Abstract
Human single nucleotide polymorphisms (SNPs) represent the most frequent type of human population DNA variation. One of the main goals of SNP research is to understand the genetics of the human phenotype variation and especially the genetic basis of human complex diseases. Non-synonymous coding SNPs (nsSNPs) comprise a group of SNPs that, together with SNPs in regulatory regions, are believed to have the highest impact on phenotype. Here we present a World Wide Web server to predict the effect of an nsSNP on protein structure and function. The prediction method enabled analysis of the publicly available SNP database HGVbase, which gave rise to a dataset of nsSNPs with predicted functionality. The dataset was further used to compare the effect of various structural and functional characteristics of amino acid substitutions responsible for phenotypic display of nsSNPs. We also studied the dependence of selective pressure on the structural and functional properties of proteins. We found that in our dataset the selection pressure against deleterious SNPs depends on the molecular function of the protein, although it is insensitive to several other protein features considered. The strongest selective pressure was detected for proteins involved in transcription regulation.
INTRODUCTION
A considerable effort is underway to relate human phenotypes to variation at the DNA level. Most human genetic variation is represented by single nucleotide polymorphisms (SNPs) and many of them are believed to cause phenotypic differences between individuals. However, identifying SNPs responsible for specific phenotypes appears to be a problem that is very difficult to solve.
The concept of association studies has been proposed as an experimental technique to identify SNPs underlying complex phenotypes, mostly human multifactorial disorders (1). The question of study design is, however, disputable. Linkage disequilibrium-based whole genome scanning (2,3) has the advantage of being a completely hypothesis-free approach, though possibly too demanding because of the extraordinary number of markers to be screened. Candidate gene studies (2,4) try to reduce the number of SNPs to those from genes most likely to constitute the genetic basis of the disease. Although, even in the latter case, especially if large sets of candidate genes are considered, multiple testing of hundreds and even thousands of SNPs makes detection of the association difficult.
A possible way to overcome the problem of testing overwhelming numbers of SNPs, especially in the case of candidate gene studies, would be to prioritise SNPs according to their functional significance (4,5). As a priori biological knowledge can be used to reduce the number of SNPs by focusing on specific genomic regions or gene sets, bioinformatics expertise may help to discriminate between neutral SNPs, which constitute the majority of genetic variation, and SNPs of likely functional importance. Below, we specifically focus on non-synonymous SNPs (nsSNPs), i.e. SNPs located in coding regions and resulting in amino acid variation in the protein products of genes. It has been shown in several recent studies (6–11) that the impact of amino acid allelic variants on protein structure and function can be predicted by analysis of multiple sequence alignments and protein 3D structures. As we demonstrated in an earlier work, these predictions correlate with the effect of natural selection seen as an excess of rare alleles (7,12). Therefore, predictions at the molecular level reveal SNPs affecting actual phenotypes.
Here we present: (i) a Web server for annotation of functional nsSNPs (www.bork.embl-heidelberg.de/Poly Phen); (ii) a dataset of nsSNPs extracted from a public SNP database, HGVbase (13) (www.bork.embl-heidelberg.de/PolyPhen/data); (iii) an analysis of these data with regard to predicted effect on protein structure and function.
Prioritisation of SNPs in the candidate gene approach is not the only suggested use of the PolyPhen (polymorphism phenotyping) server and the collection of nsSNPs. The server could also be useful to reveal the structural basis of disease mutations and explain the molecular cause of a disease. This might help in some cases to identify the causative allelic variant (14) after a disease has been linked to a particular locus.
On the other hand, since numerous disease associations published recently could not be confirmed by subsequent independent studies (2,4), the independent evidence of functionality of a nsSNP could be an additional argument to discriminate true associations from false positives.
Analysis of the database of nsSNPs enabled us to test whether certain characteristics of proteins are associated with accumulation of nsSNPs (especially slightly deleterious nsSNPs).
MATERIALS AND METHODS
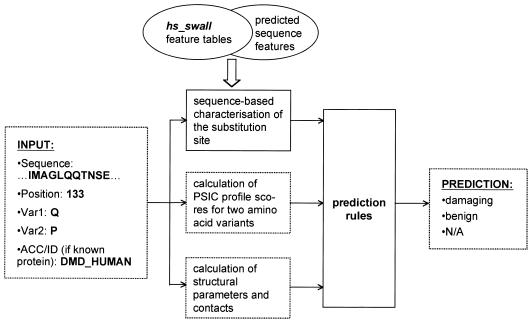
PolyPhen is a World Wide Web server devoted to automated functional annotation of coding nsSNPs. PolyPhen input is the amino acid sequence of a protein or the SWALL database (14) ID or accession number together with sequence position and two amino acid variants characterising the polymorphism. Given the input, PolyPhen starts a fully automated pipeline of several programs described step by step in this section. The pipeline is schematically presented in Figure 1. The server was used to annotate all SNPs deposited in the HGVbase database and the resulting dataset of annotated SNPs is available at http://www.bork.embl-heidelberg.de/PolyPhen/data.
