7240 | 27 790 |
| Tissue classification database | 5871 | 20 737 |
| (At least three ESTs observed in one tissue) | 4352 | 14 045 |
| Tissue-specific alternative splicing (HC) | 454 (10.4%) | 667 (4.7%) |
| (At least two ESTs observed in one tissue) | 5132 | 17 288 |
| Tissue-specific alternative splicing (LC) | 1572 (30.6%) | 2873 (16.6%) |
Detection of tissue-specific alternative splicing
To identify tissue-specific alternative splicing automatically and with statistical robustness, we developed a tissue specificity (TS) scoring function. This calculation measures the percent confidence that a specific splice S1 is preferred in a given tissue T1 (i.e. that S1 is found in a larger proportion of transcripts there than the alternative splice S2), minus the same confidence calculated for the pool of ESTs from all other tissues (see Materials and Methods for details). TS > 0 means that splice S1 is preferred in tissue T1 more than it is in other tissues. The higher the TS score is, the stronger the evidence of tissue specificity. For example, if the confidence that splice S1 is the major splice form was 70% in brain and 40% in the pool of other tissues, then the TS score would be 70 – 40 = 30. If splice S1 is preferred in all tissues, it will get a low TS score. If there are insufficient EST counts to be confident about the proportion of S1 in tissue T1 or other tissues, this will also give a low TS score, both by decreasing the certainty that splice S1 is preferred in tissue T1 and increasing the possibility that splice S1 might be preferred in other tissues as well.
Table 4 illustrates the use of the TS score to distinguish tissue-specific splicing within ESTs from one gene (GAR22, UniGene cluster Hs.322852). We identified two alternative splicing relationships in this cluster (indicated in Table 4 by their splice IDs, 17571–17572 and 17577–17578) and calculated TS scores for these alternative splices in the tissues in which they were observed. Of 11 candidate splice–tissue pairs, four had a positive TS score. However, the number of EST observations supporting these scores was not large.
Table 4. TS calculations for GAR22 (UniGene cluster Hs.322852).
| Tissue | TS | Splice 1 | Splice 2 | N1T | N2T | N1∼ | N2∼ | rTS | rTS∼ |
|---|---|---|---|---|---|---|---|---|---|
| Brain | 45.27 | 17572 | 17571 | 7 | 2 | 3 | 3 | 1.092685 | 0.457746 |
| Breast | –9.23 | 17572 | 17571 | 1 | 0 | 9 | 5 | –0.428388 | 0.157909 |
| Testis | –9.23 | 17572 | 17571 | 1 | 0 | 9 | 5 | –0.428388 | 0.157909 |
| Blood | –9.23 | 17572 | 17571 | 1 | 0 | 9 | 5 | –0.428388 | 0.157909 |
| Brain | –43.78 | 17571 | 17572 | 2 | 7 | 3 | 3 | 1.121913 | 0.443241 |
| Eye_retina | 92.6 | 17571 | 17572 | 3 | 0 | 2 | 10 | 1.166472 | 2.091432 |
| Breast | 73.33 | 17577 | 17578 | 1 | 0 | 0 | 5 | 0.471644 | 1.697701 |
| Brain | –13.83 | 17578 | 17577 | 1 | 0 | 4 | 1 | –0.252673 | 0.226528 |
| Lung | 6.7 | 17578 | 17577 | 2 | 0 | 3 | 1 | –0.273215 | –0.288415 |
| Kidney | –13.83 | 17578 | 17577 | 1 | 0 | 4 | 1 | –0.252673 | 0.226528 |
| Bone_marrow | –13.83 | 17578 | 17577 | 1 | 0 | 4 | 1 | –0.252673 | 0.226528 |
To take this into account, we calculated the ‘robustness’ of the TS score, which measures how much TS drops when a single EST observation of splice S1 in tissue T1 is removed from the sample. This assesses how stable the TS value is to possible error models (e.g. the library classification may have errors) and is analogous to the ‘jack-knife’, a common statistical resampling method (36). Defining the robustness as rTS = –log10(ΔTS/TS), where ΔTS = |TS(N1T) – TS(N1T – 1)| (see Materials and Methods for details), rTS = 1 means TS drops by 10–1 = 10% and rTS = 0 means TS drops to 0. This measures the amount of EST evidence supporting the TS score; if N1T >> 1, then the robustness, rTS, can be high.
Only one candidate tissue-specific splice in GAR22 (splice ID 17571 in tissue eye_retina) passes the combined criteria of >2-fold tissue specificity (TS > 50) and good robustness (rTS > 0.9, equivalent to a 14 point drop in TS; see Materials and Methods for details). It is striking that the only other observations of this splice form are in brain, which suggests that the putative association of this form with eye_retina is real and that this form may be found exclusively in neuronal tissue.
We have computed TS scores for all alternative splices detected in the EST libraries contained in our 46 tissue classification database (Table 5). We divided those with positive TS scores into two groups: a HC group with TS > 50% and rTS > 0.9, designed to screen out false positives (but causing a high level of false negatives), and a LC group with TS > 0 and rTS > 0.5. We identified 894 tissue-specific relationships for 667 alternative splices in the HC group and a total of 2873 alternative splices showing tissue specificity in the LC group. These data suggest that by our HC criteria 10.4% of alternatively spliced genes in the human genome have tissue specificity discernible in current EST data, and 30.6% by our LC criteria. Given the very gross form of tissue specificity of the EST libraries (typically an entire organ rather than a specific cell type), this probably underestimates the true extent of tissue-specific alternative splicing.
Independent validation of our tissue specificity results
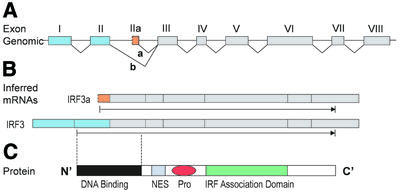
We have performed extensive validation analysis of our tissue-specific alternative splice forms. Figure 1 shows one example of brain-specific alternative splicing detected automatically by our procedure in the IRF3 gene (Hs.75254, interferon regulatory factor-3) with a TS score of 88. IRF3 is a member of the IRF family and plays an important role in the virus- and double-stranded RNA-mediated induction of interferon β (IFNβ) and RANTES (regulated upon activation normal T cell expressed and secreted) (37,38). Our automated procedure detected two alternative splice forms: a longer mRNA consisting of eight exons and a short form in which exons I and II are replaced by a new exon IIa. These match isoforms reported in the literature as IRF3 and IRF3a. In the EST data we detected five ESTs in brain, all matching the IRF3a form, and five ESTs elsewhere, of which four match the IRF3 form. According to the literature, both isoforms are expressed in multiple tissues but the ratio IRF3a:IRF3 is dramatically high in brain compared with other human tissues (39).
