Abstract
Heart disease remains the most frequent cause of death in the general population with increasing incidence in the elderly population. The pathologic failure of the aging heart may be related to structural and functional alterations in cardiac muscle cells. However, the molecular mechanisms underlying the aging-related decline in cardiac muscle function are largely unknown. To provide the first analysis of cardiac aging at the level of gene expression, we established and compared cDNA libraries from apparently healthy young and aged mouse ventricular cardiac muscle cells. We report the identification of genes that exhibit aging-related changes of mRNA levels. Aging expression profiles in aged hearts indicate decreased cellular adaptation and protection against stress-induced injury together with the development of contractile dysfunction. The data suggest reduced activity of the mitochondrial electron transport system and reduced levels of cardiac-specific transcription regulators. The cardiomyocyte aging profile of gene expression displays similarities with known heart disorders. Genes whose mRNA levels change with aging in cardiomyocytes might profoundly affect pathological changes in the heart.
INTRODUCTION
The genetics of aging is one of the biggest challenges to genomic and medical research. The ultimate genomic goals are the understanding of the dynamic network of genes, its effect on the aging process, interaction with disease, and organ specificity (1). It has been proposed that the genetic component of aging is small (2), however, studies with centenarians have provided evidence that genes most likely play a prominent role in the ability to achieve older age (3).
Although the heart is the focus of major problems in the aging population and aging-related heart disease is the most frequent cause of death, at present, genetic analysis of cardiac aging is not available. Cellular aging is commonly associated with the instability of the nuclear and mitochondrial genomes and oxidative protein damage because of long-term exposure to reactive oxygen species. Cardiac myocytes, however, are highly specialized high-oxygen-content cells that generally do not divide. Therefore, errors in nuclear DNA duplication and replicative senescence should not be critical in their aging. The cardiac myocytes, however, house a large number of mitochondria that are dividing. Somatic mutations in the mitochondrial genome are documented with aging. These mutations have the potential to influence mitochondrial functions, which in turn may influence functions in the nuclear genome. Mitochondrial electron transport abnormalities because of DNA deletion mutations can result in oxidative cellular protein damage. Oxidative protein damage may directly cause changes in transcription factors, chromatin structure and function. With aging, these changes can result in alterations of nuclear gene expression, mRNA stability or both.
Searching for cardiac muscle cellular genes that change expression levels during aging would lead to a better understanding of decreased cardiac performance with aging. We applied a combination of four different methods to screen directly for differences in gene expression levels in cardiac myocytes from young and aged mouse hearts. We found 43 genes that accumulate at different levels with age. The levels of these genes did not change significantly from animal to animal. Our results clearly indicate the presence of cardiac-specific aging gene candidates. We also found that the cardiomyocyte aging profile of gene expression displays similarities with known heart disorders.
MATERIALS AND METHODS
Adult mice hearts
Hearts were obtained from C57BL/6 mice with no clinical evidence of heart failure such as labored breathing, ascites, and significant weight gain or weight loss.
Adult mouse cardiomyocytes
Left ventricular cardiac myocytes (LVCs) were enzymatically dissociated from the left ventricle according to a previously published protocol (4) with slight modifications. Hearts from young animals were perfused for 20 min and old hearts were perfused for 25 min. Each heart from young animals yielded approximately 2–3 × 106 cells and from aged animals 0.7–1.0 × 106 contracting rod-shaped cells. The purity, the shape and the contractility of the myocyte preparations were inspected by light microscopy. Cells were also inspected for sarcolemma structure by confocal microscopy with FITC-labeled rabbit antibodies (Sigma), which are specific to cardiac alpha actinin. The accumulation of autofluorescent material with aging was monitored by confocal microscopy (excitation at 500 nm). Freshly isolated cardiomyocytes typically contained >90% rod-shaped cells and <1% of non-myocytes. Cells were used immediately for preparation of RNA or for microscopy. Young C57BL/6 adult male mice were 4 months old and the aged C57BL/6 adult male mice were 20 months old. Retired C57BL/6 male breeders (Jackson laboratory) were used after reaching the age of 20 months or delivered from the National Institute of Aging at the age of 20 months.