Figure 1.
PolyPhen query processing flowchart. PolyPhen combines information on sequence features, multiple alignment with homologous proteins and structural parameters and contacts to make a prediction of nsSNP effect on protein function. hs_swall is the abbreviation for the Homo sapiens subset of the SWALL database (also known as SPTR, i.e. SwissProt + TrEMBL). Var1,2, two amino acid variants; ACC/ID, SWALL accession number or ID.
Identifying nsSNPs in known genes
The necessary first step in the analysis of nsSNPs is to identify whether a given SNP is indeed non-synonymous. For this purpose we map SNPs onto known proteins on the basis of SNP DNA flanking sequences. Flanking genomic sequences of SNPs from HGVbase (13) with length 25 bp each have been translated in all six possible frames and searched for in the proteins in the human proteins subset of the SWALL database (15). Protein sequences and genomic fragments were pre-processed with the SEG (16), XNU (17), RepeatMasker (18) and DUST programs, which are used to filter out areas of low compositional complexity, regions containing internal repeats of short periodicity and known human genomic repeat sequences. ALU subfamily proteins were also excluded from the set. We required that at least one translated flanking sequence should have an exact match with a database protein sequence. If this match was detected, we further required that the second flanking sequence had either an exact match with the protein sequence or matched the protein sequence in all positions until the end of the protein or a conventional exon/intron border is observed. The resulting mapping of a SNP onto a protein sequence is always unique.
The above procedure is available as a stand alone World Wide Web-based program snp2prot. The link to this program is provided from the main PolyPhen page. We also provide a link to the SNP annotation tool HNP (Y.Yuan, unpublished results).
After processing HGVbase v.12 (983 589 SNP entries), we obtained a set of 20 462 coding SNPs. Of these, 11 152 were non-synonymous, whereas 9310 were synonymous SNPs and do not produce any change in the amino acid sequence. The nsSNPs formed our dataset, which can be downloaded as one text file or searched against with a straightforward World Wide Web-based engine. The search results contain links to the other databases that provide additional information, e.g. chromosomal location of a nsSNP.
PolyPhen analysis of nsSNPs
Sequence-based characterisation of the substitution site. The substitution may occur at a specific site, e.g. active or binding, or in a non-globular, e.g. transmembrane, region. A query identifies the protein by its SWALL accession number or ID or by the sequence itself. In the latter case, PolyPhen tries to find the given sequence in the human subset of the SWALL database and use the FT (feature table) section of the corresponding entry. If the sequence cannot be found in the human subset of SWALL, this step is skipped. PolyPhen checks if the amino acid replacement occurs at a site that is annotated in the SWALL database feature table as DISULFID, THIOLEST or THIOETH bond, BINDING, ACT_SITE, LIPID, METAL, SITE or MOD_RES site or as a site located in a TRANSMEM, SIGNAL or PROPEP region.
PolyPhen also uses the TMHMM (19) algorithm to predict transmembrane regions, the Coils2 (20) program to predict coiled coil regions and the SignalP (21) program to predict signal peptide regions of the protein sequences.
For a substitution in a transmembrane region, PolyPhen uses the PHAT (22) transmembrane-specific matrix score to evaluate possible functional effect of a nsSNP in the transmembrane region.
At this step PolyPhen memorises all positions that are annotated in the query protein as BINDING, ACT_SITE, LIPID or METAL. At a later stage, if the search for a homologous protein with known 3D structure is successful, it is checked whether the substitution site is in spatial contact with these critical residues.
Profile analysis of homologous sequences. The amino acid replacement may be incompatible with the spectrum of substitutions observed at that position in a family of homologous proteins. PolyPhen identifies homologues of the input sequences via a BLAST (23) search of the NRDB database. The set of aligned sequences with sequence identity to the input sequence in the range 30–94% (inclusive) is used by the new version of the PSIC (position-specific independent counts) software (24) to calculate the so-called profile matrix (http://strand.imb.ac.ru/PSIC/). Elements of the matrix (profile scores) are logarithmic ratios of the likelihood of a given amino acid occurring at a particular site to the likelihood of this amino acid occurring at any site (background frequency). PolyPhen computes the absolute value of the difference between profile scores of both allelic variants in the polymorphic position. PolyPhen also shows the number of aligned sequences at the query position; this may be used to assess the reliability of profile score calculations.