Figure 1.
Brain-specific alternative splicing of IRF3. (A) Genomic structure of the IRF3 gene. Exons are shown as boxes and colors show alternative exons. Splice a is specific to brain. (B) The two alternative forms of IRF3 mRNA inferred from the expressed sequence data. The protein coding region is indicated by an arrow beneath each form. (C) Schematic representation of the IRF3 protein. The DNA-binding domain, the NES element, the proline-rich region and the C-terminal IRF association domain are indicated. Dashed lines mark the boundaries of the DNA-binding domain.
This example also illustrates the functional interpretability of the large structural changes that alternative splicing often causes. We performed a series of bioinformatics analyses to predict the protein isoform sequences and protein structural domains (see Materials and Methods). Prediction of the protein products identified an ORF in both cases, revealing a replacement AUG start site in exon IIa that encodes 22 amino acids before entering exon III in the same coding frame as in the long IRF3 form. Protein domain prediction using SMART (34) and PFAM (35) showed that the brain-specific splice disrupts the 110 amino acid DNA-binding domain (PFAM id 00605) at the IRF3 N-terminus, by replacing the first 55 amino acids with the 22 amino acids from exon IIa (Fig. 1B). On the basis of this simple domain analysis, we would predict that in brain the role of IRF3 in regulating gene expression (e.g. activating IFNβ and RANTES) would be dramatically altered.
This is strongly validated by the published literature. IRF3 has been shown to bind to the IFNβ promoter and up-regulate the transcription of IFNβ with other enhancers after virus infection (40,41). The brain-specific splice eliminates the ability of IRF3 to bind to the IFNβ promoter in vitro (10) and seems to play a protective role in brain, reducing the toxic effect of IFNβ (42,43) by suppressing its expression in brain (10).
To assess the overall accuracy of our automated tissue specificity detection method for many genes, we followed this same procedure of testing for independent validation of our results. We used two tissues, brain and muscle, as the test samples for this validation, since many studies of alternative splicing have been done for genes in these tissues (44). For a random sample of 37 tissue-specific alternative splices identified in the HC group for these tissues, we searched the published literature to see whether tissue specificity was independently reported for those genes. To count a splice form as validated, we required that a complete sequence matching our splice form be found in the literature and be demonstrated to be specific to the tissue reported by our procedure. Previously unknown genes (e.g. Hs.7973) were excluded from this analysis, since no studies of their tissue-specific alternative splicing have been published.
We found that 80% (8/10) of our brain- and muscle-specific splices were validated by the existing literature (Table 6). In the two cases where the splice forms were not validated by a matching isoform in the literature, published papers reported a different isoform that matched our tissue specificity. For example, in Hs.57435 we detected a brain-specific alternative splice. This splice was not validated by a matching sequence in the literature, but another isoform was reported and shown to be brain-specific. Unfortunately, there are no ESTs from brain that align to this region of the gene, so there was no possibility of our detecting this form. Similarly, our muscle-specific alternative splice for Hs.239069 was not validated by the literature, but another isoform of this gene was reported to be muscle-specific. These coincidences suggest that even the tissue specificities we detected for these genes may not actually be incorrect. There were five additional cases where the splice form we detected was validated by the literature but no tissue specificity studies were reported. Within the limits of available published data, these results indicate that the reliability of our database is likely to be high.
We have also used these data to estimate the fraction of our results that are novel discoveries of tissue-specific alternative splicing. These come from two categories: detecting previously unknown tissue specificity for a known alternative splice (in a known gene); detecting tissue specificity for a previously unknown alternative splice (in a known gene or in a novel gene). Combining these categories, 78% (29/37) of our brain- and muscle-specific alternative splices appear to be novel. It seems likely that our database can be a valuable source of interesting discoveries for biologists who study these genes, as well as researchers who study tissue-specific regulation of splicing.
Distribution and enrichment of tissue-specific alternative splicing
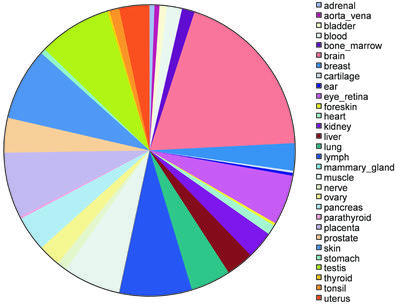
We analyzed the distribution of tissue-specific alternative splicing over the 46 human tissues in our classification (Fig. 2). We identified tissue-specific alternative splicing in the HC group for 30 of the 46 human tissues. The largest category by total number of tissue-specific splice forms was brain, which represented 18% of all tissue-specific alternative splicing events we observed. The tissues that each accounted for at least 4% of observed tissue-specific forms were brain, eye_retina, lung, liver, pancreas, placenta, ovary, uterus, testis, lymph, muscle and skin. This is consistent with results from a survey of the alternative exons from the published literature, in which brain and neurons were ranked highest among human tissues (26).
Figure 2.
Tissue distribution of human tissue-specific alternative splicing. Areas on the pie chart are proportional to the total number of alternative splices with high confidence tissue specificity for a particular tissue.
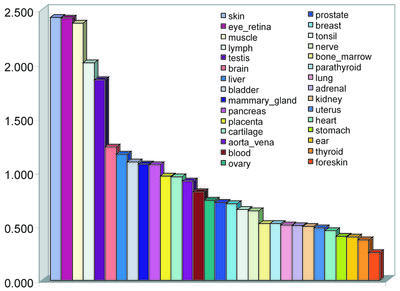
Since the number of ESTs sampled is very different for each tissue, we have also normalized the rate of detection of tissue-specific alternative splices within each tissue by its total number of ESTs. We defined the enrichment factor (EF) for a tissue as the proportion of total tissue-specific alternative splicing that it represents divided by the proportion of total ESTs that it represents. Figure 3 compares the enrichment factors for our classified human tissues. Skin, eye_retina and muscle ranked highest (2.4 times more tissue-specific splice forms than average), with lymph (2.0) and testis next (1.8), followed by brain (1.2) and liver (1.1). Other tissues with above-average tissue specificity were bladder, mammary_ gland and pancreas. Overall, this is consistent with previous studies indicating the immune and nervous systems as major loci of alternative splicing (45,46) and a common focus on regulation of alternative splicing in neuronal tissues (44). It is also striking that at the high end of the ranking, more sharply defined tissue categories (e.g. retina, muscle, skin, lymph) returned higher yields of tissue specificity detection than broadly defined categories such as brain. This does not necessarily mean that there is more tissue-specific alternative splicing in retina or muscle than in the many tissues in the brain. Instead, it may simply reflect serious limitations in the nature of the library samples (that most of them are grossly defined, combining all the different tissues that compose an entire organ) and of our TS scoring calculation.
