Abstract
iceA1 in Helicobacter pylori is a homolog of nlaIIIR, which encodes the CATG-specific restriction endonuclease NlaIII in Neisseria lactamica. Analysis of iceA1 sequences from 49 H.pylori strains shows that a full-length NlaIII-like ORF is present in 10 strains, including CH4, but in other strains, including strain 60190, the ORFs are truncated due to a variety of mutations. Our goal was to determine whether iceA1 can encode a NlaIII-like endonuclease. Overexpression in Escherichia coli of iceA1 from CH4, but not from 60190, yielded NlaIII-like activity, indicating that the full-length iceA1 is a functional endonuclease gene. Repair of the iceA1 frameshift mutation in strain 60190 and its expression in E.coli yielded functional NlaIII-like activity. We conclude that iceA1 in CH4 is a functional restriction endonuclease gene, while iceA1 in 60190 is not, due to a frameshift mutation, but that its repair restores its restriction endonuclease activity.
INTRODUCTION
Helicobacter pylori is a gram-negative bacterium that colonizes the stomach of more than half of the world’s population, and their presence increases the risk of developing peptic ulcers and gastric adenocarcinomas (1–7). In Western populations, H.pylori strains of particular genotypes (such as cagA+,vacA s1m1) are more virulent than other strains (8–11). Recently, two unrelated genes, designated iceA1 and iceA2, have been identified, which are present at the same genomic locus among various H.pylori strains (12). iceA1 expression is up-regulated by contact with epithelial cells (12) and in some (12,13), but not all, populations (14) the iceA1 genotype is associated with peptic ulcers. However, it is not known whether the gene is functional and whether or how the iceA1 product is involved in H.pylori colonization of the human stomach.
DNA analysis indicates that iceA1, but not iceA2, has strong homology to nlaIIIR (15), which encodes a CATG-specific endonuclease in Neisseria lactamica. A nlaIIIM-like methylase gene, hpyIM (16), is located immediately downstream of either iceA1 or iceA2. Therefore, the iceA1–hpyIM gene locus is a homolog of the CATG-specific type II restriction–modification system in N.lactamica (15,17). Our previous studies indicate that hpyIM is well conserved and encodes a CATG-specific DNA methylase in H.pylori (16), and that promoters for hpyIM expression vary between iceA1 and iceA2 strains (18). Whether or not iceA1 encodes a functional endonuclease gene remains to be determined.
Studies on H.pylori strains from different geographic origins demonstrated that most iceA1 genes, including those in strains 60190 (12) and 26695 (19), have frameshifts or nonsense mutations leading to early termination in their nlaIIIR-like ORF (14,20), whereas iceA1 from a few strains, like CH4, have an intact nlaIIIR-like ORF. To determine whether iceA1 encodes a NlaIII-like endonuclease and whether the polymorphisms in iceA1 affect the function of its product, in this study we examined the function of iceA1 from H.pylori strains CH4 and 60190, which represent full-length and truncated forms of iceA1, respectively.
MATERIALS AND METHODS
Nucleotide sequence accession number
The sequence of iceA1 from H.pylori strain CH4 has been submitted to GenBank with the accession no. AF459446.
Strains, plasmids, growth conditions and reagents
Helicobacter pylori strains, Escherichia coli strains and plasmids used in this study are listed in Table 1, and growth conditions of E.coli and H.pylori strains were as described (16). Restriction enzymes, digestion buffers and substrate DNAs were from New England Biolabs (Beverly, MA) and columns for protein purification were from Biosepra (Marlborough, MA). Oligonucleotides used in this study (Table 1) were synthesized using a Milligen 7500 DNA synthesizer at the Vanderbilt University Cancer Center DNA Core Facility.
Table 1. Strains, plasmids and oligonucleotides used in this study.
| Name | Relevant genotype or sequence (5′→3′) | Reference or orientation |
|---|---|---|
| Escherichia coli | ||
| ER2169 | mcrBC, λDE3, pLysS, pSYX20-NlaIIIM, CmR, KanR | (15) |
| Helicobacter pylori | ||
| 60190 | iceA1, cagA+, vacA s1a m1 | (9,10,12) |
| CH4 | iceA1, cagA+, vacA s1a m2 | (9,10,12) |
| Plasmids | ||
| pSYX22 | pET11-derived vector, AmpR | (15) |
| pQXCH4 | CH4 iceA1 in pSYX22 | This study |
| pQX60A | 60190 iceA1 starting from ATG site, in pSYX22 | This study |
| pQX60T | 60190 iceA1 starting from TTG site, in pSYX22 | This study |
| pQX60R | The repaired iceA1 of 60190 in pSYX22 | This study |
| Primers | ||
| IceA60A-F | CACACATATGTGTGGTGTGCGTGGCAACTC | Forward |
| IceA60T-F | TAGCCATATGGCTAAAGAATTTAATTTGGAG | Forward |
| IceACH4-R | GTTGGATCCGTCGACGTAGTTCATTGCAACCG | Reverse |
| IceA-F | GTTGGATCCTTAAGGAGGTTTAACATATGAGTAAAAGTAAAAAG | Forward |
| IceA-R | GTTGGATCCGTCGACGTAGTTCATTGCAACC | Reverse |
| Fix-F | GGTCTGCAGCTAGGTAATGGGGGGAGTTGGTGCAGG | Forward |
| Fix-R | TAGCTGCAGTCCTTGGTATTTTCC | Reverse |
DNA techniques
Preparation of plasmid and chromosomal DNA, digestion of DNA with restriction endonucleases, automated DNA sequencing and PCR were performed as described (16). Computer analysis of DNA and amino acid sequences was performed with the GCG program (21).
