Abstract
Microarrays traditionally have been used to analyze the expression behavior of large numbers of coding transcripts. Here we present a comprehensive approach for high-throughput transcript discovery in Escherichia coli focused mainly on intergenic regions which, together with analysis of coding transcripts, provides us with a more complete insight into the organism’s transcriptome. Using a whole genome array, we detected expression for 4052 coding transcripts and identified 1102 additional transcripts in the intergenic regions of the E.coli genome. Further classification reveals 317 novel transcripts with unknown function. Our results show that, despite sophisticated approaches to genome annotation, many cellular transcripts remain unidentified. Through the experimental identification of all RNAs expressed under a specific condition, we gain a more thorough understanding of all cellular processes.
INTRODUCTION
Genome sequence information has accumulated at a fast pace in recent years and the generation of whole genome sequences is now commonplace. However, the number of uncompleted genome projects significantly exceeds the number of completely annotated and published sequences (http://www.tigr.org and http://www.ncbi.nlm.nih.gov/PMGifs/Genomes/micr.html). One of the primary reasons for this gap between sequence generation and public release is the still difficult task of sequence annotation, of interpreting raw sequence data into useful biological information. Most of the genome annotation information is generated using bioinformatics approaches. These in silico methods used for gene prediction in combination with homology searches are applied to the primary genome sequence. However, coding sequences, those portions of the genome that are transcribed and ultimately translated, are not the only elements of the genome which are transcribed into RNA. Transcribed but untranslated regions (UTRs) are common at the 5′ and 3′ ends of genes since transcription initiation and termination sites generally extend beyond translation start and stop sites. In prokaryotes such as Escherichia coli, operons may be described as consecutive genes, which are transcribed into a single polycistronic mRNA molecule. In the case of these operons, the intergenic regions are transcribed but not translated. In addition, at least 34 untranslated small RNA molecules, which may have regulatory functions, have been reported (1–3). Further, there is evidence that current annotation algorithms have limitations which can cause errors in the annotation process (4–14). By investigating observed transcripts, which are distinct from previously annotated genes, we are able to identify new potential genes.
Several in silico approaches, based largely on primary sequence analysis, have proven successful at identifying many of these transcript elements, including promoter regions (15,16), transcription termination sites (17,18), operons (19) and small RNAs (1–3). In addition to these transcribed elements, a number of intergenic repeats in E.coli have been computationally identified and documented (20,21). However, these computational approaches rely on primary sequence analyses and cross-species sequence comparisons. Genome-wide experimental identification of transcripts, such as with microarrays, has been limited primarily to coding sequences. For identifying transcribed intergenic regions, we present an orthogonal approach to in silico primary sequence analysis methods that is based on high density oligonucleotide probe arrays, which interrogate the sense strand of coding sequences and both strands in the intergenic regions of the genome. Using E.coli RNA from cells grown on different media, we have identified over 1100 transcripts corresponding to intergenic regions. We proceeded to classify these transcripts using sequence analysis, expression clustering, sequence homology and information collected from the literature and public databases.
MATERIALS AND METHODS
Strain and growth conditions
Escherichia coli strain MG1655 cells were grown in Luria–Bertani broth or on solid medium and used for inoculation of liquid cultures. Cells were grown in 50-ml batch cultures in 250-ml Erlenmeyer flasks at 37°C with aeration by rotary shaking (300 r.p.m.). The culture media used were Luria–Bertani (LB) or M9 minimal medium as described elsewhere (22) supplemented with glucose (0.2%) or glycerol (0.2%). Anaerobic growth was performed at 37°C in the same flasks fitted with butyl rubber stoppers and the air in the dead space replaced with argon. Growth was monitored at 600 nm on a Hitachi U-2000 spectrophotometer. Cells were harvested in mid log phase, midway between beginning log phase and stationary phase, early stationary phase or deep stationary growth phase (24 h after the culture reached stationary phase) (Table 1).