Figure 3.
Enrichment of tissue-specific alternative splicing in 30 human tissues. The y-axis shows the enrichment factor for each tissue, defined as the ratio of the number of tissue-specific alternative splices observed in a tissue divided by the total number of ESTs observed in that tissue, normalized to have an average value of 1 (see text).
Bioinformatics analysis of a novel tissue-specific splice form
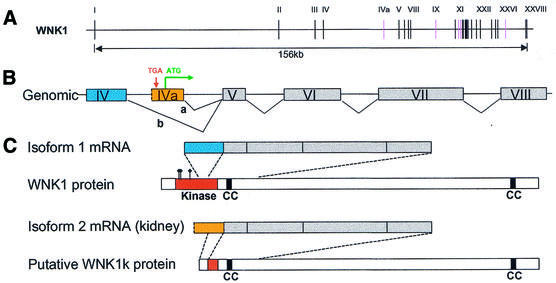
To demonstrate the value of our database for biological discovery, Figure 4 shows our analysis of a representative example (Hs.184592) in which we detected a novel kidney-specific alternative splicing event (TS score 94). As part of our genome-wide analysis of alternative splicing, we generated predicted protein isoforms and analyzed their domain composition by searching against protein domain databases. This gene encodes a serine/threonine protein kinase, WNK1 (with no K = lysine), which has only recently been described (47). This gene has 28 exons and encodes a huge protein with a kinase domain near its N-terminus and two coiled-coil conserved domains (Fig. 4) (48). Its name refers to the surprising replacement of an active site lysine residue with a cysteine, which leaves kinase activity intact.
Figure 4.
Kidney-specific alternative splicing of WNK1. (A) Genomic structure of WNK1. The genomic segment spanning WNK1 is represented by a horizontal line and exons by numbered vertical lines. Pink vertical lines indicate the alternative exons, IVa, IX, XI, XII and XXVI. (B) Gene structure for exons IV–VIII of the WNK1 gene. Exons are shown as boxes and colors show alternative exons. Splice a is specific to kidney. The putative in-frame stop codon UGA and start codon AUG are indicated. (C) The two alternative forms of WNK1 mRNA inferred from the expressed sequence data and the schematic representation of WNK1 protein sequences. The conserved kinase domain, two coiled-coil (CC) domains and the corresponding protein regions of mRNA forms are indicated. Three amino acids (K233, C250 and D368) that are required for the kinase activity of WNK1 (48) are marked by flags on the WNK1 protein.
Our automated procedure identified a novel kidney-specific alternative splicing event between exons IV and V (Fig. 4B). For the isoform we detected outside kidney our deduced protein sequence is identical to the reported protein sequence (49). Exons IV, V and VI encode the second half of the kinase domain of this enzyme (Fig. 4C). In the kidney-specific isoform exon IV is replaced by exon IVa, drastically altering the protein sequence. It contains a 63 nt upstream in-frame stop codon (UGA), as well as a subsequent start codon (AUG) for an ORF that extends in-frame into the rest of the normal protein sequence in exons V onwards. Thus it is likely that exon IVa encodes an alternative 30 amino acid N-terminus of the WNK1 protein, replacing 384 amino acids of the usual WNK1 protein N-terminus. This appears to disrupt the kinase domain and to eliminate WNK1 kinase activity specifically in kidney. Our data show eight ESTs aligning with exon IVa and extending up to 42 nt upstream of the AUG start codon. However, because ESTs are short fragments, in this case they do not extend to a 3′ splice site and thus do not show where the beginning of exon IVa might be. However, in the 21 nt between the in-frame stop codon and the start of the first EST alignment there is no consensus 3′ splice site (polypyrimidine tract + AG). Thus, even if exon IVa is spliced to upstream exons, the in-frame stop codon would evidently be included and the kinase domain would be removed from the WNK1 protein product. It is also possible that exon IVa represents an alternative promoter site.
Although this is a novel discovery, there is experimental data that support it. Previous studies of the expression of WNK1 have reported both an 11–12 kb band observed ubiquitously in many tissues and a 9.5–10 kb band expressed at high level in the kidney (47,49). This truncated transcript is consistent with the kidney-specific exon IVa alternative splice that we have identified, but the reported band has not been further characterized. In 7–10 ESTs we have also detected alternative splicing in a variety of tissues omitting exons IX, XI and/or XII (and a single EST omitting exon XXVI) in WNK1. Exons IX, XI and XII have been independently reported to be alternatively spliced (48,49).
Bioinformatics analysis of a known splice variant
To illustrate further the value of our database, Figure 5 shows our bioinformatics analysis for CDC42 (cell division cycle 42). Many studies of this gene provide insight into its splicing and isoforms (50–54). CDC42 is a member of the Rho family, which is a group of small GTPases. CDC42 plays multiple functional roles in cell regulation (51).
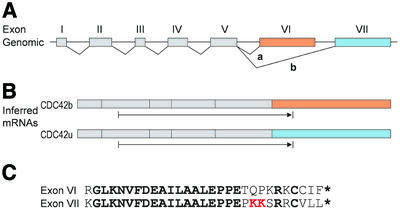
Figure 5.
Brain-specific alternative splicing of CDC42. (A) Genomic structure of the CDC42 gene. Exons are shown as boxes and colors show alternative exons. Splice a is specific to brain. (B) The two alternative forms of CDC42 mRNA inferred from the expressed sequence data. The protein coding region is indicated by an arrow beneath each form. (C) Alignment between the protein sequences encoded by the alternative exons VI and VII of CDC42. Conserved amino acids are in bold. The dilysine motif is indicated in red and the stop codons by asterisks.
Our automated analysis detected an alternative splice in CDC42, with one splice form showing brain specificity. Our forms matched the two isoforms reported in the literature, CDC42b and CDC42u. In agreement with our EST results, CDC42b has been reported to be expressed exclusively in brain, while CDC42u is expressed in a wide variety of tissues (54).