Construction of plasmids
Plasmids pQXCH4, pQX60T and pQX60A (Table 1), which carry a full-length nlaIIIR-like ORF from strain CH4, the longest ORF (starting from the TTG site) in strain 60190 iceA1 and an ORF starting from the downstream ATG site in 60190 iceA1, respectively, were constructed as follows. DNA fragments containing the strain CH4 iceA1 ORF were amplified through PCR using IceA-F (with a 5′ NdeI site) and IceACH4-R (with a 5′ SalI site) as primers and chromosomal DNA from strain CH4 as template. DNA fragments containing the two 60190 iceA1 ORFs starting from the TTG or ATG sites were created by PCR using IceA60T-F (with a 5′ NdeI site) or IceA60A-F (with a 5′ NdeI site), respectively, with iceA-R (with a 5′ SalI site) as primer pairs and 60190 chromosomal DNA as template. After digestion of the DNA fragments with NdeI and SalI, the PCR products were ligated with NdeI/SalI-digested and dephosphorylated vector pSYX22 in the presence of T4 DNA ligase to create pQXCH4, pQX60T and pQX60A. Each ligation mixture was transformed into E.coli strain ER2169 carrying a NlaIII methylase gene (nlaIIIM) in pSYX20-NlaIIIM to protect the E.coli host DNA from cleavage (15) and transformants were selected on LB plates containing ampicillin and kanamycin. Plasmid DNA was prepared from the transformants and the constructs were confirmed by DNA sequencing.
Repair of the frameshift in 60190 iceA1 and construction of pQX60R
A site-directed mutagenesis method was used to repair a point deletion mutation in H.pylori strain 60190 iceA1. Two pairs of PCR primers, iceA-F (with a 5′ NdeI site) and fix-R (with a 5′ PstI site) and fix-F (with a 5′ PstI site and an extra G residue in the region corresponding to the site of the frameshift site in iceA1) and iceA-R (with a 5′ SalI site) were designed (Table 1) to correct the frameshift of iceA1 located between the upstream (ATG) and the second (TTG) putative start codons. With primer pair iceA-F and fix-R and chromosomal DNA of H.pylori 60190 as template, a 140 bp PCR product was amplified, representing the 5′-most 105 bp region of iceA1, beginning at the first ATG site. With fix-F and iceA-R as primers and the same DNA as template, a 616 bp PCR product was produced, representing the remainder of iceA1, from bp 106 to the end of the gene, including the frameshift site where a point deletion mutation was present. Since primer fix-F has an extra G residue inserted at a position equivalent to bp 123 of 60190 iceA1 (Table 1), the iceA1 frameshift was repaired in the 616 bp PCR product. After digestion of the 140 bp PCR product with NdeI and PstI, and the 616 bp PCR product with SalI and PstI, the two PCR products were ligated with the NdeI/SalI-digested vector pSYX22 to create pQX60R. The ligation mixture was transformed into E.coli strain ER2169 carrying pSYX20-NlaIIIM (15) and transformants were selected on LB plates containing ampicillin and kanamycin. Plasmid DNA was prepared and sequenced to confirm that the point deletion mutation of iceA1 had been corrected.
Expression in E.coli of iceA1 from CH4 and from 60190
In pQXCH4, pQX60A, pQX60T and pQX60R, each iceA1 gene was inserted downstream of a ribosomal binding site, under the control of the T7 promoter (22). In E.coli strains such as ER2169, the T7 RNA polymerase gene is under the control of the lac operon. Thus, gene expression was inducible with IPTG in these E.coli strains. To express iceA1, E.coli strain ER2169/pSYX20-NlaIIIM carrying one of the plasmids was grown in LB medium with ampicillin, kanamycin and IPTG at 37°C overnight. Escherichia coli ER2169/pSYX20-NlaIIIM without an iceA1-containing plasmid was grown under the same conditions as a control. To detect expression of the iceA1 product, the cells from E.coli ER2169/pSYX20-NlaIIIM with or without each of the iceA1 plasmids were lysed by boiling at 100°C. The lysates were resolved by SDS–PAGE (4% stacking gels and 8% separating gels) and gels were stained with Coomassie brillliant blue (23).
Endonuclease activity assay
To determine whether the iceA1 products encode NlaIII-like endonucleases, the cultured cells were disrupted by sonication, followed by centrifugation, as described (24). The supernatant underwent serial dilutions in NEB buffer 4 and was then used to digest λ DNA, and the products were resolved on agarose gels. To further purify the iceA1 product, the remainder of the crude extract was applied to a Heparin Hyper D column, which was washed and eluted, as described (24). The eluted fractions from this column were assayed for endonuclease activity and the active fractions were used in parallel with NlaIII to digest pBR322, pUC19 and φX174 substrate DNA to determine whether the iceA1 protein has NlaIII-like endonuclease activity.
RESULTS
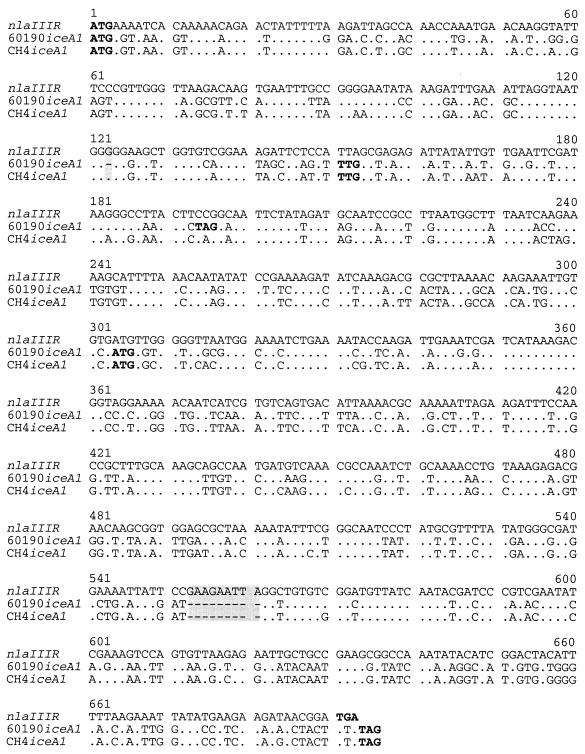
Comparison of the DNA sequences of nlaIIIR from N.lactamica and iceA1 from H.pylori strains 60190 and CH4
DNA analysis demonstrates that iceA1 from H.pylori has ∼60% similarity to nlaIIIR, which has a 690 bp ORF and encodes a 230 amino acid protein product, the restriction endonuclease NlaIII (15). However, the homology at the amino acid level is limited in most of the 49 iceA1 genes studied (14,20; GenBank accession nos AF239991–AF239994 and AF001537–AF001539), because of frameshifts and early terminations in the nlaIIIR-like ORF. Thus far, a full-length nlaIIIR-like ORF has only been observed in 10 (PO31, AU8, AU18, CH4, F38, F72, F43, India227, Alaska209 and Alaska218) of 49 iceA1 strains studied, and similarity between their deduced IceA1 proteins and NlaIII was 52–57% (14,20).