Differential gene subtraction
For differential gene expression analysis, we used the PCR-based cDNA subtraction procedure (Clontech). LVCs isolated from male and female hearts were used for the isolation of total RNA as suggested by the manufacturer (Qiagen). cDNA libraries were prepared using the SMART cDNA Synthesis Kit (Clontech) as suggested by the manufacturer. Subtraction hybridization was routinely performed with 600 ng of driver cDNA that was mixed with 20 ng of the tester cDNA. Subtracted fractions were then amplified by selective PCR using the Advantage cDNA PCR Core Kit (Clontech). The subtraction efficiency was determined by virtual northern blot analyses as suggested by the manufacturer. Differential subtractions were independently conducted with six cDNA libraries prepared from hearts from both ages (12 young C57BL/6 male and 12 old C57BL/6 male mice).
Cloning and analysis of subtracted products
The subtracted cDNAs were ligated into Zero Blunt TOPO™ cloning vector (Invitrogen) and transferred to DH5α Escherichia coli. The positive clones were identified following standard procedures. In brief, the tester and the driver cDNA fractions were 33P radioactively labeled and used for the colony hybridization. The clones that hybridize to the tester probe but not to the driver probe represent differentially expressed mRNAs. These clones were isolated and each cDNA fragment from the positive clones was used as a probe to confirm its differential expression by virtual northern blotting. Confirmed positive clones were sequenced using a 310/317 Genetic Analyzer (Applied Biosystems). Sequence data were compared with the GenBank database using the BLAST program at the National Center for Biotechnology Information (National Institutes of Health, Bethesda, MD).
Northern blotting
We routinely used 2 µg of RNA, which was separated on a 0.7% agarose gel, denatured and then blotted onto a nylon membrane (GeneScreen plus; NEN). Hybridization with the radioactively labeled probes was performed according to standard protocols in Quick Hybrid solution (Stratagene). We analyzed the RNA pool made from 10 male and 10 female C57BL/6 mouse heart LVC preparations. Using pairwise, young and respectively aged mice RNA pools, comparisons were made with [33P]dATP radioactively labeled (Prime-It II; Stratagene) individual cDNA clones. The radioactive hybridization signal was visualized after 2–6 days exposure using the PhosphorImaging system, NIH Image and Adobe PhotoShop software.
Differential gene expression analysis by DNA array
An alternative method to analyze gene expressions was performed using DNA-arrayed membranes. We developed a DNA array with immobilized oligonucleotides that recognize the 3′ end of the mRNAs of the differentially expressed genes. To ensure gene-specific hybridization for each gene, 20 fmol of two to four different 60 bp oligonucleotides (MWG Biotech) were spotted onto hybridization transfer membranes (GeneScreen plus) with a 1536-pin replicator (V and P Scientific, CA). The selected oligonucleotides satisfied the following criteria: Tm within 85–95°C; absence of secondary structure; and absence of cross-hybridization verified by querying each oligonucleotide against the expressed sequence tag (EST) database. Blank spots without oligonucleotides were included for evaluation of background caused by non-specific interactions of individual probes with the membrane. For the array analysis the cDNA libraries were labeled with [33P]dCTP (Prime-It II). Labeled cDNA was purified from free [33P]dCTP by a QiaQuick PCR column (Qiagen) and heat denatured before hybridization to the membrane array. After washing and exposure, the spot reading on the membranes was performed with the PhosphorImager. Matrix overlay maps spotted oligonucleotides on the array. In a single experiment, two or three identical membranes were hybridized with each of the radioactively labeled cDNA libraries and analyzed. Individual cDNA libraries from eight young and eight old male hearts were labeled and used separately for hybridization. The data were imported into Microsoft Excel spreadsheets. The data from different arrays were normalized using a set of highly and steadily expressed genes, and the probes with hybridization below background noise were excluded from the analysis. The readings of each oligonucleotide probe averaged among replicate arrays hybridized with the same cDNA were considered independent measurements. All such measurements obtained from old animals were pooled to make the ‘old’ sample, and all from the young made up the ‘young’ sample. The hypothesis that a gene was differentially expressed in the old as compared with the young hearts was tested by calculating the corresponding P-value using the native Excel t-test option.