Bioinformatics analysis of these forms was revealing. ORF prediction for the two splice forms produced almost identical protein sequences. The brain-specific alternative splice replaced exon VII (which encodes the protein C-terminus) with a new exon (VI) as the last exon. Remarkably, exon VI supplied an almost identical C-terminal sequence (Fig. 5C), of exactly the same length and with 20 of 29 amino acids identical. The divergent nine amino acids include a C-terminal dilysine motif of retrieval receptors that has been shown to be critical for binding to coatomer complex (COP) in the endoplasmic reticulum and to cargo receptors in the Golgi apparatus (55–57). CDC42u has this motif at positions 183–184, but our analysis indicated that CDC42b eliminates this motif, replacing it with glutamine and proline (QP). This suggests that the brain-specific splicing blocks CDC42 binding to COP in brain.
This novel hypothesis is consistent with available experimental data. In vitro site-directed mutagenesis of the CDC42u sequence, replacing lysines 183–184 with serine, eliminated binding of the γ-COP subunit (52). Moreover, it has been shown that binding of γ-COP is necessary for CDC42 to induce malignant transformation (52), suggesting that this brain-specific splice has functional importance.
DISCUSSION
Our results can be useful to biologists in several ways. First, they provide a validated, automatic method for large-scale discovery of tissue-specific alternative splicing, which can be applied to many EST and other datasets where tissue information is available. Second, we have discovered 667 tissue-specific splice forms in the human transcriptome. Our comparisons with the published literature suggest that up to 78% of our tissue specificity findings are novel. These data can furnish biologists with many new functional insights into well-studied genes (by identifying a novel tissue-specific splice form), which can be of great interest for further experimental study. Our data can also provide interesting functional suggestions for unknown genes, since observation of tissue specificity (combined with other information, such as homology) may itself suggest fruitful directions for research. Moreover, the large scale of alternative splice impact on the protein product (e.g. removal or addition of a domain) often yields interpretable functional implications (e.g. removal of a DNA-binding activity, as in IRF3). Finally, researchers who study regulation of splicing can benefit from this large, searchable database of tissue-specific alternative splicing spanning many distinct tissue types.
Benefits and deficiencies of our approach
Our use of readily available expressed sequence data has both advantages and disadvantages. One of the biggest problems with ESTs is their fragmentary character; the difficulty of interpreting individual ESTs (because they are tiny fragments rather than full-length cDNAs) and the high rate of errors in their sequencing and clustering. To address this, we perform a rigorous, comprehensive analysis of the total set of all EST data, which does not assume confidence in single ESTs. Multiple forms of evidence are required. This process depends on making a complete, clearly interpretable match between the genomic sequence of a gene and the total set of ESTs that map to that gene location, carefully considering many details such as intronic splice junction sequences and current knowledge about mechanisms of alternative splicing (7). Our procedure is conservative (designed to avoid false positives) in the sense that if a given set of ESTs does not fit its rigid model, they are simply excluded from our analysis. This procedure is more likely to give false negative errors (real alternative splices that are missed by the analysis) than false positives (reported alternative splices that are incorrect).
An advantage of using ESTs for alternative splice detection is that this furnishes exact sequence information for the novel splice form, in many cases suggesting a clearly interpretable functional effect. In contrast, northern blot, PCR-based and microarray hybridization-based methods do not directly read out the sequence of a novel form as sequencing does and instead would require indirect means to infer a sequence. For example, this can be done by using the genomic sequence to predict what splice forms are possible and then correlate the observed transcript sizes (on a northern blot) or hybridization signals (on a microarray) versus these forms. Thus these experiments are most interpretable when the set of probes has been carefully designed to distinguish the particular set of alternative forms that are expected. EST data, on the other hand, can readily detect an unexpected new form by providing a direct readout of its sequence.
This is particularly helpful for interpretation of newly detected forms. Knowing the exact sequence change that an alternative splice produces is the difference between simply having a new ‘band on a gel’ versus being able to apply the full resources of sequence analysis and available literature to interpreting its likely functional impact. For example, for WNK1, previous studies may have detected our kidney-specific WNK1 isoform (reported only as a 9.5–10 kb band; 48,49), but have not published data giving this functional significance. In contrast, detection of a kidney-specific splice form by our automatic procedure immediately suggested an interesting functional impact due to truncation of the highly conserved N-terminal kinase domain.
Our approach has many disadvantages. Our results are likely to be far from complete, in that they may not include many genes that do have tissue-specific splice forms. We have designed our approach to reduce the rate of false positives (incorrectly reporting a splice form to be tissue-specific), by accepting a much higher rate of false negatives (failing to report a splice form that actually is tissue-specific). We have tried to set the cut-off score for our HC group high enough to avoid a high rate of false positives, and the independent validation results (80% validation) support this. However, this means that a number of genes with tissue-specific alternative splicing may be missed, either because of inadequate EST data or overly conservative scoring. Our TS scoring function is based on rigorous statistical inference methods, but is far from perfect in dealing with the many possible problems in this data. For example, the TS metric quantifies the evidence that a given splice form is preferred (>50% frequency) in a specific tissue and not preferred in the pool of all other tissues. It is certainly possible that a splice form could be tissue-specific (e.g. found only in brain) and still be a minor splice form in that tissue (i.e. <50% frequency). The TS metric can miss such cases and thus is not a completely satisfactory definition of tissue specificity. This could be corrected by a simple Z-test of the null hypothesis that the proportion of splice types is the same in each tissue. More fundamentally, our procedure depends on pooling (combining many different libraries into one tissue class) to get enough counts for each class so the results will be statistically significant. Pooling can obscure real specificity signals if the libraries that are being pooled as one tissue actually have very different patterns of splice form expression. Our current metric does not address this.
However, while our tissue specificity scoring could be improved, we suspect that problems in the very nature of the EST data and libraries are more serious. These problems are as follows.
Poor coverage. There are relatively few ESTs in a given region of a gene, and the problem only gets worse when we subdivide by the 46 tissue classes. Statistically significant results for a single tissue specificity typically require 5–10 ESTs at that alternative splice point. The bottom line is that no improvement in theory will make up for the lack of sufficient experimental data.
Sample specificity. Even if we had sufficient data and theory to detect specificity at the level of individual EST libraries, they are themselves pools of many tissues. Most EST libraries represent at best an entire organ (e.g. ‘brain’). Some types of tissue (e.g. epithelium) may be present in many different organs, further confusing the picture. Is tissue specificity predominantly at the scale of an entire organ or at the scale of specific cell types and differentiation states? The latter kind of tissue specificity will generally be hard to detect in the existing EST data. Consistent with this hypothesis, the frequency with which we discovered tissue-specific splice forms in brain was less than half the frequency in a more specific neuronal tissue (retina). For these reasons, our measured rate of tissue-specific alternative splicing probably underestimates its true extent.