Table 1. The 13 different growth conditions used for the E.coli transcriptome analysis.
| Experiment | Medium | Carbon source | Aeration | Growth stage and other relevant information |
|---|---|---|---|---|
| 1 | M9 | Glucose | Aerobic | Mid log phase |
| 2 | M9 | Glucose | Aerobic | Midway between log phase and stationary phase |
| 3 | M9 | Glucose | Aerobic | Early stationary phase |
| 4 | M9 | Glucose | Aerobic | Late (24 h) stationary phase |
| 5 | M9 | Glycerol | Aerobic | Mid log phase |
| 6 | M9 | Glucose | Anaerobic, shift to aerobic | 15 min after shift |
| 7 | M9 | Glucose | Anaerobic, shift to aerobic | 30 min after shift |
| 8 | M9 | Glucose | Anaerobic, shift to aerobic | 60 min after shift |
| 9 | M9 | Glucose | Aerobic | Mid log phase, 42°C |
| 10 | M9 | Glucose | Aerobic | Mid log phase, 20°C |
| 11 | M9 | Glucose | Aerobic | Starvation, withdrawing of glucose at mid log phase |
| 12 | LB | Aerobic | Mid log phase | |
| 13 | LB | Anaerobic | Mid log phase |
RNA isolation, cDNA synthesis and target labeling
Total RNA was isolated from the cells using the protocol accompanying the MasterPure™ complete DNA/RNA purification kit from Epicentre Technologies (Madison, WI). Isolated RNA was resuspended in diethyl pyrocarbonate-treated water and quantitated based on the absorption at 260 nm. The cDNA synthesis method has been described previously (23). Briefly, 10 µg total RNA was reverse transcribed using the Superscript II system for first strand cDNA synthesis from Life Technologies (Rockville, MD). The remaining RNA was removed using 2 U RNase H (Life Technologies) and 1 µg RNase A (Epicentre) for 10 min at 37°C in 100 µl total volume. The cDNA was purified using the Qiaquick PCR purification kit from Qiagen (Valencia, CA). Isolated cDNA was quantitated based on the absorption at 260 nm and fragmented using a partial DNase I digest. The fragmented cDNA was 3′ end-labeled using terminal transferase (Roche Molecular Biochemicals, Indianapolis, IN) and biotin-N6-ddATP (DuPont/NEN, Boston, MA). The fragmented and end-labeled cDNA was added to the hybridization solution without further purification.
Genomic DNA labeling and hybridization
Escherichia coli genomic DNA (5 µg) was fragmented using 0.2 U DNase I (Roche) in one-phor-all buffer (Amersham, Piscataway, NJ), adjusted to a final volume of 20 µl and incubated at 37°C for 10 min, followed by inactivation of DNase at 99°C for 10 min. The fragmented DNA was subsequently labeled with terminal transferase (Roche) and biotin-N6-ddATP (DuPont/NEN) in accordance with the manufacturers’ protocols. Standard hybridization, wash and stain protocols were used (Affymetrix, Santa Clara, CA).
Reverse transcription–PCR (RT–PCR)
RNA isolation and cDNA synthesis was performed as described above. The PCR reaction was carried out with 70 ng cDNA as template and 1 µM forward and reverse primers. The reaction was cycled 25 times with a 55°C annealing temperature and a 2–4-min extension time at 72°C, depending on the size of the expected product. We used the same RNA in the PCR reaction as a negative control to test for genomic contamination.
Array design
A detailed description of the microarray is available (24). In summary, each array chip contains 295 936 oligonucleotide probes. Half of the probes are designed to be perfect match (PM) probes, which correspond to 25mer oligonucleotides in the E.coli genome, while the other half are designed to be mismatch (MM) probes, which correspond to the same 25mers as the PM probes except that the 13th base pair is complemented. The chip assays every annotated gene (10) with a set of probe pairs and every intergenic region in both orientations with a set of probe pairs. Probe sets generally contain 15 PM and 15 MM probes.