To determine whether iceA1 encodes a restriction endonuclease, we chose strains CH4 and 60190 for functional analysis. iceA1 from strain CH4 has a full-length nlaIIIR-like 684 bp ORF, encoding a 228 amino acid product, very similar to that of nlaIIIR, whereas the nlaIIIR-like ORF in strain 60190 is not complete and the longest ORF is only 534 bp, encoding a 178 amino acid product. Alignment of sequences with nlaIIIR (Fig. 1) reveals several mutations in the predicted iceA1 coding region, including a 9 bp in-frame deletion that is present in both CH4 iceA1 and 60190 iceA1, and a single nucleotide G deletion in the run of six G residues present in CH4, which results in a frameshift in the 60190 iceA1 coding region. This frameshift results in the ORF terminating at a TAG beginning at position 193 of 60190 iceA1, and leads to a 64 amino acid ORF representing the N-terminus of a putative restriction endonuclease. The longest predicted coding region from the 60190 iceA1 starts from a TTG site that is 150 bp downstream of the nlaIIIR-equivalent start site ATG, to form a 534 bp ORF. This would encode a 178 amino acid product corresponding to the C-terminus of a putative restriction endonuclease. An ORF starting from an ATG at position 304, an iceA1 translational start site that is conserved in various strains (20), is present within both CH4 and 60190 iceA1.
Figure 1.
Alignment of the nucleotide sequences of N.lactamica nlaIIIR (GenBank accession no. U59398) and the iceA1 regions from H.pylori strains 60190 and CH4. Deletion and insertion mutations in the iceA1 regions, relative to the full-length nlaIIIR, are shaded. Dots indicate identical nucleotides. Dashes indicate gaps in the nucleotide sequence. Potential translation start and stop sites are shown in bold.
Expression of the CH4 and 60190 iceA1 products in E.coli
To determine whether iceA1 encodes a functional NlaIII-like endonuclease, we sought to express the potential nlaIIIR-like ORFs in E.coli. The 684 bp (full-length) nlaIIIR-like ORF in iceA1 from strain CH4 was amplified from CH4 chromosomal DNA and cloned into the vector pSYX22 to create pQXCH4. The longest coding region of 534 bp for iceA1 in strain 60190 starts from a TTG site that is 150 bp downstream of the equivalent ATG start site in nlaIIIR (Fig. 1). This DNA fragment was amplified and cloned into pSYX22 to create pQX60T. A downstream 381 bp nlaIIIR-like ORF in iceA1 from strain 60190 starting from the ATG site at bp 304 (Fig. 1) was also cloned into pSYX22 to create pQX60A. The resulting constructs, pQXCH4, pQX60T and pQX60A, were then transformed into E.coli strain ER2169, which carries a functional nlaIIIM gene in pSYX20-NlaIIIM (15). After IPTG induction, E.coli produced iceA1 protein products of the expected sizes (data not shown).
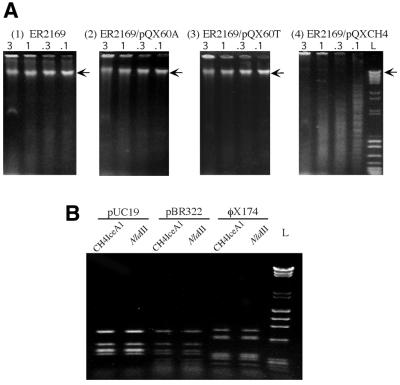
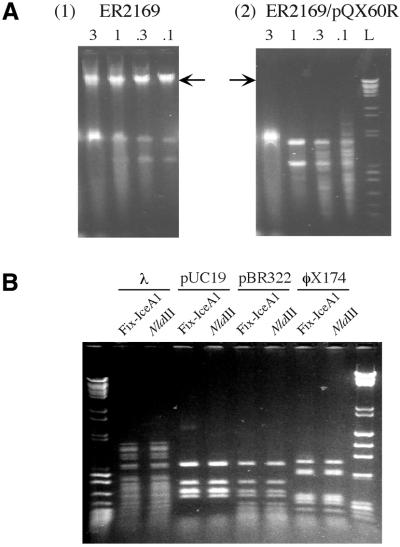
To determine whether the cloned iceA1 products had restriction endonuclease activity, serial dilutions of crude cell extracts from cultured E.coli ER2169/pSYX20-NlaIIIM, with or without pQXCH4, pQX60T or pQX60A, were used to digest λ DNA. As expected, the crude extract from E.coli ER2169/pSYX20-NlaIIIM failed to digest λ DNA (Fig. 2A). A fraction of the DNA in these lanes did not migrate from the loading wells, which may be explained by the presence of large amounts of DNA-binding proteins in the crude extract. When the crude extract from E.coli ER2169/pSYX20-NlaIIIM with pQXCH4 was used for digestion, the substrate DNA was digested into smaller DNA fragments (Fig. 2A). The lane loaded with the smallest volume (0.1 µl) had more small DNA fragments than the other lanes, which may be due to less interference by DNA-binding proteins. In contrast, crude extracts from E.coli with pQX60T or pQX60A failed to digest the λ DNA into smaller fragments. In total, these results indicate that endonuclease activities were present in the crude cell extract of E.coli ER2169/pQXCH4, but not in the control cells or cells with pQX60T or pQX60A.
Figure 2.
Endonuclease assay of the products of the iceA1 genes in CH4 and 60190. (A) Assay of crude extracts from E.coli strain ER2169/pSYX20-NlaIIIM (1) and ER2169 carrying pQX60A (2), pQX60T (3) and pQXCH4 (4). Crude extracts (0.1–3 µl) were used to digest 1 µg bacteriophage λ DNA and the digested DNA was electrophoresed on a 1% agarose gel. Arrows indicate the position of undigested λ DNA in the gels. (B) Assay of the purified CH4 iceA1 endonuclease. Parallel digestion of pUC19, pBR322 and φX174 DNA was performed with both the NlaIII and the CH4 iceA1 endonucleases. The DNA standards are labled L.