Gene expression analysis by Affymetrix gene chips
We analyzed total RNA from two sets of six mice independently. cDNA was labeled and hybridized to the gene chip (murine genome array U74Av2) in the Affymetrix facility at Beth Israel Deaconess Medical Center. The data analysis of the micro-arrays was performed using Affymetrix software. Pairwise comparisons between individual mice were made using Excel software as recommended by Affymetrix. Changes higher than 1.2-fold were considered as a difference and they were included in the tables. The data analysis of the micro-arrays was performed in the Affymetrix facility. Each gene in the Affymetrix array was represented by perfectly matched (PM) and mismatched (MM) control oligonucleotides. Fluorescence intensity was read for each oligonucleotide to calculate the average signal intensity (SI) for each gene by subtracting the intensities of the PM from the intensity of the MM control, after discarding the maximum, the minimum and any outliers beyond three standard deviations. The effects of aging were determined by comparing each young (a mixture from six young hearts) with each old (a mixture from six old hearts) cRNA, generating two pairwise comparisons. The data reported in Table S1 of the Supplementary Material, represent the average fold changes obtained through the two pairwise combinations. The calculations were performed by an Affymetrix algorithm.
RESULTS
To find cardiac-specific ‘aging genes’, we first constructed cDNA libraries from total RNA of purified LVCs from C57BL/6 mouse male hearts at age 4 months (young) and at age 20 months (aged). The isolated LVCs contained typically >90% rod-shaped cardiomyocyte cells (Fig. 1). Confocal microscopy revealed the expected presence of the autofluorescent cytoplasmic lipofuscin inclusions in the aged cells (Fig. 1B) (5,6). Significantly fewer fluorescent inclusions were observed in LVCs from young mice (Fig. 1A) (5). We first compared RNA levels from both ages by the cDNA differential subtraction method. Three hundred and sixty individual clones from the subtracted cDNA libraries were sequenced to identify the differentially expressed genes. We found clones that were present in the young cDNA libraries at a significantly higher level than in the aged libraries (Table 1, D), as well as other clones that are induced in aged LVCs (Table 1, U). The proportion of mitochondrial RNAs was relatively high (48%) in the libraries from both groups, most likely because of the high copy number of mitochondrial genomes per cardiac muscle cell and the presence of tandem A-run sequences.
Figure 1.
Confocal microscopy of LVCs from healthy young and aged mouse hearts. Anti-alpha actinin–FITC conjugated antibodies (green) were used to label the alpha actinin distribution in the contractile apparatus. The autofluorescent lipofuscin inclusions (pink and yellow) were visualized with the Texas red filter. Nuclei are labeled with arrows. Pictures were taken at magnification ×600 and simultaneously with three of the filters. (A) LVCs from young animals. (B) LVCs from aged animals.
Table 1. mRNAs that decrease or increase in LVCs from aging mouse hearts compared with those from young mouse hearts.
The table lists mRNAs that were selected first in differential gene subtraction and the differences were next confirmed by northern blot analysis (1), by DNA array (2) and by Affymetrix gene chip (3) as indicated in the last right column. Identity of columns, from left to right: GenBank accession number; name of gene encoding mRNA; change in mRNA steady state level with aging, decrease (D), increase (U). Some of the differentially expressed genes are not presented in the Affymetrix gene chip. For these clones ‘3’ is not added to the last column. For clones that are not present in the DNA array we developed, ‘2’ is not added to the last column.
To further validate the data, we analyzed mRNA expression in young and aged LVCs applying northern blotting. Equal amounts of total RNA from young and aged LVCs were probed for signals from 34 of the differentially expressed genes. Examples of the northern blot assays are displayed in Figure 2. To confirm results from the cDNA subtraction experiments and to verify the reproducibility of aging-related differential gene expression in individual mouse preparations, we performed expression analysis experiments using an oligonucleotide array. We developed an array for differentially expressed genes. In addition, we included probes for 120 genes previously identified to play roles in the cardiac muscle specialized functions (Fig. 3). We performed eight sets of independent hybridization experiments with identical array membranes and radioactively labeled cDNA libraries from individual hearts. We also conducted two sets of analyses with an independent gene array containing 6000 mouse genes and ESTs. The results of the gene array comparative assays are presented in Table S1 of the Supplementary Material. To further validate the data, we analyzed mRNA expression in young and aged LVCs applying northern blotting. Equal amounts of total RNA from young and from aged LVCs were probed for signals from 34 of the differentially expressed genes. Examples of the northern blot assays are shown in Figure 2.