Transcript identification
The GeneChip® Software analysis program MAS 4.1 and DMT 2.0 (Affymetrix) were used for the analysis of gene expression and expression clustering, respectively. To identify transcripts within intergenic regions, we developed an algorithm for the analysis of the .cel file generated by MAS 4.1. The .cel file contains the probe locations and the individual intensities of the PM and corresponding MM probes on the microarray. In order to identify transcripts, we looked for sets of adjacent probes (two or more probes) in which PM – MM for each adjacent probe exceeds an expression threshold in both replicates (based on empirical results, we used a difference threshold of 200). We prefer reasonably strict criteria for transcript identification to ensure a high specificity for transcript detection. For each duplicate experiment, we searched for all possible transcripts which met these criteria in all interrogated intergenic regions. In order to correct for possible cross-hybridization effects, labeling inconsistencies or hybridization variations, we combined neighboring transcripts in the same intergenic region into a single transcript if they were separated by a single probe, which failed to meet our expression criteria. We applied this approach to all interrogated intergenic regions genome wide and then proceeded to classify the identified transcripts.
RESULTS
Many approaches that quantify the expression level of a gene based on oligonucleotide array data operate under the assumption that all (or at least most) oligonucleotide probes for a given gene are essentially independent measurements of the same transcript expression (24–27). This is a reasonable and convenient assumption for genes, where the existence and the exact position of the transcript are known a priori, but when we search for new transcripts, such as in intergenic regions, we do not have the luxury of this assumption. Rather, we perform RNA expression analysis at a sub-transcript resolution. Initial analysis of the data across all experiments showed a range of hybridization affinities for different probes. We removed 2671 probes in the intergenic regions from the analysis for which there was evidence of significant cross-hybridization or other non-specific hybridization. These probes were determined by hybridizing E.coli genomic DNA labeled directly with terminal transferase to the probe array and removing the probes that failed to meet our difference threshold. The remaining probes were studied by hybridizing biotin-labeled cDNA (23) derived from 13 different growth conditions in duplicate for a total of 26 arrays (Table 1).
For transcript discovery a stringent difference model was developed, which is based on evidence that an average difference model can linearly approximate actual expression levels (25,26). A probe had to meet the difference requirement in both duplicate experiments before we considered the probe ‘expressed’ (see Materials and Methods). After identifying a conservative set of potential transcripts in intergenic regions, we then proceeded with their classification based on their genome location as operon elements, 5′-UTRs, 3′-UTRs or as transcripts of unknown function (Table 2). For additional validation of our classification, we determined the co-regulation of the identified transcripts with their flanking ORFs using the self-organizing map (SOM) algorithm (28). Transcripts that are co-regulated across many conditions are likely to be from the same transcript (28). In addition, we performed a homology search against the complete genome sequence of Salmonella typhimurium (the closest fully sequenced relative to E.coli) to identify conserved regions (329). Sequences can be conserved for many different reasons, including coding regions, complex promoters or leader sequences, transcriptional and post-transcriptional regulatory signals, small RNAs, transcriptional terminators and sequences of as yet unknown function. We used the cluster and homology analyses together with annotation programs (11,12,30) and information collected from the literature and public databases to further characterize the transcripts and to classify them as potential new ORFs or RNA transcripts that serve as small regulatory RNAs (sRNA) (Table 2).
Table 2. Summary of all detected transcripts and their classification and characterization.
| Transcript classification | Annotated operonsa | New operonsb | Unknown transcriptsb | 5′-UTRb | 3′-UTRb | Total | |
|---|---|---|---|---|---|---|---|
| Predicted | Documented | ||||||
| Transcript characterization | |||||||
| Transcripts detected | 189 | 100 | 4 | 334 | 353 | 122 | 1102 |
| ORFs | 11 | 2 | 0 | 31 | 49 | 11 | 104 |
| sRNA | 0 | 0 | 0 | 17 | 2 | 0 | 19 |
| Regulatory region | 0 | 0 | 1 | 0 | 15 | 0 | 16 |
| Homology | n.d. | n.d. | n.d. | 183 | 250 | 69 | 502 |
| Cluster | 135 | 81 | 3 | n.a. | 139 | 30 | 388 |
| Cluster analysis | |||||||
| Operon | 33 | 44 | 1 | n.a. | n.a. | n.a. | 78 |
| Gene | 27 | 13 | 1 | n.a. | n.a. | n.a. | 41 |
| 5′-UTR | 35 | 8 | 1 | n.a. | n.a. | n.a. | 44 |
| 3′-UTR | 40 | 16 | 0 | n.a. | n.a. | n.a. | 56 |
n.a., not applicable; n.d., not determined.
bThis study.