To investigate whether the endonuclease activities are NlaIII-like, the endonuclease from the crude cell extract of E.coli ER2169 with pQXCH4 was purified using a Heparin Hyper D column. The eluted fractions that contained the endonuclease activities were used to digest substrate (pBR322, pUC19 and φX174) DNAs in parallel with NlaIII. The digestion pattern of DNA by the pQXCH4 iceA1 product is identical to that by NlaIII (Fig. 2B), which indicates that CH4 iceA1 encodes a NlaIII-like endonuclease.
Fixing the frameshift in 60190 iceA1 and examining the endonuclease activity of the repaired iceA1 product
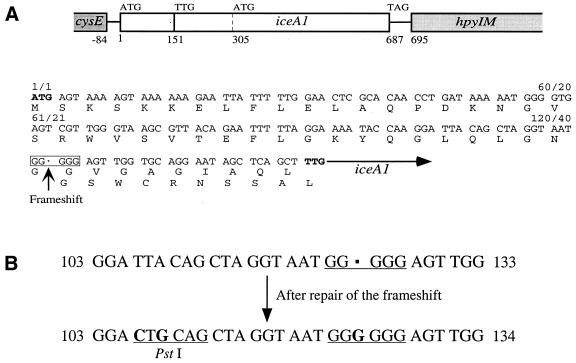
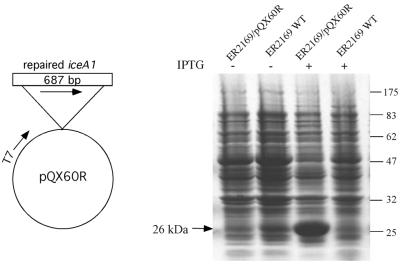
For H.pylori strain 60190, a protein beginning at the TTG site (Fig. 3) would not include a 50 amino acid region that is equivalent to the N-terminal region of NlaIII, which could explain why no endonuclease activity was detected when the iceA1 protein from strain 60190 was examined. To test whether the frameshift caused by the missing G residue at position 123 was the main problem, it was repaired by inserting an extra G residue into the poly(G) tract and the repaired iceA1 was inserted into the vector pSYX22, to create pQX60R. In the repaired iceA1, in addition to the G insertion, a T→C and an A→G point mutation were also introduced in the repaired region, to create a PstI recognition site (Fig. 3B). These substitutions are silent, since both TTA and CTG encode leucine as amino acid 36 of the repaired protein. The repaired iceA1 is a 684 bp ORF, encoding a 228 amino acid protein product, with 60% identity with NlaIII. To express the repaired iceA1 in E.coli, pQX60R was then transformed into E.coli strain ER2169 carrying pSYX20-NlaIIIM. After induction by IPTG, a 26 kDa protein was produced, as expected (Fig. 4).
Figure 3.
A frameshift is present in H.pylori 60190 iceA1 when compared to NlaIII. (A) Schematic representation of the iceA1–hpyIM region from H.pylori 60190. The locations of three potential translation start sites (ATG, TTG and ATG) and a potential stop site (TAG) in iceA1 are indicated. The DNA sequence and its deduced N-terminal NlaIII-like protein sequence from the first (ATG) to the second translation start site (TTG) in the iceA1 region with the frameshift site also are presented. The ATG and TTG sites are shown in bold. The boxed region indicated by an arrow shows the frameshift site. (B) The 60190 iceA1 sequences near the site of the frameshift, before and after the repair, are presented. The new PstI site created and the polyguanosine tract are underlined. Residues changed by the repair process are shown in bold.
Figure 4.
Expression of iceA1 in E.coli strain ER2169 after repair of its frameshift. 10% SDS–PAGE with Coomassie blue staining of lysates from E.coli strain ER2169 without (WT) or with pQX60R. +, induction with IPTG; –, no induction. The 26 kDa arrow indicates the protein product of the repaired iceA1. Migration (in kDa) of each protein standard is indicated on the left.
To determine whether the repaired iceA1 product has endonuclease activity, serial dilutions of crude cell extracts from cultured E.coli strain ER2169/pSYX20-NlaIIIM, with or without pQX60R, were used to digest λ DNA. As expected, the crude extract from E.coli ER2169/pSYX20-NlaIIIM failed to digest λ DNA (Fig. 5A). However, the migration of λ DNA was shifted to a lower position in the lanes with 3 and 1 µl of the crude extract, because of the presence of large amounts of DNA-binding proteins in the crude extract. When the crude extract from E.coli ER2169/pSYX20-NlaIIIM with pQX60R was used for digestion, the substrate DNA was digested into smaller DNA fragments (Fig. 5A). Overall, these results indicate that restriction endonuclease activities were present in the crude cell extract of E.coli ER2169/pQX60R, but not in the control cells.
Figure 5.
Endonuclease assay of the product of the repaired iceA1. (A) Assay of crude extracts from E.coli strain ER2169 (1) and ER2169 carrying pQX60R (2). Crude extracts (0.1–3 µl) were used to digest 1 µg bacteriophage λ DNA. The digested DNA was electrophoresed on a 1% agarose gel. Arrows indicate the position of undigested λ DNA in the gels. (B) Assay of the purified product of repaired iceA1 from strain 60190. Parallel digestion of pUC19, pBR322 and φX174 DNA was performed with both NlaIII and the repaired iceA1 product. The DNA standards are labeled L.
To investigate whether the activities are NlaIII-like, the restriction endonuclease from the crude cell extract of E.coli ER2169 with pQX60R was purified using a Heparin Hyper D column. The eluted fractions that contained the endonuclease activities were used to digest substrate (pBR322, pUC19 and φX174) DNAs in parallel with NlaIII. The digestion pattern of DNA by the repaired iceA1 product is identical to that by NlaIII (Fig. 5B), which indicates that iceA1 from strain 60190, after repair of the frameshift, encodes a NlaIII-like endonuclease.