Mapping of the substitution site to known protein 3-dimensional structures. Mapping of an amino acid replacement to a known 3D structure reveals whether the replacement is likely to destroy the hydrophobic core of a protein, electrostatic interactions, interactions with ligands or other important features of a protein. If the spatial structure of a query protein is unknown, one can use a homologous protein of known structure.
PolyPhen carries out a BLAST query of a sequence against a protein structure database [PDB (25) or PQS (26), see below] and retains all hits that meet the given criteria. For instance, the default sequence identity threshold is set to 50%, since this value guarantees the conservation of basic structural characteristics. Minimal hit length and maximal length of gaps are by default set to 100 and 20, respectively. The position of the substitution is then mapped onto the corresponding positions in all retained hits. By default, a hit with 3D structure is rejected if its amino acid at the position under study differs from the amino acid in the input sequence. Hits are sorted according to the sequence identity or E-value of the sequence alignment with the input protein.
Structural parameters used to evaluate the effect of amino acid substitution. Structural analysis performed by PolyPhen is based on the use of several structural parameters, as suggested previously (7–9). Importantly, although all parameters are reported in the output, only some of them are used in the final decision rules.
PolyPhen uses the DSSP (27) database to obtain the following structural parameters for the mapped amino acid residues: secondary structure (according to the DSSP nomenclature); solvent accessible surface area (absolute value in Å2); φ–ψ dihedral angles.
The following values are also calculated by PolyPhen: normalised accessible surface area [the absolute value divided by the maximal area defined as the 99% quantile of surface area distribution for this particular amino acid type in PDB (25)]; change in accessible surface propensity (knowledge-based hydrophobic ‘potentials’) resulting from the substitution; change in residue side chain volume (in Å3); region of the φ–ψ map (Ramachandran map) derived from the dihedral angles (9); normalised B factor (temperature factor) for the residue [following Chasman and Adams (9)]; loss of a hydrogen bond [following Wang and Moult (8)] according to the HBplus program (28).
By default, the parameters above are calculated for the first hit only.
Contacts with ‘critical sites’, ligands and other polypeptide chains. The presence of specific spatial contacts of a residue may reveal its role in protein function. PolyPhen checks three types of contacts for a variable amino acid residue. First, contacts with ligands (defined as all heteroatoms excluding water and ‘non-biological’ crystallographic ligands). Second, interactions between subunits of the protein molecule. Technically these are defined as contacts of a polymorphic residue with residues from other polypeptide chains present in the PDB (PQS) file. For this particular type of interaction, it is more advantageous to use the PQS (Protein Quaternary Structure) database (26) rather than PDB, since PQS entries are supposed to provide a more adequate picture of protein quaternary structure architecture.
The third type of contact analysed by PolyPhen is represented by contacts with ‘critical’ residues, where the latter are derived from the sequence annotation. The suggested default threshold for all contacts to be displayed in the output is 6 Å. However, a value of 3 Å is used in the decision rule. For evaluation of a contact between two residues or between a residue and a ligand molecule, PolyPhen finds the minimal distance amongst all possible between atoms of two residues. By default, contacts are calculated for all hits with structure. This is essential for cases where several structures correspond to one protein but carry different information about complexes with other macromolecules and ligands (see for example figure 2 in ref. 7).
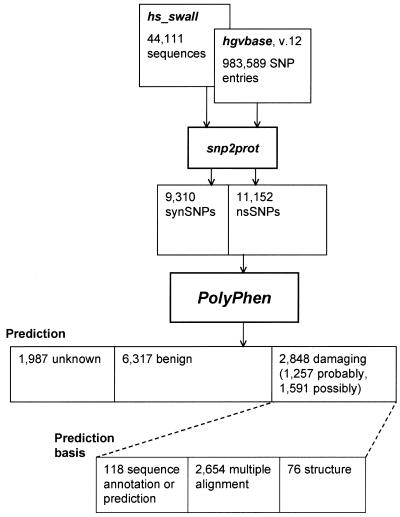
Figure 2.
Results of the PolyPhen analysis of the HGVbase database v.12. hs_swall denotes the Homo sapiens subset of the SWALL database. snp2prot is an in-house command line tool to map HGVbase SNPs onto sequences of known human proteins. 11 152 nsSNPs were identified. 1591 of them have been predicted as possibly damaging for protein structure and function and an additional 1257 as probably damaging. The number of structure-based predictions is much lower compared with the number of sequence-based predictions because structural information was available in only 1026 cases.