Operon elements
A gene can be described as belonging to an operon if it is one of two or more adjacent genes which are transcribed into a single transcript (Fig. 1). Similarly, an intergenic region is part of an operon if it is transcribed in the same transcript as both its flanking genes. The ability to identify operons can be very useful in understanding gene function, since genes that are members of the same operon generally code for proteins which have functional roles in the same cellular pathways. While correlated expression of two neighboring genes may be a reasonable indication that the genes are co-transcribed, this correlated expression coupled with evidence of similar intergenic expression between the two genes provides a much stronger signal. Intergenic transcripts are classified as part of an operon if the orientation of the intergenic region matches that of the flanking genes, if both genes are expressed and if the expressed intergenic transcript extends across the entire intergenic region. Using these parameters we identified 289 of these intergenic regions which have been previously documented or predicted as being part of an operon (21,31; http://kinich.cifn.unam.mx:8850/db/regulondb_intro. frameset) (Supplementary Material, Table S1). Based on this comparison the false positive rate for transcript detection was estimated to be <1%. Characterizing the false negative rate proves to be problematic because many of these regions showed little or no expression under our 13 conditions and we are limited to drawing conclusions only from elements which are expressed in our experiment set. In addition to the transcript analysis described above, we also performed an expression cluster analysis using the SOM algorithm (28) to investigate correlation between the expression of an intergenic region and the expression of its neighboring genes. The average difference values for the flanking genes and the intergenic region under all 13 conditions were used to identify co-regulated expression. Of our predicted operons, 71% showed co-regulation in at least two of the three transcripts (flanking genes and intergenic region), while 81% of the documented operons offered this evidence of co-regulation. Figure 2 shows the expression levels for individual probes interrogating the predicted hnr–galU operon. RT–PCR confirmed a single RNA transcript for these two genes and the intergenic region (Fig. 3). We confirmed a single RNA transcript using RT–PCR for six additional predicted operons. Of the four intergenic regions which have not previously been documented or predicted as part of an operon but for which we observe operon evidence, we found that two were co-regulated with flanking genes (rpsM/rpmJ and rplN/rpsQ), which code for 30S and 50S ribosomal subunit proteins. Based on our findings and the close functional relationship of the gene products, they are strong candidates for new, previously unidentified operons.
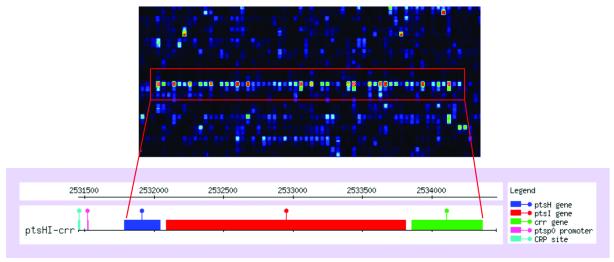
Figure 1.
The pts operon in E.coli. The enlarged pictures of a microarray show the high expression level of the neighboring genes ptsH, ptsI and crr in the pts operon. The bottom picture (from RegulonDB; http://kinich.cifn.unam.mx:8850/db/regulondb_intro.frameset) shows the location of the genes within the genome.
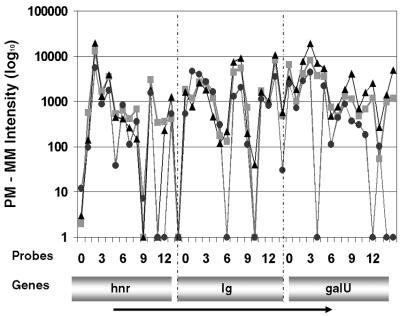
Figure 2.
Operon detection using oligonucleotide probe intensities. Individual oligonucleotide probe intensities (PM – MM) from three conditions are shown to validate the microarray-predicted hnr–galU operon. Intensities for individual probes interrogating hnr, the 200 bp intergenic region and galU are shown. This operon was independently confirmed using RT–PCR (Fig. 3).