Fragmentation. Because EST sequences represent fragments of a transcript rather than a full-length sequence, these data can detect individual alternative splicing events but cannot necessarily distinguish whether they are combined in a single transcript molecule. Indeed, this is a general problem for most methods of detecting alternative splicing, such as probe hybridization (e.g. splicing arrays; 28–30) or protein fragment mass spectrometry. Only full-length sequencing of a carefully subcloned mRNA can resolve with certainty the exact combination of splices in a single transcript molecule. For this reason, in this paper we have avoided the term ‘isoform’ (implying a particular full-length transcript form) and instead use the term ‘splice form’ to indicate the set of transcripts containing a particular alternative splice S1 (see Materials and Methods for a detailed definition). This is what the EST data can show, not full-length isoforms.
Working with EST data, our method can handle cases where more than two alternative splice forms are detected in a gene, but it cannot distinguish whether they are combined in a coordinated way. Since multiple alternative splicing events are observed as independent events in the EST fragment data, our method treats them as independent events in its scoring, i.e. it calculates TS scores independently for each alternative splicing event (i.e. pair of mutually exclusive splices) in the gene. If multiple alternative splices were combined in a coordinated way (revealed by full-length transcript sequences), our method would still correctly detect their tissue specificity patterns individually. However, it would be up to the user to notice that they all had the same pattern and indeed were all observed in the same transcripts.
A database of novel biological discoveries and functional implications
We believe our database of 667 tissue-specific splice forms can be a rich source of discovery for researchers studying human biology and disease. While there is growing interest in alternative splicing and tissue-specific splice forms, there are relatively few large-scale information resources for this field, compared with other areas such as genome annotation/gene discovery (databases such as Ensembl), polymorphism (databases such as dbSNP) or the study of transcriptional regulation (databases such as TRANSFAC). In addition to several databases of alternative splicing from the literature or ESTs (2–4,6,7,58–63), there are databases of known tissue-specific alternative splicing. The Alternative Exon Database (http://cgsigma.cshl.org/new_alt_exon_db2) includes 379 human alternative splices, of which 30 exons (in 19 genes) are reported to be specific to brain, muscle or two tissues (26). As part of a computational analysis of candidate intronic splice regulatory elements, Brudno et al. (25) created a collection of 25 brain-specific alternative spliced exons. However, at this time there is still no single resource where one can go to reliably find all splice form specificities reported in the literature. For example, to seek validation for our brain- and muscle-specific forms we had to perform extensive manual literature searches. Our database can make some contribution to this, since it contains approximately 167 tissue specificities that are likely to be already known.
However, its major value is providing previously unknown alternative splice forms that show tissue specificity: approximately 500 with high confidence and 2200 that show evidence of tissue specificity with low confidence. Furthermore, the EST data are continuing to grow. From February 2001 to January 2002, for example, the human EST data grew from 2.4 million to 3 million, a 27% increase over 11 months. Furthermore, ESTs from many other organisms are also being sequenced. Thus, the method presented in this paper can be applied in the future to much more EST data, to greatly expand the database of tissue-specific alternative splice forms.
Our database can provide biologists with valuable bioinformatics analyses that suggest hypotheses about function. As we have showed in a number of cases, the large-scale changes in the protein product produced by alternative splicing often make them interpretable enough to suggest exciting ideas that merit further experimental tests. Our database provides biologists with essential information for interpreting functional impact, such as inferred protein isoform sequences and predicted changes in protein domain composition based on conserved domain databases such as SMART and PFAM. We will continue to add useful analyses, such as transmembrane domain prediction (64), localization signal analysis, etc.
Our data can also be useful for studies of the regulation of mRNA splicing. Given the relatively small number of genes and tissues in which the mechanistic details of splice regulation have been studied carefully (for a review see 65), it seems likely that a large new database of tissue-specific alternative splicing can be a valuable resource for the field. First of all, most of our tissue-specific splice form data appear to be novel, providing researchers with many new cases of tissue-specific splicing to work on. Second, it spans a large number of genes (454 in the HC group, 1572 in the LC group) with different functions and gene structures, giving researchers a very diverse set to study. Third, it spans a large number of tissues, providing many more examples for tissue specificities that have been previously studied (e.g. neuronal), as well as many examples of tissue specificities that have not been studied in full mechanistic detail before. Finally, our database provides a lot of useful information for mechanistic studies, including the genomic sequence for each gene and the detailed evidence from expressed sequences for each exon– intron junction and alternative splice. In principle this provides the information needed not only for designing appropriate experimental strategies to study the regulation of these splice forms (e.g. probe sequences, PCR primers, etc.), but also for searching for possible binding sites for known or novel splice regulatory factors. The dataset is large enough (e.g. 213 HC brain- and retina-specific alternative splices) that statistical analysis might be useful for detecting novel tissue specificity motifs.
Observation of tissue specificity also adds valuable information to de novo alternative splice discovery databases. An extremely challenging problem for the field is how to validate novel splice forms efficiently. What fraction of the enormous new datasets of alternative splicing discovery (e.g. 27 790 in this paper) represent real biological forms of functional regulation, as opposed to experimental or bioinformatics artifacts (27,66)? This is not an easy question to answer. High throughput technologies such as microarrays can help address part of this, by providing much more experimental data indicating that these individual forms really are abundant in cells. However, simply showing that a form is present does not prove that it has functional importance for biology.
Observation of tissue specificity is one good starting point for answering this harder functional question. First of all, the very fact of observing tissue specificity demonstrates a non-random pattern in the putative alternative splicing data. This makes it much less likely that a given splice form (e.g. detected by an automated procedure such as ours) is simply an experimental or bioinformatics error. Secondly, evidence of biological regulation (i.e. that a form is tissue-specific) can itself be taken as evidence of participation in a functional process. Thus, one way to look at our data is to assert that the most interesting and most reliable alternative splices from our automatic detection procedure (out of the total of 27 790) are simply those for which we found a tissue specificity. These are probably the most fruitful starting point for further experimental studies, both by individual researchers and by high throughput technologies such as microarrays.
Medical interest of tissue-specific alternative splicing: WNK1
For example, there is some evidence that the kidney-specific alternative splicing of WNK1 discovered in this paper could have medical importance. A genome scan of hypertension patients in a family study identified WNK1 as a genetic cause of pseudohypoaldosteronism type II (PHAII) hypertension and found a specific mutation of WNK1 in a large proportion of the patients (49). The homologous gene WNK4 shares the same key features (conserved kinase domain and two coiled-coil domains), and mutations in WNK4 have also been linked to PHAII hypertension. Consistent with this hypothesis and the chloride-dependent character of PHAII hypertension, the WNK1 and WNK4 proteins have been localized to the distal renal tubules of the kidney, which play a key role in maintaining the body’s electrolyte balance (49). WNK4 appears to be expressed exclusively in kidney (49).
