Abstract
An RNA affinity tag was incorporated into the RNA subunit of human nuclear RNase P. The tagged RNA assembled with the protein components of RNase P inside HeLa cells to generate an active enzyme. Because of the specificity of the RNA tag to streptavidin, the reconstituted complex could be separated from the native enzyme and other ribonucleoproteins (particularly RNase MRP) by streptavidin agarose chromatography and could be recovered by the eluting agent, biotin. A mutant, tagged RNase P RNA, whose P3 domain was partially replaced, could not reconstitute with the proteins to yield an active enzyme. The P3 domain, therefore, is critical for the structure and function of RNase P.
INTRODUCTION
Ribonuclease P (RNase P) is an essential enzyme required for the processing of the 5′ termini of precursor tRNAs in all living organisms (1). The eubacterial RNase P consists of one RNA subunit and one protein subunit. The RNA subunit retains its catalytic function in a high concentration of salt in vitro in the absence of the protein subunit (2). However, the protein component facilitates catalysis under physiological salt concentrations and is required for efficient tRNA processing in vivo (3).
The currently characterized eukaryotic (either Saccharomyces cerevisiae or Homo sapiens) nuclear RNase P possesses numerous protein subunits and one RNA subunit (4,5). Unlike the eubacterial enzyme, the combination of human RNase P RNA transcribed in vitro and its purified recombinant proteins expressed in Escherichia coli when mixed together did not achieve an active enzyme (data not shown). Total, reliably reproduced, reconstitution in vitro of the RNA and protein subunits of the eukaryotic RNase P has not been reported so far.
An RNA aptamer (S1) has been developed recently to bind specifically to streptavidin (6). The RNA subunit of yeast nuclear RNase P was tagged with the streptavidin-binding element and the tagged RNA subunit was shown to be able to reconstitute an active enzyme with the protein subunits in vivo. The enzyme could be isolated from column chromatography by elution with biotin because the S1-binding ability to streptavidin agarose beads is much lower than that of biotin (6). In this study, we have shown that the RNA affinity tag also works well in a mammalian system, and the aptamer-tagged human nuclear RNase P could be immobilized to streptavidin agarose from crude cell extracts and eluted with biotin under mild conditions. In addition, we have applied this method to the study of an essential protein-binding domain (P3) of the RNase P RNA from HeLa cells.
MATERIALS AND METHODS
Materials
Restriction and modification enzymes were obtained from New England Biolabs. Radiochemicals were obtained from Amersham. Oligonucleotides were synthesized by the Keck Facility at Yale University.
Cloning and mutagenesis. Four oligonucleotides, oligo 1 (AAAAG GGAGT CGACC GACCA GAATC ATGCA AGTGC GTAAG ATAGT CGCGG GCCGG GGGCG), oligo 2 (TATTA TGTGC GTCTA CATCT AGACT CATAA AAGGC CCCgg cc), oligo 3 (GGGGC CTTTT ATGAG TCTAG ATGTA GACGC ACATA ATACG CCCCC GGCCC GCG), oligo 4 (TATCT TACGC ACTTG CATGA TTCTG GTCGG TCGAC TCCCT TTTgg cc) were annealed and ligated with pUC/H1 cut by ApaI. The bases represented by lower case letters facilitate cloning into the sticky ends of the restriction enzymes. The resulting plasmid was then digested by EcoRI and SalI, and the ∼400 bp fragment was ligated to the plasmid pΔEGFP which is derived from pEGFP-N1 (Clontech) after removal of the gene encoding EGFP. The final plasmid was named as pΔEGFP–H1S1 containing CMV IE promoter, the gene for H1S1 (H1 RNA with S1 tag), kanamycin and neomycin selection markers, and colE1 and SV40 replication origins.
To generate the deletion of P3 domain of H1 RNA, two oligonucleotides, P3forward (AGAAT TCATA GGGCG GAGGG AAGCT CATCA GTGGG GATGT CCCTT GGGAA GGT) and P3reverse (AAGGA TCCAA TGGGC GGAGG AGAGT AGT) were used to perform PCR with pΔEGFP–H1S1 as template. The PCR fragment was cloned into pΔEGFP after being digested by EcoRI to get the plasmid pΔEGFP–H1S1ΔP3 (Fig. 1). The coding regions for H1S1 and H1S1ΔP3 in the plasmids pΔEGFP–H1S1 and pΔEGFP–H1S1ΔP3 were confirmed by DNA sequencing with T7 Sequenase (Amersham).