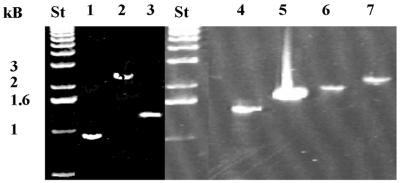
Figure 3.
RT–PCR for seven predicted operons. The DNA bands represent the PCR products for the following operons, including the expected size of the PCR product in parentheses (see also Supplementary Material, Table S1): lane 1, rpsR–rplI (0.9 kb); lane 2, ytfQ–ytfR (2.4 kb); lane 3, yaeR–mesJ (1.4 kb); lane 4, rplA–rplL (1.5 kb); lane 5, ptsH–ptsJ (1.7 kb); lane 6, purA–yjeB (1.8 kb); lane 7, hnr–galU (2 kb); lane St, 1 kb standard DNA ladder.
5′-Untranslated region (5′-UTR)
As with the operons described above, experimental evidence for 5′ expressed regions can supplement computational approaches by identifying not only transcription start sites for genes, but also multiple start sites when different promoters are employed under different conditions, as well as cis-regulatory sites upstream of known genes. In order for an intergenic transcript to be classified as a 5′-UTR in our analysis, we required the transcript to be in the same orientation as its downstream gene and to be expressed under the same growth conditions. We made the assumption that the transcript must be ≥70 nt to encode a 5′-UTR, slightly longer than the expected 50–60 nt of a promoter, and that the transcript extends close to the downstream gene translational start site, i.e. the transcript must extend to the penultimate or ultimate probe in the probe set of the intergenic region. Figure 4 shows an example for the microarray detection of the transcribed but not translated leader sequence of the ompA mRNA (32). The PM – MM probe intensities and the probe locations were used to determine the transcriptional start site, which was found to be close to the predicted promoter location for the ompA gene. We identified a conservative set of 353 transcripts which met our expression criteria for 5′-UTRs (Supplementary Material, Table S2). Of these transcripts, 294 either showed concordant expression with their downstream ORF in all 13 experiments or else showed homology to S.typhimurium with an E value < 0.01 (29) and an over all identity of >65%. Fifteen 5′-UTRs contain conserved regulatory sequences (http://kinich.cifn.unam.mx:8850/db/regulondb_intro.frameset), one of which matches a previously identified small RNA (sraB) (3) and an additional one (crpT) a potential small RNA (33). However, based on the signal location and co-expression with its downstream gene, our study suggests that crpT is the 5′-untranslated leader of yhfA as originally reported (34) and not an independent transcript as speculated by Carter et al. (35). An additional 49 other transcripts are classified as potential small ORFs (Supplementary Material, Table S2).
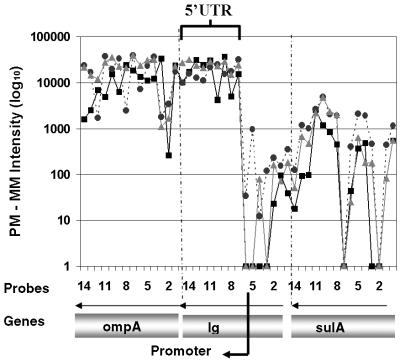
Figure 4.
5′-UTR detection upstream of ompA. Individual oligonucleotide probe intensities (PM – MM) from three conditions are shown to validate the microarray-detected 5′-UTR upstream of ompA. Intensities for individual oligonucleotide probes interrogating ompA, the 356 bp intergenic region and sulA are shown. The arrows above the indicated genes show the direction of transcription.
3′-Untranslated region (3′-UTR)
The classification of transcripts as 3′-UTRs is analogous to that of the 5′-UTRs. The intergenic transcript must be in the same orientation as its upstream gene and was required to be expressed under the same growth conditions. In addition, we restricted the transcripts to be at least 70 bp in length and to extend close to the upstream gene predicted translational stop site. With these criteria we identified 122 potential 3′-UTRs, of which 69% are either expressed concordant with their upstream gene in all 13 experiments or have sequence homology to S.typhimurium with an E value < 0.01 and an overall identity of >65% (Supplementary Material, Table S3). Eleven of the 122 transcripts are classified as potential novel small ORFs.