Our discovery of a kidney-specific disruption of the kinase domain of WNK1 by alternative splicing suggests a possible hypothesis about the pathogenesis of PHAII hypertension. Normal WNK1 function (including the kinase) should be expressed only outside the kidney. Within kidney, WNK1 function is restricted or altered by disruption of its kinase domain, whose activity is replaced by that of WNK4. An unusual feature of WNK1 is that the WNK1 mutation observed in PHAII patients is a deletion in the intron between exons I and II, which ordinarily would have no effect on the protein. On the other hand, this could have an important effect on regulation of alternative splicing. Deletion of intronic splice regulatory elements could lead to misregulation (67) or even loss of the tissue-specific splicing of the WNK1 transcript. For example, if PHAII patients produce the normal isoform of WNK1 in kidney, they will express fully functional WNK1 in their kidneys in addition to WNK4. This dosage effect could alter regulatory balances in the kidney and cause hypertension.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank D. Black, G. Chanfreau, A. Resch and Y. Xing for their helpful discussions and comments on this work. This material is based upon work supported by the National Science Foundation under grant no. 0082964. C.L. was supported by Department of Energy grant DEFG0387 ER60615. B.M. is a pre-doctoral trainee supported by NSF IGERT Award DGE-9987641.
REFERENCES
- 1.Gilbert W. (1978) Why genes in pieces? Nature, 271, 501. [DOI] [PubMed] [Google Scholar]
- 2.Mironov A.A., Fickett,J.W. and Gelfand,M.S. (1999) Frequent alternative splicing of human genes. Genome Res., 9, 1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brett D., Hanke,J., Lehmann,G., Haase,S., Delbruck,S., Krueger,S., Reich,J. and Bork,P. (2000) EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett., 474, 83–86. [DOI] [PubMed] [Google Scholar]
- 4.Croft L., Schandorff,S., Clark,F., Burrage,K., Arctander,P. and Mattick,J.S. (2000) ISIS, the intron information system, reveals the high frequency of alternative splicing in the human genome. Nature Genet., 24, 340–341. [DOI] [PubMed] [Google Scholar]
- 5.International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 6.Kan Z., Rouchka,E.C., Gish,W.R. and States,D.J. (2001) Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res., 11, 889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Modrek B., Resch,A., Grasso,C. and Lee,C. (2001) Genome-wide analysis of alternative splicing using human expressed sequence data. Nucleic Acids Res., 29, 2850–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joseph R., Dou,D. and Tsang,W. (1995) Neuronatin mRNA: alternatively spliced forms of a novel brain-specific mammalian developmental gene. Brain Res., 690, 92–98. [DOI] [PubMed] [Google Scholar]
- 9.Chen C.D., Kobayashi,R. and Helfman,D.M. (1999) Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat beta-tropomyosin gene. Genes Dev., 13, 593–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karpova A.Y., Ronco,L.V. and Howley,P.M. (2001) Functional characterization of interferon regulatory factor 3a (IRF-3a), an alternative splice isoform of IRF-3. Mol. Cell. Biol., 21, 4169–4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Philips A.V. and Cooper,T.A. (2000) RNA processing and human disease. Cell. Mol. Life Sci., 57, 235–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gunthert U., Hofmann,M., Rudy,W., Reber,S., Zoller,M., Haussmann,I., Matzku,S., Wenzel,A., Ponta,H. and Herrlich,P. (1991) A new variant of glycoprotein CD44 confers metastatic potential to rat carcinoma cells. Cell, 65, 13–24. [DOI] [PubMed] [Google Scholar]
- 13.Mottes J.R. and Iverson,L.E. (1995) Tissue-specific alternative splicing of hybrid Shaker/lacZ genes correlates with kinetic differences in Shaker K+ currents in vivo. Neuron, 14, 613–623. [DOI] [PubMed] [Google Scholar]
- 14.Wilson C.A., Payton,M.N., Elliott,G.S., Buaas,F.W., Cajulis,E.E., Grosshans,D., Ramos,L., Reese,D.M., Slamon,D.J. and Calzone,F.J. (1997) Differential subcellular localization, expression and biological toxicity of BRCA1 and the splice variant BRCA1-delta11b. Oncogene, 14, 1–16. [DOI] [PubMed] [Google Scholar]
- 15.Crook R., Verkkoniemi,A., Perez-Tur,J., Mehta,N., Baker,M., Houlden,H., Farrer,M., Hutton,M., Lincoln,S., Hardy,J. et al. (1998) A variant of Alzheimer’s disease with spastic paraparesis and unusual plaques due to deletion of exon 9 of presenilin 1. Nature Med., 4, 452–455. [DOI] [PubMed] [Google Scholar]
- 16.Jiang Z.H. and Wu,J.Y. (1999) Alternative splicing and programmed cell death. Proc. Soc. Exp. Biol. Med., 220, 64–72. [DOI] [PubMed] [Google Scholar]
- 17.Dreyfuss G., Matunis,M.J., Pinol-Roma,S. and Burd,C.G. (1993) hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem., 62, 289–321. [DOI] [PubMed] [Google Scholar]
- 18.Fu X.D. (1995) The superfamily of arginine/serine-rich splicing factors. RNA, 1, 663–680. [PMC free article] [PubMed] [Google Scholar]
- 19.Manley J.L. and Tacke,R. (1996) SR proteins and splicing control. Genes Dev., 10, 1569–1579. [DOI] [PubMed] [Google Scholar]
- 20.Krecic A.M. and Swanson,M.S. (1999) hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol., 11, 363–371. [DOI] [PubMed] [Google Scholar]
- 21.Spingola M., Grate,L., Haussler,D. and Ares,M.J. (1999) Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA, 5, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis C.A., Grate,L., Spingola,M. and Ares,M.J. (2000) Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast. Nucleic Acids Res., 28, 1700–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen K.B., Dredge,B.K., Stefani,G., Zhong,R., Buckanovich,R.J., Okano,H.J., Yang,Y.Y. and Darnell,R.B. (2000) Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron, 25, 359–371. [DOI] [PubMed] [Google Scholar]