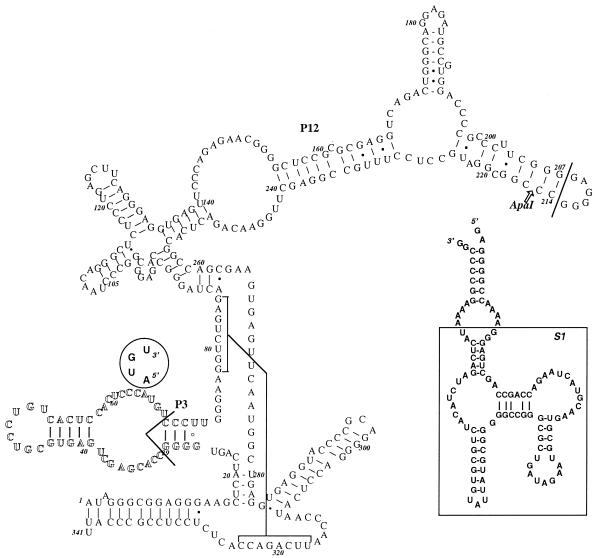
Figure 1.
The secondary structure of human nuclear RNase P from HeLa cells. The P3 domain nucleotides are shown in hollow letters. The restriction cleavage site for ApaI is indicated as an arrow. The S1 aptamer (rectangle) with an extra stem–loop was inserted into the gene for H1 RNA between position +207 and +214 in the original sequence in the construct of pΔEGFP–H1S1. The main part of the P3 domain (from +30 to +67) was replaced additionally by a tetraloop AUGU (see circle) in the construct of pΔEGFP–H1S1ΔP3. P12 extends from position 155 to position 240 (7).
Transfection of HeLa cells
Transfection of the plasmids into HeLa S3 cells was done with SuperFect transfection reagent (Qiagen) according to the manufacturer’s handbook. The cells were incubated for 24 h after transfection, washed with PBS, harvested and stored at –80°C.
Purification of S1-tagged human RNase P
The purification was performed at 4°C as described (6) with minor modifications. The pellets of cells (∼5 × 107) were resuspended in 0.5 ml of lysis buffer (50 mM HEPES, pH 8.0, 10 mM MgCl2, 100 mM KCl, 1 mM DTT, 10% glycerol) with the Complete protease inhibitor cocktail (Roche). The mixture was homogenized by passing it 15 times through a 25G7/8 needle fitted into a 1 ml syringe. The lysate was centrifuged at 16 000 g for 30 min. The S16 cell extract was incubated and rotated with 50 µl of streptavidin agarose (Gibco BRL) overnight in a 15 ml tube. The beads were spun down to remove the supernatant and washed five times with 5 ml of lysis buffer, for 15 min each. The beads were then transferred to an Ultrafree-MC centrifugal filter unit, 0.45 µm in size (Millipore), and further washed three times with 400 µl of lysis buffer. Elution was accomplished by incubating with 100 µl of lysis buffer containing 0.5 or 5 mM biotin for 1 h.
Northern blot analysis
Aliquots for the streptavidin agarose purification were extracted from column fractions by phenol and precipitated by ethanol. The samples were separated by a 3% agarose gel and then transferred onto a positively charged nylon membrane (Roche). The northern blot was performed with Rapid-hyb buffer (Amersham Pharmacia Biotech), using a 5′-terminally [32P] phosphorylated DNA oligonucleotide complementary to positions 319–340 of human RNase P RNA or 245–265 of human RNase MRP RNA.
Western blot analysis
Samples were separated in 13% polyacrylamide/0.1% SDS gels, electrotransferred to a nitrocellulose filter and immunoblotted with a 1:100 dilution of the primary antibody serum. As a secondary antibody, a 1:5000 dilution of goat anti-rabbit IgG antibody was used. Blots were washed and antibody–antigen complexes were visualized with ECL-Plus detection reagent (Amersham Pharmacia Biotech), following the manufacturer’s instructions. The membrane was stripped of bound antibodies and reprobed with others.
Enzyme activity assays
RNase P enzyme activities were assayed in 1× PA buffer (50 mM Tris–HCl, pH 7.5, 10 mM MgCl2 and 100 mM NH4Cl) at 37°C. The substrate RNA, E.coli precursor tRNATyr (pTyr), was transcribed in vitro (2) in the presence of [α-32P]GTP, purified on a 7 M urea 5% polyacrylamide gel, and used at a final concentration of 100 nM (2000 c.p.m. per reaction).