Transcripts of unknown function
Finally, we identified 334 transcripts longer than 70 bp that were expressed but which could not be classified as operon elements, 5′-UTRs or 3′-UTRs (Supplementary Material, Table S4). This group of transcripts has a hybridization signal separate from and discontinuous with the signals from neighboring ORFs. Over 200 transcripts in this group showed sequence homology with S.typhimurium or considerable expression levels (more than three times background). Of the 34 reported E.coli sRNA molecules interrogated in intergenic regions on the array (1–3), we detected transcripts for 19 on the reported strand (Supplementary Material, Table S5) and an additional five on the opposite strand in this group. Several of the known sRNAs are expressed only under specific growth conditions and we cannot draw conclusions from a negative result. We predict an additional nine transcripts as being good candidates for new small RNAs based on their homology to S.typhimurium, their average transcript intensity and their expression in either late log, early stationary or stationary growth phase. In addition, their sizes range from 50 to 400 nt (Supplementary Material, Table S5). These are conditions under which most of the other sRNA transcripts were identified. Furthermore, we detected transcripts for 31 predicted but not experimentally confirmed ORFs from the Colibri and EcoGene databases (http://genolist.pasteur.fr/Colibri/ and http://bmb.med.miami.edu/EcoGene/EcoWeb/) (Supplementary Material, Table S5). This is the most challenging group of transcripts to characterize and understand. Further sequence analysis to identify promoters, transcriptional terminators and secondary structure is necessary to say how many of these transcripts qualify as small RNA molecules, new ORFs, regulatory transcripts such as antisense RNAs or transcripts of as yet unknown function. Additional protein sequence homology, functional annotation information and experimental validation is also needed to confirm the predicted ORFs. We have intentionally not assigned any new E.coli gene names to our transcripts awaiting further characterization.
DISCUSSION
While the molecular biology community has met with great success in developing computational approaches for genome wide transcriptome analysis, experimental evidence to support this analysis tends to be either on a gene-by-gene level or else, in the case of microarrays, only targeted at transcripts that are also translated (24,36,37). A recent study of chromosomes 21 and 22 in human cells identified novel RNA transcripts not detected by sequence analysis (38). For the analysis of gene expression in an organism and the interpretation of the generated data we will increasingly rely on complete and accurate catalogs of genes, mRNAs, proteins and untranslated but transcribed regions. With the recent advances in oligonucleotide probe array technology and the availability of complete genome sequences we are now in a position to assay the complete transcriptome of many organisms, as opposed to only their coding subset. By assaying the E.coli transcriptome under a range of conditions, we identified multiple non-coding transcript elements, including 5′-UTRs, 3′-UTRs, small RNA molecules and polycistronic elements (operons). In addition, we were able to detect transcripts that escaped gene prediction programs due to non-conforming characteristics. The arrays can be constructed without a priori knowledge of genome annotation and can provide the experimental foundation for a complete transcriptome analysis. By interrogating both strands of a genomic sequence on one array, valuable information on possible antisense gene regulation can be obtained and can provide the basis for a more accurate understanding of gene translation. Our experimental approach to transcriptome analysis in E.coli could be extended to the genomes of other organisms with complete sequence data but incomplete annotation information. Using conservative criteria, we identified a set of 1102 transcripts in the intergenic regions of E.coli which we classified as operon elements, 5′-UTRs, 3′-UTRs, small RNAs, new ORFs or transcripts of unknown function (Table 2). Using the experimental approach of detecting transcripts under different growth conditions gives us the opportunity to detect >95% of all possible transcripts as judged by the total number of genes detected (data not shown). In fact, we validated most of the reported sRNAs, operons and ORFs by using secondary analysis tools.
Our analysis will be most effective if applied in combination with other evidence, such as computational approaches, which predict operons (19), promoters and ORFs (11,12,15,16), sRNAs (1–3), transcription termination sites (17,18), etc. Whenever possible, we attempted to validate our observed intergenic transcripts with independent means of analysis, such as sequence homology, expression clustering or ORF identification programs. The data presented show that a large portion of the E.coli transcriptome remains to be characterized. We also expect that experimental evidence of transcripts will be useful for developing the next generation of gene prediction algorithms, and only with a complete understanding of transcription for both coding regions and intergenic regions can we fully comprehend cellular processes.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank J. Dai for running the homology analysis, Alex Picone for technical assistance, G. Storz and S. Gottesman for comments on the manuscript and Leroy Hood for support. This work was supported in part by DOE Microbial Cell Program grant DE-FG08-01ER63218 to E.K.