Figure 2.

Representative northern blot analyses confirming changes in mRNA levels. Total cellular RNA samples (2 µg) from young and aged LVCs were examined by northern blot analysis. Genes to which the probes correspond are identified to the left of the autoradiograms. Y, young LVCs; A, aged LVCs. Probes were radioactively labeled differentially expressed cDNA fragments. The level of mRNA specific for beta actin did not change significantly with aging and was used as an internal control.
Figure 3.
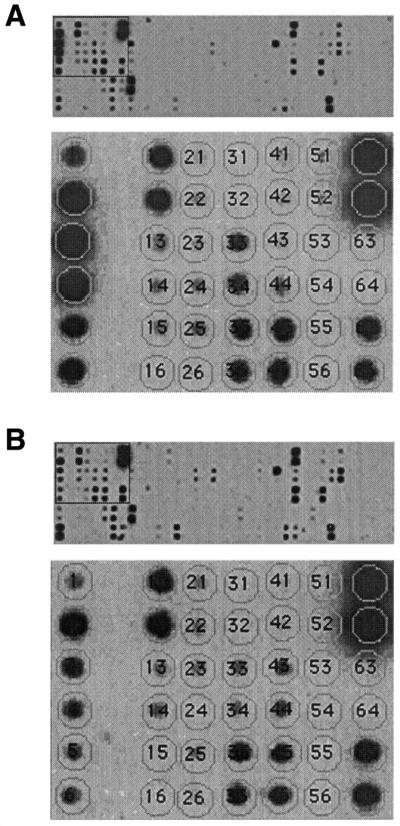
DNA arrays were used to confirm differential gene expression profiles. Identical DNA array membranes were probed with individual 33P-labeled cDNA libraries prepared from (A) young and (B) aged LVCs. Arrays contain oligonucleotide probes that are specific for the 45 differentially expressed clones and for 120 additional genes with known function in the cardiac muscle cells. Bottom panels of (A) and (B) show an enlargement of the boxed portions in the top panels. Matrix overlay maps the individual oligonucleotides for each of the genes. Hybridization position of some of the differentially expressed genes in the magnified region: 1 and 2, cytochrome NADH dehydrogenase subunit 1; 3 and 4, cytochrome c oxidase, subunit 3; 5 and 6, cytochrome b; 33 and 34, cardiac myosin light-chain; 41 and 42, α B-crystallin; 65 and 66, Hsp25.
We found 43 RNAs that accumulate at different levels with male mouse strain C57BL/6 age. Their levels did not change significantly from animal to animal (Table 1). Five of these clones have not been reported previously in the gene bank.
DISCUSSION
Our results indicate that the induction of an oxidative stress-induced transcriptional response may be a common feature of cellular aging in cardiac and in skeletal muscle of rodents and primates, but the extent to which aging modifies these responses may be cell specific.
Our results indicate that the expression of several members of the stress-responsive heat-shock protein (Hsp) family is modulated by aging (Table 1, Figs 2 and 3, Table S1 in Supplementary Material). Changes in the oxidative stress-induced transcriptional response may be a common feature of skeletal (7,8) and cardiac muscle aging. The mRNA for the inducible Hsp70 and Hsp25 decreased in aged LVCs. A similar aging pattern was observed in mouse and monkey skeletal muscle cells (7,8). The accumulation of Hsp70 in cardiac cells after heat stress is known to protect the myocardium from ischemic injury (9). We found that the aged LVCs expressed less hsc70 and α B-crystallin mRNAs. One of the roles of the constitutively expressed hsc70 is to bind to misfolded polypeptides to deter inappropriate interactions, which may lead to aggregation and loss of function (10,11). α B-crystallin has been shown to protect against hyperthermia, hypertonic stress and various cytotoxic agents, and its ectopic expression in adult rat cardiomyocytes leads to increased protection against ischemic injury (12). We found that Hsp32 mRNA was present in young LVCs and undetectable in cDNA libraries from aged LVCs. Hsp32, also known as heme oxygenase 1 (HO-1), is inducible in many tissues by various agents, including heme compounds, heavy metals, sulfhydryl reagents and hydrogen peroxide (13). The heart is rich in cytochromes and is a site of heme synthesis, the substrate for HO-1 (14). The reduced expression of Hsp32 mRNA might significantly contribute to changes in myocardial mechanisms of cellular adaptation and sensitivity to environment.