RESULTS
Cloning and transfection of the RNA subunit
Helix P12 of H1 RNA, the RNA subunit of human nuclear RNase P, is not conserved in vertebrates (7). Accordingly, the S1 aptamer (shown in the rectangle in Fig. 1) with an extra stem–loop was incorporated between the nucleotides at position +207 and +214 of the native H1 RNA (8) to allow for function of the whole H1 RNA molecule. The first nucleotide cloned is the 5′ G at position 208 and the last is the 3′ G at position 213 (see Fig. 1 for tag sequence). The S1-tagged H1 (H1S1) gene was placed into a mammalian expression vector pΔEGFP which is a truncated version of pEGFP-N1 (see Materials and Methods). The mutant, H1S1ΔP3, which has a deletion of the main part of the P3 domain of H1 RNA (Fig. 1), was also cloned into the pΔEGFP vector.
After transfection, either with pΔEGFP–H1S1 or pΔEGFP–H1S1ΔP3, the HeLa cells had a growth phenotype different from cells without transfection or transfected with pEGFP-N1. The new cells transfected with H1S1 RNA or H1S1ΔP3 RNA grew quite slowly (doubling time much longer than 24 h, the usual doubling time of these cells) and ∼10% of them died and were washed away by PBS washes (see Materials and Methods). The slow growth may be caused by the streptavidin tag in H1 RNA.
Purification by streptavidin agarose
The crude extracts (S16) of transfected cells were bound to streptavidin agarose and extensively washed with lysis buffer (Materials and Methods), and then eluted with lysis buffer with 0.5 mM and then 5 mM biotin. The eluates were analyzed for the presence of H1 RNA or MRP RNA by northern blot hybridization as well as for the activity of RNase P.
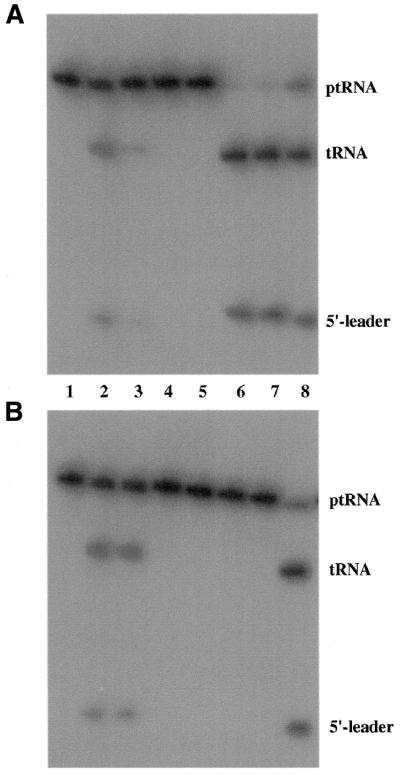
The H1S1 transfected cell extract contained the S1-tagged enzyme. The enzyme bound to streptavidin agarose and was eluted with 5 mM biotin (Fig. 2). The one-step purification fold resulted in an ∼410-fold enrichment of the RNase P activity, which is close to the 510-fold purification achieved by the classic three-step purification [DEAE Sepharose, glycerol gradient and FPLC monoQ column (9)]. However, for the mutant H1S1ΔP3 RNA, the RNA was isolated after chromatography (see below) but no active enzyme was detected after the elution.
Figure 2.
RNase P activity assay of fractions from the purification step of streptavidin agarose chromatography. Reaction conditions are detailed in Materials and Methods. (A) HeLa cells transfected with pΔEGFP–H1S1. Lane 1, no enzyme. Lane 2, S16 extract. Lane 3, the supernatant after rotation with streptavidin agarose overnight. Lane 4, the streptavidin column flow through washed with lysis buffer. Lane 5, the eluate (0.5 mM biotin). Lane 6, the eluate (5 mM biotin). Lane 7, the agarose beads assayed after eluates. Lane 8, RNase P purified by conventional methods. (B) The same procedures except extracts of HeLa cells were transfected with pΔEGFP–H1S1ΔP3.
Subsequent to the streptavidin agarose column, the RNase P with the tagged RNA is still impure because of contaminating proteins that might still bind to H1 RNA or to its protein subunits. The tagged RNA enzyme was then subjected to further chromatography as indicated by the conventional method for RNase P purification (9). The tagged RNA enzyme purification behavior is shown in Table 1, which shows a purification of 12 000-fold. The final steps, DEAE Sepharose, FPLC mono Q and Superose 6 and the streptavidin column can all be achieved in 1 week, though the enzyme is not yet pure as judged by SDS gel electrophoresis of the proteins (data not shown). The conventional method, including glycerol gradients, without the streptavidin column, results in a purification factor of 2300 and takes 1 month (9).