REFERENCES
- 1.Wassarman K.M., Repoila,F., Rosenow,C., Storz,G. and Gottesman,S. (2001) Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev., 15, 1637–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rivas E., Klein,R.J., Jones,T.A. and Eddy,S.R. (2001) Computational identification of noncoding RNAs in E. coli by comparative genomics. Curr. Biol., 11, 1369–1373. [DOI] [PubMed] [Google Scholar]
- 3.Argaman L., Hershberg,R., Vogel,J., Bejerano,G., Wagner,E.G., Margalit,H. and Altuvia,S. (2001) Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol., 11, 941–950. [DOI] [PubMed] [Google Scholar]
- 4.Hogenesch J.B., Ching,K.A., Batalov,S., Su,A.I., Walker,J.R., Zhou,Y., Kay,S.A., Schultz,P.G. and Cooke,M.P. (2001) A comparison of the Celera and Ensembl predicted gene sets reveals little overlap in novel genes. Cell, 106, 413–415. [DOI] [PubMed] [Google Scholar]
- 5.Claverie J.M. (1997) Computational methods for the identification of genes in vertebrate genomic sequences. Hum. Mol. Genet., 6, 1735–1744. [DOI] [PubMed] [Google Scholar]
- 6.Guigo R., Agarwal,P., Abril,J.F., Burset,M. and Fickett,J.W. (2000) An assessment of gene prediction accuracy in large DNA sequences. Genome Res., 10, 1631–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boguski M.S. (1999) Biosequence exegesis. Science, 286, 453–455. [DOI] [PubMed] [Google Scholar]
- 8.Wheelan S.J. and Boguski,M.S. (1998) Late-night thoughts on the sequence annotation problem. Genome Res., 8, 168–169. [DOI] [PubMed] [Google Scholar]
- 9.Powledge T.M. (2000) Genomics annotation. Beyond the first draft. Sci. Am., 283, 16, 18. [DOI] [PubMed] [Google Scholar]
- 10.Blattner F.R., Plunkett,G.,III, Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F., Gregor,J., Davis,N.W., Kirkpatrick,H.A., Goeden,M.A., Rose,D.J., Mau,B. and Shao,Y. (1997) The complete genome sequence of Escherichia coli K-12 [comment] [see comments]. Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- 11.Lukashin A.V. and Borodovsky,M. (1998) GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res., 26, 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delcher A.L., Harmon,D., Kasif,S., White,O. and Salzberg,S.L. (1999) Improved microbial gene identification with GLIMMER. Nucleic Acids Res., 27, 4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serres M.H., Gopal,S., Nahum,L.A., Liang,P., Gaasterland,T. and Riley,M. (2001) A functional update of the Escherichia coli K-12 genome. Genome Biol., 2, RESEARCH0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skovgaard M., Jensen,L.J., Brunak,S., Ussery,D. and Krogh,A. (2001) On the total number of genes and their length distribution in complete microbial genomes. Trends Genet., 17, 425–428. [DOI] [PubMed] [Google Scholar]
- 15.Audic S. and Claverie,J.M. (1998) Visualizing the competitive recognition of TATA-boxes in vertebrate promoters [letter]. Trends Genet., 14, 10–11. [DOI] [PubMed] [Google Scholar]
- 16.Fickett J.W. and Hatzigeorgiou,A.G. (1997) Eukaryotic promoter recognition, Genome Res., 7, 861–878. [DOI] [PubMed] [Google Scholar]
- 17.Abe H., Abo,T. and Aiba,H. (1999) Regulation of intrinsic terminator by translation in Escherichia coli: transcription termination at a distance downstream. Genes Cells, 4, 87–97. [DOI] [PubMed] [Google Scholar]
- 18.Ermolaeva M.D., Khalak,H.G., White,O., Smith,H.O. and Salzberg,S.L. (2000) Prediction of transcription terminators in bacterial genomes. J. Mol. Biol., 301, 27–33. [DOI] [PubMed] [Google Scholar]