Prediction rules. PolyPhen uses empirically derived rules (Table 1) to predict that an nsSNP is damaging, i.e. is supposed to affect protein function, or benign, i.e. most likely lacking any phenotypic effect. The rule is based on the analysis of the ability of various structural parameters and profile scores to discriminate between disease mutations and substitutions between human proteins and closely related mammalian orthologues (7). We introduced two categories of prediction: nsSNPs possibly damaging protein function/ structure and nsSNPs probably damaging protein function/structure. The scheme presented in Table 1 successfully predicts ∼82% (∼57% for the more stringent set of rules) of disease-causing mutations annotated in SwissProt database 14 and produces ∼8% (∼3% for the more stringent set of rules) false positives given the control set of between-species substitutions. We note that many parameters, though computed by the server, were excluded from the decision rule. Due to correlation with other parameters they did not help to increase sensitivity without significant loss of specificity of predictions. Multiple alignment-based profile scores provided the major contribution to the prediction. Therefore, even in the case of proteins with no homologue with known 3D structure, predictions remain reasonably reliable.
Table 1. Rules used by PolyPhen to predict effect of nsSNPs on protein function and structure.
| Rules (connected with logical AND) | Prediction | ||
|---|---|---|---|
| PSIC score difference Δ | Substitution site properties | Substitution type properties | |
| Arbitrary | Annotated as a functionala or bond formationb site | Arbitrary | Probably damaging |
| Not considered | In a region annotated or predicted as transmembrane | PHAT matrix difference resulting from substitution is negative | Possibly damaging |
| Δ ≤ 0.5 | Arbitrary | Arbitrary | Benign |
| Δ > 1.0 | Atoms are closer than 3.0 Å to atoms of a ligand or residue annotated as BINDING, ACT_SITE, LIPID, METAL | Arbitrary | Probably damaging |
| 0.5 < Δ ≤ 1.5 | Normed accessibility ACC ≤ 15% | Absolute change of accessible surface propensity is ≥0.75 or absolute change of side chain volume is ≥60 | Possibly damaging |
| Normed accessibility ACC ≤ 5% | Absolute change of accessible surface propensity is ≥1.0 or absolute change of side chain volume is ≥80 | Probably damaging | |
| 1.5<Δ ≤ 2.0 | Arbitrary | Arbitrary | Possibly damaging |
| Δ > 2.0 | Arbitrary | Arbitrary | Probably damaging |
One row corresponds to one rule, which may consist of several parts connected by logical AND. For a given substitution, all rules are tried one by one, resulting in prediction of functional effect: benign, possibly damaging or probably damaging. If no evidence for a damaging effect is seen, substitution is considered benign.
aBINDING, ACT_SITE, SITE, MOD_RES, LIPID, METAL, SE_CYS (SwissProt feature table terms).
bDISULFID, THIOLEST, THIOETH (SwissProt feature table terms).
RESULTS
Retrieval of nsSNPs
HGVbase v.12 (13), a comprehensive public database with extensive curation, was chosen as a source of SNP data. The database had 983 589 SNP entries, which represented SNPs from various sources. Importantly, SNPs in the database are classified according to reliability. Namely, SNPs confirmed by independent and solid experimental verification are marked as ‘Proven’, whereas other SNP candidates are marked as ‘Suspected’. Version 12 of the database contained 984 093 entries, 983 589 of these being SNPs, while the rest represent other types of genetic variants. Only 14 986 SNPs, however, appeared in the ‘Proven’ category. We mapped all available SNPs onto known proteins and found 9310 of them to be synonymous and 11 152 non-synonymous, causing amino acid changes in protein sequences. 1276 of these identified nsSNPs were ‘Proven’.
Only 1026 nsSNPs were mapped to proteins with at least 50% sequence identity to a protein with known 3D structure. The analysis for the rest of the nsSNPs was performed on the basis of multiple alignment information only.
The database of these nsSNPs and their analysis using PolyPhen is available at http://www.bork.embl-heidelberg.de/PolyPhen/data. PolyPhen analysis was only possible for 9165 (82%) of these nsSNPs, as the remainder have been mapped to proteins with no applicable site annotation and no reasonably close homologous sequences available in the SWALL database for multiple alignment or structural analysis.
The results of the PolyPhen analysis are presented in Figure 2.