Table 1. Purification profile of H1S1-tagged human RNase P from HeLa cells transfected with pΔEGFP–H1S1.
| Purification | Total proteins (µg) | Total activities (U) | Specific activities (U/mg) | Purification fold |
|---|---|---|---|---|
| S16 | 110 000 | 65 | 0.59 | 1 |
| Streptavidin agarose | 158 | 38 | 240 | 410 |
| DEAE Sepharose | 34 | 22 | 650 | 1100 |
| Mono Q | 5.3 | 14.8 | 2800 | 4800 |
| Superose 6 | 1.4 | 9.9 | 7100 | 12 000 |
One unit of RNase P is defined as the amount of protein required to produce 1 pmol product/min at 37°C. The specific activity is defined as the amount of RNase P units in 1 mg of protein.
Northern blot hybridization
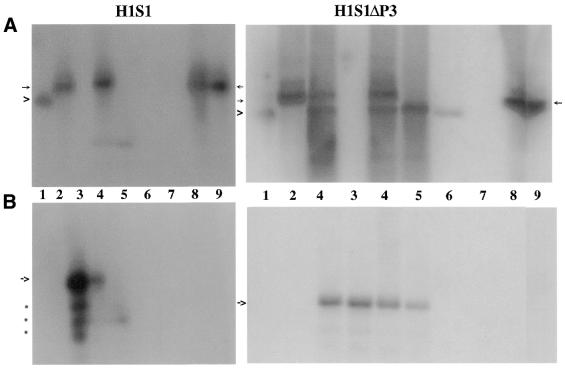
Northern blot analysis of the aliquots from the purification step revealed that both H1S1 and H1S1ΔP3 were found in their eluates of 5 mM biotin from the agarose beads, respectively. MRP RNA, which normally associates with RNase P in the early steps of its purification, was not present in the eluates. It should be noted that the expression of internal H1 RNA was almost completely inhibited in the cells transfected with pΔEGFP–H1S1 (Fig. 3, lane 3), but not in those transfected with pΔEGFP–H1S1ΔP3.
Figure 3.
Northern blot analysis of fractions from the purification step of streptavidin agarose (Materials and Methods). (A) Northern blot with an oligonucleotide probe against RNase P RNA. The arrowhead indicates H1 RNA and the arrow indicates H1S1 RNA. (B) Northern blot with an oligonucleotide probe against MRP RNA. The left part of each panel is from HeLa cells transfected with pΔEGFP–H1S1 whereas the right part is from cells transfected with pΔEGFP–H1S1ΔP3. Lane 1, H1 RNA transcribed in vitro by T7 RNA polymerase. Lane 2, H1S1 RNA transcribed in vitro by T7 RNA polymerase. Lane 3, MRP RNA transcribed in vitro by T7 RNA polymerase. Lane 4, S16 extract of HeLa cells transfected with pΔEGFP–H1S1 (left) or pΔEGFP–H1S1ΔP3 (right). Lane 6, flow through washed with lysis buffer. Lane 7, the eluate (0.5 mM biotin). Lane 8, the eluate (5 mM biotin). Lane 9, agarose beads after eluates. The arrow in (B) indicates MRP RNA and the dots indicate fragments of MRP RNA.
Western blot hybridization
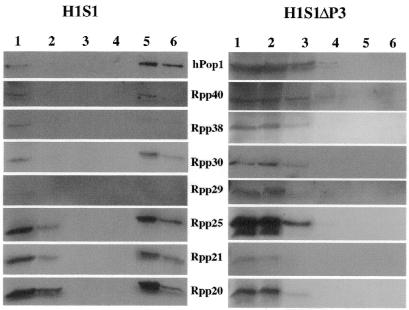
To determine whether there are protein subunits in the samples eluted from streptavidin columns, western blots were done to detect eight of the known protein components (hPop1, Rpp40, Rpp38, Rpp30, Rpp29, Rpp25, Rpp21 and Rpp20) of human nuclear RNase P by their cognate antibodies (9–14). As shown in Figure 4, all the proteins could be detected by their cognate antibodies in the 5 mM biotin eluate of cells transfected with pΔEGFP–H1S1, though there were two proteins (Rpp38 and Rpp29) that showed weak signals. This result is anticipated if the tagged RNA is part of an active enzyme. However, none of the proteins could be detected in the corresponding sample from the cells transfected with pΔEGFP–H1S1ΔP3.