- 24.Markovtsov V., Nikolic,J.M., Goldman,J.A., Turck,C.W., Chou,M.Y. and Black,D.L. (2000) Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol., 20, 7463–7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brudno M., Gelfand,M.S., Splengler,S., Zorn,M., Dubchak,I. and Conboy,J.G. (2001) Computational analysis of candidate intron regulatory elements for tissue-specific alternative pre-mRNA splicing. Nucleic Acids Res., 29, 2338–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stamm S., Zhu,J., Nakai,K., Stoilov,P., Stoss,O. and Zhang,M.Q. (2000) An alternative-exon database and its statistical analysis. DNA Cell Biol., 19, 739–756. [DOI] [PubMed] [Google Scholar]
- 27.Modrek B. and Lee,C. (2002) A genomic view of alternative splicing. Nature Genet., 30, 13–19. [DOI] [PubMed] [Google Scholar]
- 28.Hu G.K., Madore,S.J., Moldover,B., Jatkoe,T., Balaban,D., Thomas,J. and Wang,Y. (2001) Predicting splice variant from DNA chip expression data. Genome Res., 11, 1237–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark T.A., Sugnet,C.W. and Ares,M.J. (2002) Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science, 296, 907–910. [DOI] [PubMed] [Google Scholar]
- 30.Yeakley J.M., Fan,J.B., Doucet,D., Luo,L., Wickham,E., Ye,Z., Chee,M.S. and Fu,X.D. (2002) Profiling alternative splicing on fiber-optic arrays. Nat. Biotechnol., 20, 353–358. [DOI] [PubMed] [Google Scholar]
- 31.Schuler G. (1997) Pieces of the puzzle: expressed sequence tags and the catalog of human genes. J. Mol. Med., 75, 694–698. [DOI] [PubMed] [Google Scholar]
- 32.Lee C. and Irizarry,K. (2001) The GeneMine system for genome/proteome annotation and collaborative data-mining. IBM Syst. J., 40, 592–603. [Google Scholar]
- 33.Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schultz J., Milpetz,F., Bork,P. and Ponting,C.P. (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl Acad. Sci. USA, 95, 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sonnhammer E.L., Eddy,S.R. and Durbin,R. (1997) Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins, 28, 405–420. [DOI] [PubMed] [Google Scholar]
- 36.Kunsch H.R. (1989) The jackknife and the bootstrap for general stationary observations. Ann. Stat., 17, 1217–1241. [Google Scholar]
- 37.Wathelet M.G., Lin,C.H., Parekh,B.S., Ronco,L.V., Howley,P.M. and Maniatis,T. (1998) Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol. Cell, 1, 507–518. [DOI] [PubMed] [Google Scholar]
- 38.Lin R., Heylbroeck,C., Genin,P., Pitha,P.M. and Hiscott,J. (1999) Essential role of interferon regulatory factor 3 in direct activation of RANTES chemokine transcription. Mol. Cell. Biol., 19, 959–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karpova A.Y., Howley,P.M. and Ronco,L.V. (2000) Dual utilization of an acceptor/donor splice site governs the alternative splicing of the IRF-3 gene. Genes Dev., 14, 2813–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weaver B.K., Kumar,K.P. and Reich,N.C. (1998) Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol. Cell. Biol., 18, 1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoneyama M., Suhara,W., Fukuhara,Y., Fukuda,M., Nishida,E. and Fujita,T. (1998) Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J., 17, 1087–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akwa Y., Hassett,D.E., Eloranta,M.L., Sandberg,K., Masliah,E., Powell,H., Whitton,J.L., Bloom,F.E. and Campbell,I.L. (1998) Transgenic expression of IFN-alpha in the central nervous system of mice protects against lethal neurotropic viral infection but induces inflammation and neurodegeneration. J. Immunol., 161, 5016–5026. [PubMed] [Google Scholar]
- 43.Campbell I.L., Krucker,T., Steffensen,S., Akwa,Y., Powell,H.C., Lane,T., Carr,D.J., Gold,L.H., Henriksen,S.J. and Siggins,G.R. (1999) Structural and functional neuropathology in transgenic mice with CNS expression of IFN-alpha. Brain Res., 835, 46–61. [DOI] [PubMed] [Google Scholar]
- 44.Grabowski P.J. and Black,D.L. (2001) Alternative RNA splicing in the nervous system. Prog. Neurobiol., 65, 289–308. [DOI] [PubMed] [Google Scholar]
- 45.Seya T., Hirano,A., Matsumoto,M., Nomura,M. and Ueda,S. (1999) Human membrane cofactor protein (MCP, CD46): multiple isoforms and functions. Int. J. Biochem. Cell Biol., 31, 1255–1260. [DOI] [PubMed] [Google Scholar]
- 46.Smith C.W.J. and Valcarcel,J. (2000) Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci., 25, 381–388. [DOI] [PubMed] [Google Scholar]
- 47.Xu B., English,J.M., Wilsbacher,J.L., Stippec,S., Goldsmith,E.J. and Cobb,M.H. (2000) WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. J. Biol. Chem., 275, 16795–16801. [DOI] [PubMed] [Google Scholar]
- 48.Verissimo F. and Jordan,P. (2001) WNK kinases, a novel protein kinase subfamily in multi-cellular organisms. Oncogene, 20, 5562–5569. [DOI] [PubMed] [Google Scholar]
- 49.Wilson F.H., Disse-Nicodeme,S., Choate,K.A., Ishikawa,K., Nelson-Williams,C., Desitter,I., Gunel,M., Milford,D.V., Lipkin,G.W., Achard,J.M. et al. (2001) Human hypertension caused by mutations in WNK kinases. Science, 293, 1107–1112. [DOI] [PubMed] [Google Scholar]
- 50.Nicole S., White,P.S., Topaloglu,H., Beigthon,P., Salih,M., Hentati,F. and Fontaine,B. (1999) The human CDC42 gene: genomic organization, evidence for the existence of a putative pseudogene and exclusion as a SJS1 candidate gene. Hum. Genet., 105, 98–103. [DOI] [PubMed] [Google Scholar]
- 51.Erickson J.W. and Cerione,R.A. (2001) Multiple roles for Cdc42 in cell regulation. Curr. Opin. Cell Biol., 13, 153–157. [DOI] [PubMed] [Google Scholar]
- 52.Wu W.J., Erickson,J.W., Lin,R. and Cerione,R.A. (2000) The gamma-subunit of the coatomer complex binds Cdc42 to mediate transformation. Nature, 405, 800–804. [DOI] [PubMed] [Google Scholar]
- 53.Mott H.R., Owen,D., Nietlispach,D., Lowe,P.N., Manser,E., Lim,L. and Laue,E.D. (1999) Structure of the small G protein Cdc42 bound to the GTPase-binding domain of ACK. Nature, 399, 384–388. [DOI] [PubMed] [Google Scholar]