It is not known exactly how changes in Hsp transcript levels contribute to the decline of cardiac performance with age. The precise mechanism by which heat-shock proteins confer LVC protection remains to be elucidated. This knowledge ultimately would help in developing pharmacological methods to improve the aging heart’s ability to respond to environmental and oxidative stresses.
We also found age-related changes in RNA levels in mitochondrially encoded transcripts. Constituents of the mitochondrial respiratory chain such as the mitochondrially encoded cytochrome b and cytochrome c oxidase subunit 3 were decreased in aged LVCs. Cytochrome b, a catalytic subunit of complex III, is directly involved in electron transfer. A decrease in both of these enzymatic activities with aging has been previously demonstrated (15). In mouse and monkey skeletal muscle cells mRNAs levels for some of the nuclear encoded subunits of these enzymes decline with aging (8,16). Although LVCs house a large number of mitochondria, the ATP consumption in these cells is high and the loss of metabolic activity in a certain number of mitochondria might result in cardiac life span shortening.
We observed less mitochondrial creatine kinase (Mi-CK) mRNA in aged LVCs. The Mi-CK enzymatic activity also declines with aging as does the steady state level of the Mi-CK mRNA (data not shown). Mi-CK octamer–substrate complexes have a stabilizing and protective effect against mitochondrial permeability and pore opening (17). In mouse skeletal muscle, the steady state level of this enzyme increases with aging (8,16). The cytosolic creatine kinase mRNA and enzymatic activity in LVCs (data not shown), however, were found to increase slightly with aging. Cytosolic creatine kinases play a crucial role in the energetics of Ca2+-homeostasis (18). It is possible that with aging, cardiac muscle cells switch on compensatory mechanisms that cause elevated activity of the cytosolic creatine kinase.
Aging is associated with changes in mRNA level from genes whose proteins play a role in the LVC contractile apparatus. For example, the mRNA level for dystrophin was reduced with aging. In humans, the cardiomyopathies associated with Duchenne muscular dystrophy (19), Becker muscular dystrophy (20) and X-linked dilated cardiomyopathy are all caused by defects in the dystrophin gene (21). Since dystrophin-deficient mice have an increased vulnerability to acute pressure overload in vivo the aging-associated reduced dystrophin mRNA level might contribute to a reduced threshold for the development of contractile dysfunction upon exposure to increased levels of mechanical stress (22). There was aging-specific decreases in mRNA for α tropomyosin, α skeletal actin and sarcoplasmic reticulum Ca2+-ATPase (SERCA2) transcripts in aged LVCs (Table 1, Figs 2 and 3, Table S1 in Supplementary Material). SERCA2 plays a key role in the contraction–relaxation cycle of the myocardium by controlling the intracellular Ca2+ concentration. A reduction in the SERCA2 mRNA level because of transcription repression has been documented in all models of heart failure, including human failing ventricles (23,24). None of these contractile apparatus protein genes has been associated with aging of mouse and rhesus monkey skeletal muscle (7,8). The LVC contractile apparatus most likely ages differently from that of skeletal muscle because of its significant functional difference, namely the continuous necessity of cardiac myocytes to produce force and motion.
The mechanism of aging modulation of mRNA levels remains to be determined. Aging could at least in part modulate the stability of certain mRNAs by a heat-shock protein–ubiquitin–proteasome pathway through the reduced level of Hsp70 (25). Aging might either induce or fail to trigger a pathway that leads to repression or activation of the transcription promoters. The levels of transcription factors could also be modulated. The Nkx2.5, GATA4, c-jun and Jun B mRNA levels declined with aging (Table 1, Figs 2 and 3, Table S1 in Supplementary Material). Jun homo- and hetero-dimers regulate transcription in response to stimuli of several genes by interacting with the AP-1, phorbol 12-O-tetradecanoate 13-acetate- and cAMP-response promoter elements (26). Concerted changes in the SERCA2 (27), cytochrome c (28), actin isoforms (29) and some of the heat-shock protein mRNA levels (30) may be a direct effect of the reduced levels of c-jun and junB with aging. AP-1 and GATA4 are both involved in the response to the cardiac pressure overload (29). AP-1 and GATA4 are targets of signaling cascades (31,32). Differences in response to stress may occur because of aging-related changes in protein phosphorylation or oxidative protein damage of cascade members. GATA4 fine-tunes cardiac myocyte-specific gene expression together with Nkx2.5 (33,34). The reduced Nkx2.5 mRNA level in aged LVCs coincides with the reduction of connexin-43 and cardiac myosin light-chain mRNAs. Nkx2.5 was previously shown to regulate the expression of both genes (35,36). The cardiac phenotype of mice expressing the mutant Nkx2.5 are in some ways similar to the aging heart (36). Both result in progressive cardiac conduction defects that may lead to heart failure.