DISCUSSION
This study demonstrates that the full-length iceA1 from strain CH4 encodes a functional restriction endonuclease, which is consistent with our recent observation that NlaIII-like endonuclease activity is present in the cells of this strain (24). Although iceA1 from 60190 has strong homology to nlaIIIR in N.lactamica, the predicted iceA1 ORF is much smaller than that in nlaIIIR, because of a frameshift mutation (Fig. 1). Thus, not surprisingly, the iceA1 ORF from strain 60190 expressed in E.coli lacked NlaIII-like activity, which is also consistent with the absence of NlaIII-like activity in cells of strain 60190 (24). However, after restoring a full-length (nlaIIIR-like) ORF in iceA1 from strain 60190 by repairing the frameshift, the ‘repaired’ iceA1 encodes an NlaIII-like endonuclease in E.coli. These results explain why there was no NlaIII-like activity in this H.pylori strain (24), even though iceA1 is transcribed (25). Compared to the repaired iceA1 product, the protein transcribed from the predicted start codon (TTG) in iceA1 lacks a 50 amino acid N-terminal region (Fig. 3). The lack of endonuclease activity in the native product of iceA1 from strain 60190 indicates that the 50 amino acid N-terminal region is required for NlaIII-like function.
One question is whether this type of mutation may be repaired within H.pylori populations to produce a functional NlaIII-like endonuclease. In iceA1 from strains like 60190, only a single nucleotide correction (inserting a G residue) is needed to restore the full-length nlaIIIR-like ORF. The frequency of single transition or transversion mutations in the ribosomal RNA region of H.pylori has been measured in strain 60190 by monitoring the frequency of spectinomycin resistance (26–28) and found to be 7.8 × 10–9 (Q.Xu and M.J.Blaser, unpublished data). If the frequency of spontaneous mutation in the iceA1 region is similar to that in the ribosomal RNA region, the frequency of frameshift repair in 60190 iceA1 would be <7.8 × 10–9, assuming that production of point insertion mutations is more difficult than transition or transversion mutations (29,30). However, the frameshift in iceA1 from strain 60190 occurs in a polyguanosine tract (six G residues in strain CH4, five in 60190), suggesting that slipped strand mispairing could occur in this region during DNA replication (31,32). Thus, the addition of one guanosine residue could occur at a much higher frequency than the background mutation rate (33–36).
It can be beneficial to bacterial populations to contain genes that undergo phase variation, allowing them to be ready for environmental changes. Examples of such contigency genes in H.pylori have been found in Lewis antigen expression (37,38), which exhibits phase variation at a 0.2–0.5% rate in vitro (39). Analysis of the whole genomic sequences (19,40,41) has revealed the presence of 10 potential phase-variable restriction–modification genes, not including iceA1, in H.pylori. These genes carry either homopolymeric tracts or dinucleotide repeats with their length varying from 5 to 15 (40,41). Within different H.pylori populations, changes in the lengths of specific homopolymeric tracts have been found in some of the putative phase-variable restriction–modification genes (40). Contigency restriction–modification genes have also been identified in other bacteria (42–44), including Haemophilus influenzae, Neisseria meningitidis and Mycoplasma pulmonis. A recent study demonstrated that a contingency restriction–modification system in M.pulmonis was turned on after the bacteria contacted the lower respiratory tract in infected rats (45), suggesting that contingency restriction–modification genes may play important roles in host–bacteria interactions. If iceA1 in strain 60190 functions as a contingency gene, H.pylori could adapt to environmental changes, such as contacting epithelial cells in the stomach, or to phage infection, by encoding a NlaIII-like restriction endonuclease. However, whether iceA1 is actually a contigency gene remains to be determined.
Another question is how often iceA1 mutations in various strains are located in similar homopolymeric regions. Analysis of iceA1 sequences from 36 H.pylori strains (which are among the 49 strains referred to previously) (Table 2) showed that a total of 14 strains (including 60190) carry iceA1 genes with frameshift mutations in homopolymeric tracts, compared to CH4 iceA1. Among these homopolymeric tracts, 12 are in the same poly(G) tract of five or seven residues, ±1 nt at bp 123 (Table 2), and two in the poly(A) tract of eight residues near the beginning of the iceA1 ORF, –1 nt at bp 19 (Table 2). A +1 or –1 nt frameshift in these tracts would repair the frameshift mutations. However, additional mutations are also present in most of these other strains (Table 2), which makes repair very difficult.
Table 2. Summary of the frameshift or nonsense mutations in the nlaIIIR-equivalent coding region of iceA1 from 36 H.pylori strains.
| Gene name | Description of mutation | GenBank accession no. or reference |
|---|---|---|
| CH4iceA1 | Full-length | AF459446 |
| 60190iceA1 | –1 nt (bp 123)a | U43917 |
| 26695iceA1 | G→A (bp 70) | AE000626 |
| J166iceA1 | –1 nt (bp 58); +1 nt (bp 446) | (12,20) |