Structural characterisation of nsSNPs
As has been noted by Wang and Moult (9), most disease mutations and supposedly deleterious nsSNPs affect protein stability rather than functionality. Various structural parameters have been proposed (6–9,11) to detect the effects of amino acid substitutions. We selected a group of structural parameters and evaluated their impact through a comparison of disease mutations, nsSNPs and substitutions between human proteins and closely related mammalian orthologues [datasets from Sunyaev et al. (7)]. We also selected three characteristics responsible for functionality: annotation of the site as BINDING, ACT_SITE, LIPID or METAL (SwissProt feature table terms); proximity to an annotated site; proximity to a co-crystallised ligand. The data presented in Table 2 confirm that functionality parameters have a smaller impact on the molecular origin of disease mutations and deleterious nsSNPs than protein stability characteristics. Among the structural characteristics presented in Table 2, hydrophobic core stability parameters are the best predictors.
Table 2. Structural characteristics of disease mutations, nsSNPs and amino acid substitutions between species.
| Disease mutations (%) | nsSNPs (%) | Substitutions between species (%) | |
|---|---|---|---|
| At a functional site | 4 | 0 | 0 |
| Closer than 4 Å to a critical site | 7 | 2 | 1 |
| Closer than 3 Å to a critical site | 2 | 1 | 0 |
| Closer than 4 Å to a ligand | 11 | 8 | 4 |
| Closer than 3 Å to a ligand | 4 | 3 | 0.6 |
| Normalised accessibility (ACC) ≤ 5% | 41 | 23 | 17 |
| ACC ≤ 5% and change in hydrophobic propensity >0.75 | 21 | 6 | 2 |
| ACC ≤ 5% and change in volume >60 Å3 (overpacking) | 6 | 2 | 0 |
| ACC ≤ 5% and change in volume >–60 Å3 (cavity creation) | 2 | 2 | 0.6 |
| ACC ≤ 5% and change in electrostatic charge | 12 | 4 | 1 |
| ACC < 25% | 67 | 45 | 34 |
| ACC < 25% and absolute change in hydrophobic propensity >0.75 | 33 | 14 | 5 |
| ACC < 25% and change in volume >60 Å3 (overpacking) | 9 | 4 | 1 |
| ACC < 25% and change in volume >–60 Å3 (cavity) | 5 | 4 | 1 |
| ACC < 25% and change in electrostatic charge | 22 | 9 | 4 |
| Normalised crystallographic B-factor <–0.5 | 44 | 29 | 24 |
| Loss of a hydrogen bond | 13 | 8 | 5 |
| Loss of a disulphide bridge | 1.3 | 0.6 | 0 |
| Proline in α-helix | 3 | 1.2 | 0.4 |
| Substitution of Gly with torsion angles forbidden for other amino acids | 1.2 | 0.9 | 0.4 |
Interestingly, for all parameters analysed we observed the same pattern in Table 2. The fraction of SNPs that affect a structural parameter is always much lower than that of disease-causing mutations. At the same time, it is always higher than the corresponding number of substitutions between species. This observation suggests that all effects associated with these structural parameters are responsible for the accumulation of deleterious alleles in the human genome. Disease-causing mutations are subject to very strong selective pressure and are eliminated from the population very quickly. In contrast, slightly deleterious SNPs detected in panels of healthy individuals are supposedly under lower selective pressure and therefore have a much longer persistence time in the population. As suggested by Table 2, we did not observe any structural feature responsible solely for strong or solely for weak selection, as all parameters display the same pattern.
Although many structural parameters can serve as reasonably reliable predictors of the effect of a substitution, a strong correlation within structural parameters and especially between structural parameters and long-term selective pressure signals seen from multiple sequence alignment made exclusion of many parameters from the combined prediction rule necessary. For the set of nsSNPs predicted to be damaging, based on the combined set of rules that incorporate both multiple alignment and structural information (available for these cases), structural parameters worked as predictors in 40% of cases. However, the prediction cannot be made solely at the sequence level in 22% of cases (28% if the ‘probably damaging’ category only is considered).
Protein structural and functional characteristics and selective constraints
As has been shown by systematic studies on cSNP (coding SNPs) discovery (29–31), the distribution of nsSNP density over human genes is highly non-uniform. Apart from differences in the coalescent history of loci, this notable difference in the rate of nsSNPs is likely to be caused by variations in selective pressure against deleterious variants. We expected that the difference in selective pressure might be caused by structural properties because the number of sites important for stability or functionality might depend on the protein structure type. Also, extracellular proteins can be expected to have higher stability compared with intracellular ones and this may affect selective constraints. On the other hand, selective pressure may depend on the impact of the gene on the overall fitness of the organism (32). In order to test whether the above properties of proteins have an effect on the density of nsSNPs (considered for genes with the same number of synonymous SNPs to correct for various sources of bias), we subdivided genes from our database into groups according to the SCOP (33) and GO (34) classifications. Contrary to our expectations, we did not detect a significant correlation of selective pressure against deleterious nsSNPs with secondary structure class, localisation or biological process. This might be because we grouped genes into very large classes and the effect might be detected if a finer classification were considered. Alternatively, we have to conclude that there is no strong impact of these characteristics on the selective constraints.