Figure 4.
Western blot analysis (Materials and Methods) of fractions from the purification step of streptavidin agarose. The blots have been probed with antibodies to the various proteins listed between the columns of figures. The left part is from HeLa cells transfected with pΔEGFP–H1S1 whereas the right part is from cells transfected with pΔEGFP–H1S1ΔP3. Lane 1, S16 extract. Lane 2, supernatant after rotation with streptavidin agarose overnight. Lane 3, flow through washed with lysis buffer. Lane 4, the eluate (0.5 mM biotin). Lane 5, the eluate (5 mM biotin). Lane 6, agarose beads after eluates.
DISCUSSION
We have shown that the S1-tagged H1 RNA could reconstitute with RNase P proteins in vivo to form an active enzyme. The mutant, H1S1ΔP3, the P3 domain of which was deleted, did not yield any active enzyme. The S1-tagged enzyme could easily be separated from the native enzyme by a one-step streptavidin agarose column with a 410-fold purification. This purification achieves what a three-step conventional method does for human nuclear RNase P (9). Eight of the protein components could be detected in the eluted sample. Further purification could be conducted by DEAE Sepharose, FPLC monoQ and Superose 6 (Table 1).
At least five protein subunits of human RNase P, namely Rpp29, Rpp30, Rpp38, hPop1 and hPop5, have been shown to be associated with RNase MRP (15–18). The overlap of subunit composition and the difficulty in separating easily RNase P from RNase MRP raised the possibility that the two ribonucleoproteins might be in the same complex in vivo. The eluate that contains RNase P from streptavidin agarose did not contain the MRP RNA, indicating that the RNase P and MRP were very likely not associated with each other in one complex. (We assume that lack of MRP RNA is an indication that no functional RNase MRP is in the fractions we show in the figure.) We could not exclude completely the possibility that the S1 tag affects the interaction between RNase P and MRP in vivo, if there is any.
The P3 domain is conserved in nuclear RNase P and MRP in eukaryotic cells (7,19). This domain may play a key role in the entry of RNase P RNA and MRP RNA to the nucleolus in normal rat kidney epithelial cells (20,21). The P3 domain of yeast nuclear RNase P RNA is an essential protein-binding domain (22). The P3 domain of human nuclear RNase P RNA interacts with Rpp21, Rpp29, Rpp30 and Rpp38 in vitro by UV cross-linking (23), suggesting that these proteins are bound to the RNA subunit of the enzyme. The H1S1ΔP3 RNA was unable to assemble with the protein subunits of RNase P to produce an active enzyme. This result indicated the importance of the P3 domain of RNase P in the assembly and function of nuclear RNase P.
There are some limits on the tagged RNA purification method. The prime concern is that a stable HeLa cell strain could not be obtained by transfection with pΔEGFP–H1S1. A similar event happened with plasmid pmU6-H1S1 using a mouse U6 promoter and pEGFP–H1S1 (data not shown). This phenomenon might be connected to the fact that H1S1 RNA expression can inhibit native H1 RNA expression and, therefore, must have effects on cell survival.
The streptavidin tag method offers further opportunities for studying the roles of particular segments of the RNA in enzyme function, and for purification of RNase P free of RNase MRP.
CONCLUSION
A streptavidin tag has been inserted into a segment of H1 RNA, the RNA component of human nuclear RNase P, and has facilitated the purification of an intact enzyme from crude cell extracts. This simple procedure has yielded an enzyme that is free of human RNase MRP and allows further steps in purification. A streptavidin tag of H1 RNA that is missing the deleted P3 helix does not yield RNase P. The method is therefore useful for testing different parts of H1 RNA for function and as a part of a purification procedure.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the members of our laboratory for helpful suggestions. The research was supported by a grant from the National Institutes of Health (GM19422) to S.A.