The down regulation of these transcription factors may be important for the transition and establishment of an ‘aged mode’ of LVC transcription regulation of gene expression.
Although we might find some additional expression differences by applying less stringent conditions and testing a larger number of animals, our results suggest that aging alters expression in the ventricular cardiac muscle of the mouse. Aging influences gene expression in different cell types, some of which respond differently than the ventricular cardiac myocytes suggesting that aging gene expression pattern might differ according to the specialized cellular functions. It is not known exactly how the subset of mRNAs is selected for expression changes with LVC aging. It is not known if the LVC aging gene expression profile in cardiac myocytes from C57BL/6 mice displays similarities with the profiles of other mouse strains and of other mammalian species. It is not known if there is a connection between the pathways that lead to accumulation of the autofluorescent lipofuscin inclusions and to the LVC aging gene expression profile. We propose that several cellular genes whose mRNA levels change with LVC aging might profoundly influence the age-related pathologic changes in the heart.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Figure 1.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Xuesong Gu (Beth Israel Deaconess Medical Center) for her assistance in Affymetrix array analyses and J. Goodhaus (Princeton University, Department of Molecular Biology) for his assistance in confocal microscopy. We also thank Jeanne Wei, Simon Robson, Mark Gray, Tony Hollenberg, Roumen Pankov and Chu Choi for helpful discussions. This work was supported by the American Heart Association grant DC03299 and National Institutes of Health grant HL62458 to A.U. and AG18536 to K.K.
REFERENCES
- 1.Vijg J. and Wei,J.Y. (1995) Understanding the biology of aging: the key to prevention and therapy. J. Am. Geriatr. Soc., 43, 426–434. [DOI] [PubMed] [Google Scholar]
- 2.Finch C.E. and Tanzi,R.E. (1997) Genetics of aging. Science, 278, 407–411. [DOI] [PubMed] [Google Scholar]
- 3.Perls T.T., Bubrick,E., Wager,C.G., Vijg,J. and Kruglyak,L. (1998) Siblings of centenarians live longer. Lancet, 351, 1560. [DOI] [PubMed] [Google Scholar]
- 4.Kang P.M., Haunstetter,A., Aoki,H., Usheva,A. and Izumo,S. (2000) Morphological and molecular characterization of adult cardiomyocyte apoptosis during hypoxia and reoxygenation. Circ. Res., 87, 118–125. [DOI] [PubMed] [Google Scholar]
- 5.Terman A. and Brunk,U.T. (1998) Lipofuscin: mechanisms of formation and increase with age. APMIS, 106, 265–276. [DOI] [PubMed] [Google Scholar]
- 6.Sohal R.S. and Wolfe,L.S. (1986) Lipofuscin: characteristics and significance. Prog.Brain Res., 70, 171–183. [DOI] [PubMed] [Google Scholar]
- 7.Lee C.K., Klopp,R.G., Weindruch,R. and Prolla,T.A. (1999) Gene expression profile of aging and its retardation by caloric restriction. Science, 285, 1390–1393. [DOI] [PubMed] [Google Scholar]
- 8.Kayo T., Allison,D.B., Weindruch,R. and Prolla,T.A. (2001) Influences of aging and caloric restriction on the transcriptional profile of skeletal muscle from rhesus monkeys. Proc. Natl Acad. Sci. USA, 98, 5093–5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lepore D.A., Knight,K.R., Anderson,R.L. and Morrison,W.A. (2001) Role of priming stresses and Hsp70 in protection from ischemia- reperfusion injury in cardiac and skeletal muscle. Cell Stress Chaperones, 6, 93–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connell P., Ballinger,C.A., Jiang,J., Wu,Y., Thompson,L.J., Hohfeld,J. and Patterson,C. (2001) The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nature Cell Biol., 3, 93–96. [DOI] [PubMed] [Google Scholar]