| India227iceA1 | Full-length | AF239991 |
| India34AiceA1 | –1 nt (bp 53) | AF239992 |
| India18AiceA1 | –94 nt (bp 296) | AF239993 |
| India44AiceA1 | C→T (bp 292); –94 nt (bp 296) | AF239994 |
| Alaska209iceA1 | Full-length | AF001537 |
| Alaska214iceA1 | +1 nt (bp 460) | AF001538 |
| Alaska218iceA1 | Full-length | AF001539 |
| F13iceA1 | –5 nt (bp 188); C→T (bp 278) | AF157527 |
| F15iceA1 | –5 nt (bp 188); C→T (bp 278) | AF157528 |
| F16iceA1 | –1 nt (bp 19); –5 nt (bp 187); –7 nt (bp 288); C→T (bp 399) | AF157529 |
| F36iceA1 | G→T (bp 103) | AF157530 |
| F37iceA1 | –1 nt (bp 168) | AF157531 |
| F38iceA1 | Full-length | AF157532 |
| F43iceA1 | Full-length | AF157534 |
| F70iceA1 | –1 nt (bp 123)a; –5 nt (bp 187); –7 nt (bp 288); C→T (bp 253) | AF157533 |
| F72iceA1 | Full-length | AF157535 |
| F73iceA1 | –1 nt (bp 123)a; –5 nt (bp 187); –7 nt (bp 288) | AF157536 |
| F79iceA1 | –1 nt (bp 19)a; –5 nt (bp 187); –7 nt (bp 288); C→T (bp 399) | AF157537 |
| F82iceA1 | –1 nt (bp 123)a; –5 nt (bp 187); –7 nt (bp 288) | AF157538 |
| F83iceA1 | –1 nt (bp 123)a; –5 nt (bp 187); –7 nt (bp 288) | AF157539 |
| F84iceA1 | –1 nt (bp 123)a; –5 nt (bp 187); –7 nt (bp 288); C→A (bp 399) | AF157540 |
| OK104iceA1 | –1 nt (bp 123)a; –5 nt (bp 187); –5 nt (bp 547) | AF157541 |
| OK106iceA1 | –1 nt (bp 123)a; –5 nt (bp 187); –7 nt (bp 288) | AF157542 |
| OK107iceA1 | A→T (bp 10); +1 nt (bp 448); +1 nt (bp 479); –5 nt (bp 564) | AF157543 |
| OK111iceA1 | –1 nt (bp 58); +1 nt (bp 438) | AF157544 |
| OK115iceA1 | +1 nt (bp 123) | AF157545 |
| OK129iceA1 | –1 nt (bp 140) | AF157546 |
| OK99iceA1 | –1 nt (bp 123)a; –5 nt (bp 187); –7 nt (bp 288) | AF157547 |
| OK102iceA1 | –1 nt (bp 123)a; –5 nt (bp 187); –7 nt (bp 288) | AF157548 |
| OK108iceA1 | –1 nt (bp 584)a; –1 nt (bp 673) | AF157549 |
| OK113iceA1 | C→T (bp 289) | AF157550 |
| OK134iceA1 | –1 nt (bp 123)a; –5 nt (bp 187); –7 nt (bp 288) | AF157551 |
–, frameshift deletion; +, frameshift insertion; →, transition or transversion which causes nonsense mutation. The location of each mutation is represented as its position in each iceA1 gene, which is indicated in parentheses, and the predicated coding region of all the iceA1 genes analyzed start from the nlaIIIR-equivalent ATG start site.
aThe mutation is in a homopolymeric tract.
Among the 36 strains analyzed, only seven (including CH4) carry a full-length nlaIIIR-like ORF (Table 2). In the remaining strains, the iceA1 region has various mutations, including insertions, deletions and nonsense mutations. Only 10 (including 60190) have mutations at a single location in iceA1 when compared to nlaIIIR (Table 2), whereas others have mutations at multiple locations, making them more difficult to repair. For example, corrections of both a deletion and an insertion mutation are needed to restore the full-length nlaIIIR-like ORF in strain J166. Biochemical approaches failed to detect NlaIII-like activity in strain J166 (24), although the iceA1 region is transcribed (25). Thus, it appears that there are no special mechanisms, such as ribosomal frameshifting, that could permit translation to bypass these mutations. Furthermore, unless there is a special mechanism that would greatly increase the rate of mutation repair in non-homopolymeric tracts, it seems unlikely that the iceA1 gene serves as an ordinary contingency gene, analogous to the fucosyltransferase genes controlling synthesis of the Lewis antigens (37–39).
Since H.pylori is naturally competent for the uptake of chromosomal DNA and plasmid DNA, when more than one iceA1 strain is present in the gastric mucosa, horizontal exchange may occur. In such an exchange, an alternative way for H.pylori to repair iceA1 mutations and restore NlaIII-like activity is to acquire DNA from strains with the entire gene or complementary regions by gene conversion. However, restriction–modification systems are highly diversified among various H.pylori strains (24,46–49) and form barriers for transformation of DNA fragments between strains (50), which would limit the frequency of such repair.
Overall, we have shown that iceA1 of CH4 is a functional NlaIII-like endonuclease gene and the iceA1 gene of 60190 can be repaired to code a functional restriction enzyme. In a majority of H.pylori strains, iceA1 appears to be a degenerate gene that was once part of an restriction-modification system. However, expression of truncated iceA1, such as that from strain J166, is up-regulated by contact with epithelial cells (12). It remains to be determined whether iceA1 plays a role other than encoding a NlaIII-like endonuclease or not.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported in part by a Dissertation Enhancement Grant from the Vanderbilt Graduate School, a Vanderbilt Cancer Center Core Grant, and National Institutes of Health grants DK53707, GM56534 and GM63270.
DDBJ/EMBL/GenBank accession no. AF459446
REFERENCES
- 1.Marshall B.J. and Warren,J.R. (1984) Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet, 1, 1311–1315. [DOI] [PubMed] [Google Scholar]
- 2.Nomura A., Stemmermann,G.N., Chyou,P.-H., Kato,I., Perez-Perez,G.I. and Blaser,M.J. (1991) Helicobacter pylori infection and gastric carcinoma in a population of Japanese-Americans in Hawaii. N. Engl. J. Med., 325, 1132–1136. [DOI] [PubMed] [Google Scholar]