In contrast, molecular function of the protein showed a statistically significant association with the strength of selective pressure (the P value of the χ2 test was 0.009). The functional class showing the highest selective pressure against deleterious nsSNPs is the class of transcription factors. This class displays the greatest departure from the average level of selective constraints. Enzymes are the class of proteins with the lowest selective pressure. The fraction of nsSNPs predicted as damaging by PolyPhen is also highest for enzymes and lowest for transcription factors. This is expected and shows that low selective constraints allow for accumulation of slightly deleterious SNPs. We hypothesise that this observation can be explained in terms of the molecular basis of dominance (35). Mutations in enzymes are likely to be recessive because the flux in a metabolic pathway undergoes very minor change in response to a decrease in enzyme activity (35). In contrast, changes in the activity of transcription factors can have a high impact on the transcription level of the regulated genes. Transcription factors listed in the OMIM database (http://www.ncbi.nlm.nih.gov/Omim/) are reported to be dominant genes much more frequently than enzymes.
However, we should note that the current SNP databases are probably biased towards ‘popular’ genes, which could have affected our results. More accurate selective pressure studies will be possible in the future with larger datasets arising from large-scale systematic studies.
DISCUSSION
Server
Ideally, the end point of disease gene identification should be functional analysis of the disease-associated allele and an understanding of the molecular mechanism of causation of the disease phenotype. This functional characterisation can be facilitated by the computational analysis provided by our tool.
Unlike fully penetrant mutations causing Mendelian diseases, SNPs involved in complex human phenotypes are not a necessary and sufficient condition defining the phenotype but their effect depends on many other genetic and environmental components. In other words, SNPs may comprise risk factors of having a specific phenotype in the statistical sense. Therefore, the effect of a particular SNP on phenotype might be seen only as a frequency difference between individuals that display the phenotype and unaffected controls.
Given the very high rate of false associations recently reported, any independent evidence of the impact of the suspected allelic variant should be valued. Sequence and structure analysis of the suspected amino acid variant can increase the confidence of the finding by revealing the structural background of the disease. The PolyPhen server can be used to evaluate whether the reported/identified association can indeed have a functional meaning and therefore is less likely to represent a false positive due to statistical reasons or reasons of inappropriate study design and population choice.
Consequently, even if an association of a genomic locus with a particular phenotype is unambiguously demonstrated, it is not always clear that the identified DNA variant has a causative relationship with the disease and that statistical association is not a result of linkage disequilibrium with the true functional variant (14). In this case the PolyPhen server can be used to distinguish casual from non-casual relationships between a nsSNP and the phenotype of interest.
The database of nsSNPs annotated by PolyPhen provides a source of functionally annotated nsSNPs. The collection might be a useful resource for selection of nsSNPs for candidate gene-based association studies. The question of how to choose the set of SNPs to be screened is critical to the success of a study. The major hurdle in any model of association studies is posed by the large number of these SNPs (4,36). One side of the problem is the limitations of currently available genotyping technologies, which make studies on large SNP sets in large panels of individuals impractical. The other side, however, is of a purely statistical nature and is therefore independent of the technological progress. Multiple test correction in the case of many thousands of SNPs to be analysed makes the detection of otherwise significant allele frequency differences problematical. Possible allelic and non-allelic heterogeneity, epistatic interactions between alleles, low penetrance of the phenotype and complexity of environmental factors involved make the SNP-based detection of disease genes even more difficult (2). Without any careful pre-selection of SNPs to be screened, unrealistically large panels of individuals might be required to detect association at a reasonable level of statistical significance. Therefore, computational prediction of functional importance can be considered as one of the reasons to prioritise SNPs while looking for an association.
Survey
PolyPhen analysis of the nsSNP database confirmed earlier observations (6–9,12,37) that a significant number of human nsSNPs is represented by slightly deleterious alleles. The fraction of nsSNPs predicted to be damaging in the much larger dataset of 9165 nsSNPs is similar to the earlier result. Most predictions were computed based solely on the multiple alignment information, since structural data are available for only a very small fraction of cases.