- 54.Marks P.W. and Kwiatkowski,D.J. (1996) Genomic organization and chromosomal location of murine Cdc42. Genomics, 38, 13–18. [DOI] [PubMed] [Google Scholar]
- 55.Harter C., Pavel,J., Coccia,F., Draken,E., Wegehingel,S., Tschochner,H. and Wieland,F. (1996) Nonclathrin coat protein gamma, a subunit of coatomer, binds to the cytoplasmic dilysine motif of membrane proteins of the early secretory pathway. Proc. Natl Acad. Sci. USA, 93, 1902–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Letourneur F., Gaynor,E.C., Hennecke,S., Demolliere,C., Duden,R., Emr,S.D., Riezman,H. and Cosson,P. (1994) Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell, 79, 1199–1207. [DOI] [PubMed] [Google Scholar]
- 57.Harter C. and Wieland,F.T. (1998) A single binding site for dilysine retrieval motifs and p23 within the gamma subunit of coatomer. Proc. Natl Acad. Sci. USA, 95, 11649–11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stamm S., Zhang,M.Q., Marr,T.G. and Helfman,D.M. (1994) A sequence compilation and comparison of exons that are alternatively spliced in neurons. Nucleic Acids Res., 22, 1515–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burke J., Wang,H., Hide,W. and Davison,D.B. (1998) Alternative gene form discovery and candidate gene selection from gene indexing projects. Genome Res., 8, 276–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mangan M.E. and Frazer,K.S. (1999) An extensive list of genes that produce alternative transcripts in the mouse. Bioinformatics, 15, 170–171. [DOI] [PubMed] [Google Scholar]
- 61.Dralyuk I., Brudno,M., Gelfand,M.S., Zorn,M. and Dubchak,I. (2000) ASDB: database of alternatively spliced genes. Nucleic Acids Res., 28, 296–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kent W.J. and Zahler,A.M. (2000) The intronerator: exploring introns and alternative splicing in Caenorhabditis elegans. Nucleic Acids Res., 28, 91–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ji H., Zhou,Q., Wen,F., Xia,H., Lu,X. and Li,Y. (2001) AsMamDB: an alternative splice database of mammals. Nucleic Acids Res., 29, 260–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sonnhammer E.L., von Heijne,G. and Krogh,A. (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. ISMB, 6, 175–182. [PubMed] [Google Scholar]
- 65.Lopez A.J. (1998) Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet., 32, 279–305. [DOI] [PubMed] [Google Scholar]
- 66.Skandalis A., Ninniss,P.J., McCormac,D. and Newton,L. (2002) Spontaneous frequency of exon skipping in the human HPRT gene. Mutat. Res., 501, 37–44. [DOI] [PubMed] [Google Scholar]
- 67.Pagani F., Buratti,E., Stuani,C., Bendix,R., Dork,T. and Baralle,F.E. (2002) A new type of mutation causes a splicing defect in ATM. Nature Genet., 30, 426–429. [DOI] [PubMed] [Google Scholar]
- 68.Lin Z., Carney,G. and Rizzo,W.B. (2000) Genomic organization, expression, and alternate splicing of the mouse fatty aldehyde dehydrogenase gene. Mol. Genet. Metab., 71, 496–505. [DOI] [PubMed] [Google Scholar]
- 69.Rizzo W.B., Lin,Z. and Carney,G. (2001) Fatty aldehyde dehydrogenase: genomic structure, expression and mutation analysis in Sjogren-Larsson syndrome. Chem. Biol. Interact., 130, 297–307. [DOI] [PubMed] [Google Scholar]
- 70.Lin B., Pan,C.J. and Chou,J.Y. (2000) Human variant glucose-6-phosphate transporter is active in microsomal transport. Hum. Genet., 107, 526–529. [DOI] [PubMed] [Google Scholar]
- 71.Ihara K., Nomura,A., Hikino,S., Takada,H. and Hara,T. (2000) Quantitative analysis of glucose-6-phosphate translocase gene expression in various human tissues and haematopoietic progenitor cells. J. Inherit. Metab. Dis., 23, 583–592. [DOI] [PubMed] [Google Scholar]
- 72.He G.S. and Grabowski,G.A. (1992) Gaucher disease: a G+1----A+1 IVS2 splice donor site mutation causing exon 2 skipping in the acid beta-glucosidase mRNA. Am. J. Hum. Genet., 51, 810–820. [PMC free article] [PubMed] [Google Scholar]
- 73.Lee P.L., Gelbart,T., West,C., Halloran,C. and Beutler,E. (1998) The human Nramp2 gene: characterization of the gene structure, alternative splicing, promoter region and polymorphisms. Blood Cells Mol. Dis., 24, 199–215. [DOI] [PubMed] [Google Scholar]
- 74.Zhang L., Lee,T., Wang,Y. and Soong,T.W. (2000) Heterologous expression, functional characterization and localization of two isoforms of the monkey iron transporter Nramp2. Biochem. J., 349, 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lambrechts A., Braun,A., Jonckheere,V., Aszodi,A., Lanier,L.M., Robbens,J., Van Colen,I., Vandekerckhove,J., Fassler,R. and Ampe,C. (2000) Profilin II is alternatively spliced, resulting in profilin isoforms that are differentially expressed and have distinct biochemical properties. Mol. Cell. Biol., 20, 8209–8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Di Nardo A., Gareus,R., Kwiatkowski,D. and Witke,W. (2000) Alternative splicing of the mouse profilin II gene generates functionally different profilin isoforms. J. Cell Sci., 113, 3795–3803. [DOI] [PubMed] [Google Scholar]
- 77.Pret A.M., Balvay,L. and Fiszman,M.Y. (1999) Regulated splicing of an alternative exon of beta-tropomyosin pre-mRNAs in myogenic cells depends on the strength of pyrimidine-rich intronic enhancer elements. DNA Cell Biol., 18, 671–683. [DOI] [PubMed] [Google Scholar]
- 78.Ng E.K., Lee,S.M., Li,H.Y., Ngai,S.M., Tsui,S.K., Waye,M.M., Lee,C.Y. and Fung,K.P. (2001) Characterization of tissue-specific LIM domain protein (FHL1C) which is an alternatively spliced isoform of a human LIM-only protein (FHL1). J. Cell. Biochem., 82, 1–10. [DOI] [PubMed] [Google Scholar]
- 79.Crawford D., Hagerty,K. and Beutler,B. (1989) Multiple splice forms of ribonuclease-inhibitor mRNA differ in the 5′-untranslated region. Gene, 85, 525–531. [DOI] [PubMed] [Google Scholar]