- 3.Parsonnet J., Hansen,S., Rodriguez,L., Gelb,A.B., Warnke,R.A., Jellum,E., Orentreich,N., Vogelman,J.H. and Friedman,G.D. (1994) Helicobacter pylori infection and gastric lymphoma. N. Engl. J. Med., 330, 1267–1271. [DOI] [PubMed] [Google Scholar]
- 4.Nomura A., Stemmermann,G.N., Chyou,P.H., Perez-Perez,G.I. and Blaser,M.J. (1994) Helicobacter pylori infection and the risk for duodenal and gastric ulceration. Ann. Intern. Med., 120, 977–981. [DOI] [PubMed] [Google Scholar]
- 5.Berg D.E. and Logan,R.P.H. (1997) Helicobacter pylori, individual host specificity and human disease. Bioessays, 19, 86–90. [DOI] [PubMed] [Google Scholar]
- 6.Dooley C.P., Fitzgibbons,P.L., Cohen,H., Appleman,M.D., Perez-Perez,G.I. and Blaser,M.J. (1989) Prevalence of Helicobacter pylori infection and histologic gastritis in asymptomatic persons. N. Engl. J. Med., 321, 1562–1566. [DOI] [PubMed] [Google Scholar]
- 7.Blaser M.J. and Berg,D.E. (2001) Helicobacter pylori genetic diversity and risk of human disease. J. Clin. Invest., 107, 767–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atherton J.C., Cao,P., Peek,R.M.,Jr, Tummuru,M.K., Blaser,M.J. and Cover,T.L. (1995) Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem., 270, 17771–17777. [DOI] [PubMed] [Google Scholar]
- 9.Cover T.L., Dooley,C.P. and Blaser,M.J. (1990) Characterization of and human serologic response to proteins in Helicobacter pylori broth culture supernatants with vacuolizing cytotoxin activity. Infect. Immun., 58, 603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tummuru M.K.R., Cover,T.L. and Blaser,M.J. (1993) Cloning and expression of a high molecular weight major antigen of Helicobacter pylori: evidence of linkage to cytotoxin production. Infect. Immun., 61, 1799–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Covacci A., Censini,S., Bugnoli,M., Petracca,R., Burroni,D., Macchia,G., Massone,A., Papini,E., Xiang,Z., Figura,N. and Rappuoli,R. (1993) Molecular characterization of the 128 kDa immunodominant antigen of Helicobacter pylori association with cytotoxicity and duodenal ulcer. Proc. Natl Acad. Sci. USA, 90, 5791–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peek R.M. Jr, Thompson,S.A., Donahue,J.P., Tham,K.T., Atherton,J.C., Blaser,M.J. and Miller,G.G. (1998) Adherence to gastric epithelial cells induces expression of a Helicobacter pylori gene, iceA, that is associated with clinical outcome. Proc. Assoc. Phys., 110, 531–544. [PubMed] [Google Scholar]
- 13.van Doorn L.J., Figueiredo,C., Sanna,R., Plaisier,A., Schneeberger,P., de Boer,W.A. and Quint,W. (1998) Clinical relevance of the cagA, vacA and iceA status of Helicobacter pylori. Gastroenterology, 115, 58–66. [DOI] [PubMed] [Google Scholar]
- 14.Ito Y., Azuma,T., Ito,S., Suto,H., Miyaji,H., Yamazaki,Y., Kato,T., Kohli,Y., Keida,Y. and Kuriyama,M. (2000) Sequence analysis and clinical significance of the iceA gene from Helicobacter pylori strains in Japan. J. Clin. Microbiol., 38, 483–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgan R.D., Camp,R.R., Wilson,G.G. and Xu,S.Y. (1996) Molecular cloning and expression of the NlaIII restriction-modification system in E. coli. Gene, 183, 215–218. [DOI] [PubMed] [Google Scholar]
- 16.Xu Q., Peek,R.M.,Jr, Miller,G.G. and Blaser,M.J. (1997) The Helicobacter pylori genome is modified at CATG by the product of hpyIM. J. Bacteriol., 179, 6807–6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labbe D., Holtke,H.J. and Lau,P.C.K. (1990) Cloning and characterization of two tandemly arranged DNA methylase genes of N. lactamica: an adenine-specific M.NlaIII and a cytosine-type methylase. Mol. Gen. Genet., 224, 101–110. [DOI] [PubMed] [Google Scholar]
- 18.Xu Q. and Blaser,M.J. (2001) Promoters of the CATG-specific methyltransferase gene hpyIM differ between iceA1 and iceA2 Helicobacter pylori strains. J. Bacteriol., 183, 3875–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomb J.F., White,O., Kerlavage,A.R., Clayton,R.A., Sutton,G., Fleischmann,R.D., Ketchum,K.A., Klenk,H.P., Gill,S., Dougherty,B.A., Nelson,K., Quackenbush,J., Zhou,L., Kirkness,E.F., Peterson,S., Loftus,B., Richardson,D., Dodson,R., Khalak,H.G., Glodek,A., McKenney,K., Fitzegerald,L.M., Lee,N., Adama,M.D., Hickey,E.K., Berg,D.E., Gocayne,J.D., Utterback,T., Peterson,J.D., Kelley,J., Cotton,M.D., Weidman,J.M., Fujii,C., Bowman,C., Watthey,L., Wallin,E., Hayes,W.S., Borodovsky,M., Karp,P.D., Smith,H.O., Fraser,C. and Venter,J.C. (1997) The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature, 388, 539–547. [DOI] [PubMed] [Google Scholar]
- 20.Figueiredo C., Quint,W.G.V., Sanna,R., Sablon,E., Donahue,J.P., Xu,Q., Miller,G.G., Peek,R.M., Blaser,M.J. and van Doorn,L.J. (2000) Genetic organization and heterogeneity of the iceA locus of Helicobacter pylori.Gene, 246, 59–68. [DOI] [PubMed] [Google Scholar]
- 21.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 22.Rosenberg A.H., Lade,B.N., Chui,D.S., Lin,S.W., Dunn,J.J. and Studier,F.W. (1987) Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene, 56, 125–135. [DOI] [PubMed] [Google Scholar]
- 23.Sambook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 18.47–18.55.