It is important to note that the number of functional nsSNPs predicted for the whole database is likely to be an overestimate due to pollution of the database by erroneous SNP reports, on the one hand, and possible bias of the database towards disease-related allelic variants on the other. To test the impact of these biases on the overall conclusion of the presence of multiple slightly deleterious SNPs in individual human genomes, we compared fractions of nsSNPs predicted to be damaging (both possibly and probably) for HGVbase entries annotated as ‘Proven’ and ‘Suspected’. Additionally, we compared the prediction rate for ‘Proven’ nsSNPs originating from systematic studies (29–31) with the overall prediction rate. The overall prediction rate for the category ‘Suspected’ nsSNPs was 31.4%, for the category ‘Proven’ nsSNPs it was 28.9% and for ‘Proven’ nsSNPs from systematic studies on healthy individuals (29–31) it was 27.6%. This shows that inaccuracy and bias of the database data lead to overprediction of the fraction of deleterious nsSNPs. However, the effect of the prediction rate for nsSNPs compared with the species divergence data on a much higher fraction is seen even from the cleanest possible dataset. Similarly, trends observed in Table 2 are the same for any subset of nsSNP data.
Our analysis showed that various effects on protein stability are responsible for accumulation of slightly deleterious nsSNPs in human genes. The selection against these variants is likely to depend on the molecular function of proteins rather than on the type of structure or cellular localisation. This can possibly be explained by the relationship between molecular function and mutation dominance. Transcription factors appear to be the group with the highest selective constraints.
With the growth of public SNP data and the improvement in the quality of SNP databases, functional analysis of SNPs can possibly play a role in our understanding of the inheritance of complex human phenotypes.
Acknowledgments
ACKNOWLEDGEMENTS
The authors are thankful to Evgenia Kriventseva for her help in the work with the GO database. S.S. acknowledges Alexey Kondrashov for useful discussions.
REFERENCES
- 1.Risch N. and Merikangas,K. (1996) The future of genetic studies of complex human diseases. Science, 273, 1516–1517. [DOI] [PubMed] [Google Scholar]
- 2.Risch N.J. (2000) Searching for genetic determinants in the new millennium. Nature, 15, 847–856. [DOI] [PubMed] [Google Scholar]
- 3.Lai E., Riley,J., Purvis,I. and Roses,A. (1998) A 4-Mb high-density single nucleotide polymorphism-based map around human APOE. Genomics, 54, 31–38. [DOI] [PubMed] [Google Scholar]
- 4.Emahazion T., Feuk,L., Jobs,M., Sawyer,S.L., Fredman,D., St Clair,D., Prince,J.A. and Brookes,A.J. (2001) SNP association studies in Alzheimer’s disease highlight problem for complex disease analysis. Trends Genet., 17, 407–413. [DOI] [PubMed] [Google Scholar]
- 5.Schork N.J., Fallin,D. and Lanchbury,J.S. (2000) Single nucleotide polymorphisms and the future of genetic epidemiology. Clin. Genet., 58, 250–264. [DOI] [PubMed] [Google Scholar]
- 6.Sunyaev S., Ramensky,V. and Bork,P. (2000) Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet., 16, 198–200. [DOI] [PubMed] [Google Scholar]
- 7.Sunyaev S., Ramensky,V., Koch,I., Lathe,W.,III, Kondrashov,A.S. and Bork,P. (2001) Prediction of deleterious human alleles. Hum. Mol. Genet., 10, 591–597. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z. and Moult,J. (2001) SNPs, protein structure and disease. Hum. Mutat., 17, 263–270. [DOI] [PubMed] [Google Scholar]
- 9.Chasman D. and Adams,R.M. (2001) Predicting the functional consequences of non-synonymous single nucleotide polymorphisms: structure-based assessment of amino acid variation. J. Mol. Biol., 307, 683–706. [DOI] [PubMed] [Google Scholar]
- 10.Ng P.C. and Henikoff,S. (2001) Predicting deleterious amino acid substitutions. Genome Res., 11, 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrer-Costa C., Orozco,M. and de la Cruz,X. (2002) Characterization of disease-associated single amino acid polymorphisms in terms of sequence and structure properties. J. Mol. Biol., 315, 771–786. [DOI] [PubMed] [Google Scholar]
- 12.Sunyaev S.R., Lathe,W.C.,III, Ramensky,V.E. and Bork,P. (2000) SNP frequencies in human genes: an excess of rare alleles and differing modes of selection. Trends Genet., 16, 335–337. [DOI] [PubMed] [Google Scholar]