- 11.Strickland E., Qu,B.H., Millen,L. and Thomas,P.J. (1997) The molecular chaperone Hsc70 assists the in vitro folding of the N-terminal nucleotide-binding domain of the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem., 272, 25421–25424. [DOI] [PubMed] [Google Scholar]
- 12.Ray P.S., Martin,J.L., Swanson,E.A., Otani,H., Dillmann,W.H. and Das,D.K. (2001) Transgene overexpression of alphaB crystallin confers simultaneous protection against cardiomyocyte apoptosis and necrosis during myocardial ischemia and reperfusion. FASEB J., 15, 393–402. [DOI] [PubMed] [Google Scholar]
- 13.Choi A.M. and Alam,J. (1996) Heme oxygenase-1: function, regulation and implication of a novel stress-inducible protein in oxidant-induced lung injury. Am. J. Respir. Cell Mol. Biol., 15, 9–19. [DOI] [PubMed] [Google Scholar]
- 14.Choi A.M. (2001) Heme oxygenase-1 protects the heart. Circ. Res., 89, 105–107. [PubMed] [Google Scholar]
- 15.Lesnefsky E.J., Gudz,T.I., Moghaddas,S., Migita,C.T., Ikeda-Saito,M., Turkaly,P.J. and Hoppel,C.L. (2001) Aging decreases electron transport complex III activity in heart interfibrillar mitochondria by alteration of the cytochrome c binding site. J. Mol. Cell. Cardiol., 33, 37–47. [DOI] [PubMed] [Google Scholar]
- 16.Weindruch R., Kayo,T., Lee,C.K. and Prolla,T.A. (2001) Microarray profiling of gene expression in aging and its alteration by caloric restriction in mice. J. Nutr., 131, 918S–923S. [DOI] [PubMed] [Google Scholar]
- 17.Wallimann T., Dolder,M., Schlattner,U., Eder,M., Hornemann,T., O’Gorman,E., Ruck,A. and Brdiczka,D. (1998) Some new aspects of creatine kinase (CK): compartmentation, structure, function and regulation for cellular and mitochondrial bioenergetics and physiology. Biofactors, 8, 229–234. [DOI] [PubMed] [Google Scholar]
- 18.de Groof A.J., Fransen,J.A., Errington,R.J., Willems,P.H., Wieringa,B. and Koopman,W.J. (2002) The creatine kinase system is essential for optimal refill of the sarcoplasmic reticulum Ca2+ store in skeletal muscle. J. Biol. Chem., 277, 5275–5284. [DOI] [PubMed] [Google Scholar]
- 19.Cziner D.G. and Levin,R.I. (1993) The cardiomyopathy of Duchenne’s muscular dystrophy and the function of dystrophin. Med. Hypotheses, 40, 169–173. [DOI] [PubMed] [Google Scholar]
- 20.Ishigaki C., Patria,S.Y., Nishio,H., Yoshioka,A. and Matsuo,M. (1997) Early cardiac failure in a child with Becker muscular dystrophy is due to an abnormally low amount of dystrophin transcript lacking exon 13. Acta Paediatr. Jpn, 39, 685–689. [DOI] [PubMed] [Google Scholar]
- 21.Ferlini A., Sewry,C., Melis,M.A., Mateddu,A. and Muntoni,F. (1999) X-linked dilated cardiomyopathy and the dystrophin gene. Neuromusc. Disord., 9, 339–346. [DOI] [PubMed] [Google Scholar]
- 22.Danialou G., Comtois,A.S., Dudley,R., Karpati,G., Vincent,G., Des Rosiers,C. and Petrof,B.J. (2001) Dystrophin-deficient cardiomyocytes are abnormally vulnerable to mechanical stress-induced contractile failure and injury. FASEB J., 15, 1655–1657. [DOI] [PubMed] [Google Scholar]