REFERENCES
- 1.Gopalan V., Vioque,A. and Altman,S. (2002) RNase P: variations and uses. J. Biol. Chem., 277, 6759–6762. [DOI] [PubMed] [Google Scholar]
- 2.Guerrier-Takada C., Gardiner,K., Marsh,T., Pace,N. and Altman,S. (1983) The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell, 35, 849–857. [DOI] [PubMed] [Google Scholar]
- 3.Kole R., Baer,M.F., Stark,B.C. and Altman,S. (1980) E. coli RNAase P has a required RNA component in vivo. Cell, 19, 881–887. [DOI] [PubMed] [Google Scholar]
- 4.Chamberlain J.R., Lee,Y., Lane,W.S. and Engelke,D.R. (1998) Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev., 12, 1678–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jarrous N. (2002) Human ribonuclease P: subunits, function and intranuclear localization. RNA, 8, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Srisawat C. and Engelke,D.R. (2001) Streptavidin aptamers: affinity tags for the study of RNAs and ribonucleoproteins. RNA, 7, 632–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank D.N., Adamidi,C., Ehringer,M.A., Pitulle,C. and Pace,N.R. (2000) Phylogenetic-comparative analysis of the eukaryal ribonuclease P RNA. RNA, 6, 1895–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartkiewicz M., Gold,H. and Altman S. (1989) Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes Dev., 3, 488–499. [DOI] [PubMed] [Google Scholar]
- 9.Eder P.S, Kekuda,R., Stolc,V. and Altman,S. (1997) Characterization of two scleroderma autoimmune antigens that copurify with human ribonuclease P. Proc. Natl Acad. Sci. USA, 94, 1101–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lygerou Z., Mitchell,P., Petfalski,E., Seraphin,B. and Tollervey,D. (1994) The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev., 8, 1423–1433. [DOI] [PubMed] [Google Scholar]
- 11.Jarrous N., Eder,P.S., Guerrier-Takada,C., Hoog,C. and Altman,S. (1998) Autoantigenic properties of some protein subunits of catalytically active complexes of human ribonuclease P. RNA, 4, 407–417. [PMC free article] [PubMed] [Google Scholar]
- 12.Jarrous N., Eder,P.S., Wesolowski,D. and Altman,S. (1999) Rpp14 and Rpp29, two protein subunits of human ribonuclease P. RNA, 5, 153–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarrous N., Reiner,R., Wesolowski,D., Mann,H., Guerrier-Takada,C. and Altman,S. (2001) Function and subnuclear distribution of Rpp21, a protein subunit of the human ribonucleoprotein ribonuclease P. RNA, 7, 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guerrier-Takada C., Eder,P.S., Gopalan,V. and Altman,S. (2002) RNA, 8, 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lygerou Z., Pluk,H., van Venrooij,W.J. and Seraphin,B. (1996) hPop1: an autoantigenic protein subunit shared by the human RNase P and RNase MRP ribonucleoproteins. EMBO J., 15, 5936–5948. [PMC free article] [PubMed] [Google Scholar]
- 16.Pluk H., van Eenennaam,H., Rutjes,S.A., Pruijn,G.J. and van Venrooij,W.J. (1999) RNA–protein interactions in the human RNase MRP ribonucleoprotein complex. RNA, 5, 512–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Eenennaam H., Pruijn,G.J. and van Venrooij,W.J. (1999) hPop4: a new protein subunit of the human RNase MRP and RNase P ribonucleoprotein complexes. Nucleic Acids Res., 27, 2465–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Eenennaam H., Lugtenberg,D., Vogelzangs,J.H., van Venrooij,W.J. and Pruijn,G.J. (2001) hPop5, a protein subunit of the human RNase MRP and RNase P endoribonucleases. J. Biol. Chem., 276, 31635–31641. [DOI] [PubMed] [Google Scholar]
- 19.Schmitt M.E. (1999) Molecular modeling of the three-dimensional architecture of the RNA component of yeast RNase MRP. J. Mol. Biol., 29, 2827–2836. [DOI] [PubMed] [Google Scholar]
- 20.Jacobson M.R., Cao,L.G., Taneja,K., Singer,R.H., Wang,Y.L. and Pederson,T. (1997) Nuclear domains of the RNA subunit of RNase P. J. Cell Sci., 110, 829–837. [DOI] [PubMed] [Google Scholar]
- 21.Jacobson M.R., Cao,L.G., Wang,Y.L. and Pederson,T. (1995) Dynamic localization of RNase MRP RNA in the nucleolus observed by fluorescent RNA cytochemistry in living cells. J. Cell Biol., 131, 1649–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ziehler W.A., Morris,J., Scott,F.H., Millikin,C. and Engelke,D.R. (2001) An essential protein-binding domain of nuclear RNase P RNA. RNA, 7, 565–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang T., Guerrier-Takada,C. and Altman,S. (2001) Protein–RNA interactions in the subunits of human nuclear RNase P. RNA, 7, 937–941. [DOI] [PMC free article] [PubMed] [Google Scholar]