- 13.Fredman D., Siegfried,M., Yuan,Y.P., Bork,P., Lehvaslaiho,H. and Brookes,A.J. (2002) HGVbase: a human sequence variation database emphasizing data quality and a broad spectrum of data sources. Nucleic Acids Res., 30, 387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson G.C. and Todd,J.A. (2000) Strategies in complex disease mapping. Curr. Opin. Genet. Dev., 10, 330–334. [DOI] [PubMed] [Google Scholar]
- 15.Apweiler R. (2000) Protein sequence databases. Adv. Protein Chem., 54, 31–71. [DOI] [PubMed] [Google Scholar]
- 16.Wootton J.C. and Federhen,S. (1993) Statistics of local complexity in amino-acid-sequences and sequence databases. Comput. Chem., 17, 149–163. [Google Scholar]
- 17.Claverie J.M. and States,D.J. (1993) Information enhancement methods for large-scale sequence analysis. Comput. Chem., 17, 191–201. [Google Scholar]
- 18.Jurka J. (2000) Repbase update: a database and an electronic journal of repetitive elements. Trends Genet., 16, 418–420 [DOI] [PubMed] [Google Scholar]
- 19.Krogh A., Larsson,B., von Heijne,G. and Sonnhammer,E.L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol., 305, 567–580. [DOI] [PubMed] [Google Scholar]
- 20.Lupas A., Van Dyke,M. and Stock,J. (1991) Predicting coiled coils from protein sequences. Science, 252, 1162–1164. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen H., Engelbrecht,J., Brunak,S. and von Heijne G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng., 10, 1–6. [DOI] [PubMed] [Google Scholar]
- 22.Ng P.C., Henikoff,J.G. and Henikoff,S. (2000) PHAT: a transmembrane-specific substitution matrix. Predicted hydrophobic and transmembrane. Bioinformatics, 16, 760–766. [DOI] [PubMed] [Google Scholar]
- 23.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 24.Sunyaev S.R., Eisenhaber,F., Rodchenkov,I.V., Eisenhaber,B., Tumanyan,V.G. and Kuznetsov,E.N. (1999) PSIC: profile extraction from sequence alignments with position-specific counts of independent observations. Protein Eng., 12, 387–394. [DOI] [PubMed] [Google Scholar]
- 25.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henrick K. and Thornton,J.M. (1998) PQS: a protein quaternary structure file server. Trends Biochem. Sci., 23, 358–361. [DOI] [PubMed] [Google Scholar]
- 27.Kabsch W. and Sander,C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- 28.McDonald I.K. and Thornton,J.M. (1994) Satisfying hydrogen bonding potential in proteins. J. Mol. Biol., 238, 777–793. [DOI] [PubMed] [Google Scholar]
- 29.Cargill M., Altshuler,D., Ireland,J., Sklar,P., Ardlie,K., Patil,N., Shaw,N., Lane,C.R., Lim,E.P., Kalyanaraman,N. et al. (1999) Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nature Genet., 22, 231–238. [DOI] [PubMed] [Google Scholar]
- 30.Halushka M.K., Fan,J.B., Bentley,K., Hsie,L., Shen,N., Weder,A., Cooper,R., Lipshutz,R. and Chakravarti,A. (1999) Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nature Genet., 22, 239–247. [DOI] [PubMed] [Google Scholar]
- 31.Cambien F., Poirier,O., Nicaud,V., Hermann,S.M., Mallet,C., Ricard,S., Beague,I., Hallet,V., Blanc,H., Loucaci,V. et al. (1999) Sequence diversity in 36 candidate genes for cardiovascular disorders. Am. J. Hum. Genet., 65, 183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirsh A.E. and Fraser,H.B. (2001) Protein dispensability and rate of evolution. Nature, 411, 1046–1049. [DOI] [PubMed] [Google Scholar]
- 33.Lo Conte L., Brenner,S.E., Hubbard,T.J., Chothia,C. and Murzin,A.G. (2002) SCOP database in 2002: refinements accommodate structural genomics. Nucleic Acids Res., 30, 264–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.The Gene Ontology Consortium (2001) Creating the gene ontology resource: design and implementation. Genome Res., 11, 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kacser H. and Burns,J.A. (1981) The molecular basis of dominance. Genetics, 97, 639–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weiss K.M. and Terwilliger,J.D. (2000) How many diseases does it take to map a gene with SNPs? Nature Genet., 26, 151–157. [DOI] [PubMed] [Google Scholar]
- 37.Fay J.C., Wyckoff,G.J. and Wu,C.I. (2001) Positive and negative selection on the human genome. Genetics, 158, 1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]