- 24.Xu Q., Morgan,R.D., Roberts,R.J. and Blaser,M.J. (2000) Identification of type II restriction and modification systems in Helicobacter pylori reveals their substantial diversity among strains. Proc. Natl Acad. Sci. USA, 97, 9671–9676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donahue J.P., Peek,R.M.,Jr, van Doorn,L.J., Thompson,S.A., Xu,Q., Blaser,M.J. and Miller,G.G. (2000) Analysis of iceA1 transcripts in Helicobacter pylori. Helicobacter, 5, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris E.H., Burkhart,B.D., Gillham,N.W. and Boynton,J.E. (1989) Antibiotic resistance mutations in the chloroplast 16S and 23S rRNA genes of Chlamydomonas reinhardtii: correlation of genetic and physical maps of the chloroplast genome. Genetics, 123, 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piepersberg W. and Wittmann,B.A. (1975) Effect of different mutations in ribosomal protein S5 of Escherichia coli on translational fidelity. Mol. Gen. Genet., 140, 91–100. [DOI] [PubMed] [Google Scholar]
- 28.Sigmund C.D., Ettayebi,M. and Morgan,E.A. (1984) Antibiotic resistance mutations in 16S and 23S ribosomal RNA gene of Escherichia coli. Nucleic Acids Res., 12, 4653–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaaper R.M. and Dunn,R.L. (1991) Spontaneous mutation in the Escherichia coli lacI gene. Genetics, 129, 317–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kunz B.A., Ramachandran,K. and Vonarx,E.J. (1998) DNA sequence analysis of spontaneous mutagenesis in Saccharomyces cerevisiae. Genetics, 148, 1491–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levison G. and Gutman,G.A. (1987) Slipped-strand mispairing: mechanism for DNA sequence evolution. Mol. Biol. Evol., 4, 203–221. [DOI] [PubMed] [Google Scholar]
- 32.Bayliss C.D., Field,D. and Moxon,E.R. (2001) The simple sequence contingency loci of Haemophilus influenzae and Neisseria meningitidis. J. Clin. Invest., 107, 657–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tran H.T., Keen,J.D., Kricker,M., Resnick,M.A. and Gordenin,D.A. (1997) Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol. Cell. Biol., 17, 2859–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bichara M., Schumacher,S. and Fuchs,R.P.P. (1995) Genetic instability with monotonous runs of CpG sequences in Escherichia coli. Genetics, 140, 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strauss B.S., Sagher,D. and Acharya,S. (1997) Role of proofreading and mismatch repair in maintaining the stability of nucleotide repeat in DNA. Nucleic Acids Res., 25, 806–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Bolle X., Bayliss,C.D., Field,D., van de Ven,T., Saunders,N.J., Hood,D.W. and Moxon,E.R. (2000) The length of a tetranucleotide repeat tract in Haemophilus influenzae determines the phase variation rate of a gene with homology to type III DNA methyltransferases. Mol. Microbiol., 35, 211–222. [DOI] [PubMed] [Google Scholar]
- 37.Wang G., Ge,Z., Rasko,D.A. and Taylor,D.E. (2000) Lewis antigens in Helicobacter pylori: biosynthesis and phase variation. Mol. Microbiol., 36, 1187–1196. [DOI] [PubMed] [Google Scholar]
- 38.Appelmelk B.J., Martin,S.L., Monteiro,M.A., Clayton,C.A., McColm,A.A., Zheng,P.Y., Verboom,T., Maaskant,J.J., van den Eijnden,D.H., Hokke,C.H., Perry,M.B. and Kusters,J.G. (1999) Phase variation in Helicobacter pylori lipopolysaccharide due to changes in lengths of poly(C) tracts in α3-fucosyltransferase genes. Infect. Immun., 67, 5361–5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Appelmelk B.J., Shiberu,B., Trinks,C., Tapsi,N., Zheng,P.Y., Verboom,T., Maaskant,J., Hokke,C.H., Schiphorst,W.E.C.M., Blanchard,D., Simoons-Smit,I.M., van den Eijden,D.H. and Vandenbroucke-Grauls,C.M.J.E. (1998) Phase variation in Helicobacter pylori lipopolysaccharide. Infect. Immun., 66, 70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alm R.A., Ling,L.L., Moir,D.T., King,B.L., Brown,E.D., Doig,P.C., Smith,D.R., Guild,B.C., de Jonge,B.L., Carmel,G., Tummino,P.J., Caruso,A., Uria-Nickelsen,M., Mills,D.M., Ives,C., Gibson,R., Merberg,D., Mills,S.D., Jiang,Q., Taylor,D.E., Vovis,G.F. and Trust,T.J. (1999) Genomic sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature, 397, 176–180. [DOI] [PubMed] [Google Scholar]
- 41.Saunders N.J., Peden,J.F., Hood,D.W. and Moxon,E.R. (1998) Simple sequence repeats in the Helicobacter pylori genome. Mol. Microbiol., 27, 1091–1098. [DOI] [PubMed] [Google Scholar]
- 42.Hood D.W., Deadman,M.E., Jennings,M.P., Bisercic,M., Fleischmann,R.D., Venter,J.C. and Moxon,E.R. (1996) DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc. Natl Acad. Sci. USA, 93, 11121–11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeffries A.C., Peden,J.F., Hood,D.W., Tettelin,H., Rappuoli,R. and Moxon,E.R. (2000) Repeat-associated phase variable genes in the complete genome sequence of Neisseria meningitidis strain MC58. Mol. Microbiol., 37, 207–215. [DOI] [PubMed] [Google Scholar]
- 44.Dybvig K., Sitaraman,R. and French,C.T. (1998) A family of phase-variable restriction enzymes with differing specificities generated by high-frequency gene rearrangements. Proc. Natl Acad. Sci. USA, 95, 13923–13928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gumulak-Smith J., Teachman,A., Tu,A.T., Simecka,J.W., Lindsey,J.R. and Dybvig,K. (2001) Variation in the surface proteins and restriction enzyme systems of Mycoplasma pulmonis in the respiratory tract of infected rats. Mol. Microbiol., 40, 1037–1044. [DOI] [PubMed] [Google Scholar]
- 46.Vitkute J., Stankevicius,K., Tamulaitiene,G., Maneliene,Z., Timinskas,A., Berg,D.E. and Janulaitis,A. (2001) Specificities of eleven different DNA methyltransferases of Helicobacter pylori strain 26695. J. Bacteriol., 183, 443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin L.F., Posfai,J., Roberts,R.J. and Kong,H. (2001) Comparative genomics of the restriction-modification systems in Helicobacter pylori. Proc. Natl Acad. Sci. USA, 98, 2740–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nobusato A., Uchiyama,I. and Kobayashi,I. (2000) Diversity of restriction-modification gene homologues in Helicobacter pylori. Gene, 259, 89–98. [DOI] [PubMed] [Google Scholar]
- 49.Xu Q., Stickel,S., Roberts,R.J., Blaser,M.J. and Morgan,R.D. (2000) Purification of the novel endonuclease, Hpy188I, and cloning of its restriction-modification genes reveal evidence of its horizontal transfer to the Helicobacter pylori genome. J. Biol. Chem., 275, 17086–17093. [DOI] [PubMed] [Google Scholar]
- 50.Ando T., Xu,Q., Torres,M., Kusugami,K., Israel,A. and Blaser,M.J. (2000) Restriction-modification system differences in Helicobacter pylori are a barrier to interstrain plasmid transfer. Mol. Microbiol., 37, 1052–1065. [DOI] [PubMed] [Google Scholar]