- 23.Aoyagi T., Yonekura,K., Eto,Y., Matsumoto,A., Yokoyama,I., Sugiura,S., Momomura,S., Hirata,Y., Baker,D.L. and Periasamy,M. (1999) The sarcoplasmic reticulum Ca2+-ATPase (SERCA2) gene promoter activity is decreased in response to severe left ventricular pressure-overload hypertrophy in rat hearts. J. Mol. Cell. Cardiol., 31, 919–926. [DOI] [PubMed] [Google Scholar]
- 24.Somura F., Izawa,H., Iwase,M., Takeichi,Y., Ishiki,R., Nishizawa,T., Noda,A., Nagata,K., Yamada,Y. and Yokota,M. (2001) Reduced myocardial sarcoplasmic reticulum Ca(2+)-ATPase mRNA expression and biphasic force-frequency relations in patients with hypertrophic cardiomyopathy. Circulation, 104, 658–663. [DOI] [PubMed] [Google Scholar]
- 25.Laroia G., Cuesta,R., Brewer,G. and Schneider,R.J. (1999) Control of mRNA decay by heat shock–ubiquitin–proteasome pathway. Science, 284, 499–502. [DOI] [PubMed] [Google Scholar]
- 26.Karin M., Liu,Z. and Zandi,E. (1997) AP-1 function and regulation. Curr. Opin. Cell Biol., 9, 240–246. [DOI] [PubMed] [Google Scholar]
- 27.Du Y., Carlock,L. and Kuo,T.H. (1995) The mouse plasma membrane Ca2+ pump isoform 1 promoter: cloning and characterization. Arch. Biochem. Biophys., 316, 302–310. [DOI] [PubMed] [Google Scholar]
- 28.Xia Y., Buja,L.M. and McMillin,J.B. (1998) Activation of the cytochrome c gene by electrical stimulation in neonatal rat cardiac myocytes. Role of NRF-1 and c-Jun. J. Biol. Chem., 273, 12593–12598. [DOI] [PubMed] [Google Scholar]
- 29.Herzig T.C., Jobe,S.M., Aoki,H., Molkentin,J.D., Cowley,A.W.,Jr, Izumo,S. and Markham,B.E. (1997) Angiotensin II type1a receptor gene expression in the heart: AP-1 and GATA-4 participate in the response to pressure overload. Proc. Natl Acad. Sci. USA, 94, 7543–7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Camhi S.L., Alam,J., Otterbein,L., Sylvester,S.L. and Choi,A.M. (1995) Induction of heme oxygenase-1 gene expression by lipopolysaccharide is mediated by AP-1 activation. Am. J. Respir. Cell Mol. Biol., 13, 387–398. [DOI] [PubMed] [Google Scholar]
- 31.Lin A., Smeal,T., Binetruy,B., Deng,T., Chambard,J.C. and Karin,M. (1993) Control of AP-1 activity by signal transduction cascades. Adv. Second Messenger Phosphoprotein Res., 28, 255–260. [PubMed] [Google Scholar]
- 32.Liang Q., Wiese,R.J., Bueno,O.F., Dai,Y.S., Markham,B.E. and Molkentin,J.D. (2001) The transcription factor GATA4 is activated by extracellular signal-regulated kinase 1- and 2-mediated phosphorylation of serine 105 in cardiomyocytes. Mol. Cell. Biol., 21, 7460–7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lien C.L., Wu,C., Mercer,B., Webb,R., Richardson,J.A. and Olson,E.N. (1999) Control of early cardiac-specific transcription of Nkx2-5 by a GATA-dependent enhancer. Development, 126, 75–84. [DOI] [PubMed] [Google Scholar]
- 34.Kasahara H., Usheva,A., Ueyama,T., Aoki,H., Horikoshi,N. and Izumo,S. (2001) Characterization of homo- and heterodimerization of cardiac Csx/Nkx2.5 homeoprotein. J. Biol. Chem., 276, 4570–4580. [DOI] [PubMed] [Google Scholar]
- 35.Lyons I., Parsons,L.M., Hartley,L., Li,R., Andrews,J.E., Robb,L. and Harvey,R.P. (1995) Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev., 9, 1654–1666. [DOI] [PubMed] [Google Scholar]
- 36.Kasahara H., Wakimoto,H., Liu,M., Maguire,C.T., Converso,K.L., Shioi,T., Huang,W.Y., Manning,W.J., Paul,D., Lawitts,J. et al. (2001) Progressive atrioventricular conduction defects and heart failure in mice expressing a mutant Csx/Nkx2.5 homeoprotein. J. Clin. Invest., 108, 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