- 19.Salgado H., Moreno-Hagelsieb,G., Smith,T.F. and Collado-Vides,J. (2000) Operons in Escherichia coli: genomic analyses and predictions. Proc. Natl Acad. Sci. USA, 97, 6652–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rudd K.E. (1999) Novel intergenic repeats of Escherichia coli K-12. Res. Microbiol., 150, 653–664. [DOI] [PubMed] [Google Scholar]
- 21.Rudd K.E. (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res., 28, 60–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Rosenow C., Saxena,R.M., Durst,M. and Gingeras,T.R. (2001) Prokaryotic RNA preparation methods, useful for high density array analysis: comparison of two approaches. Nucleic Acids Res., 29, e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selinger D.W., Cheung,K.J., Mei,R., Johansson,E.M., Richmond,C.S., Blattner,F.R., Lockhart,D.J. and Church,G.M. (2000) RNA expression analysis using a 30 base pair resolution Escherichia coli genome array [In Process Citation]. Nat. Biotechnol., 18, 1262–1268. [DOI] [PubMed] [Google Scholar]
- 25.Lockhart D.J., Dong,H., Byrne,M.C., Follettie,M.T., Gallo,M.V., Chee,M.S., Mittmann,M., Wang,C., Kobayashi,M., Horton,H. and Brown,E.L. (1996) Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol., 14, 1675–1680. [DOI] [PubMed] [Google Scholar]
- 26.Li C. and Wong,W.H. (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl Acad. Sci. USA, 98, 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H. and Hong,F. (2001) Cluster-Rasch models for microarray gene expression data. Genome Biol., 2, RESEARCH0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamayo P., Slonim,D., Mesirov,J., Zhu,Q., Kitareewan,S., Dmitrovsky,E., Lander,E.S. and Golub,T.R. (1999) Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc. Natl Acad. Sci. USA, 96, 2907–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salzberg S.L., Delcher,A.L., Kasif,S. and White,O. (1998) Microbial gene identification using interpolated Markov models. Nucleic Acids Res., 26, 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huerta A.M., Glasner,J.D., Gutierrez-Rios,R.M., Blattner,F.R. and Collado-Vides,J. (2002) GETools: gene expression tool for analysis of transcriptome experiments in E. coli. Trends Genet., 18, 217–218. [Google Scholar]
- 32.Chen L.H., Emory,S.A., Bricker,A.L., Bouvet,P. and Belasco,J.G. (1991) Structure and function of a bacterial mRNA stabilizer: analysis of the 5′ untranslated region of ompA mRNA. J. Bacteriol., 173, 4578–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gottesman S., Storz,G., Rosenow,C., Majdalani,N., Rapoila,F. and Wassarman,K.M. (2001) Small RNA regulators of translation: mechanisms of action and approaches for identifying new small RNAs. Cold Spring Harbor Symp. Quant. Biol., LXVI, 353–362. [DOI] [PubMed] [Google Scholar]
- 34.Okamoto K., Hara,S., Bhasin,R. and Freundlich,M. (1988) Evidence in vivo for autogenous control of the cyclic AMP receptor protein gene (crp) in Escherichia coli by divergent RNA. J. Bacteriol., 170, 5076–5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carter R.J., Dubchak,I. and Holbrook,S.R. (2001) A computational approach to identify genes for functional RNAs in genomic sequences. Nucleic Acids Res., 29, 3928–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shoemaker D.D., Schadt,E.E., Armour,C.D., He,Y.D., Garrett-Engele,P., McDonagh,P.D., Loerch,P.M., Leonardson,A., Lum,P.Y., Cavet,G., Wu,L.F., Altschuler,S.J., Edwards,S., King,J., Tsang,J.S., Schimmack,G., Schelter,J.M., Koch,J., Ziman,M., Marton,M.J., Li,B., Cundiff,P., Ward,T., Castle,J., Krolewski,M., Meyer,M.R., Mao,M., Burchard,J., Kidd,M.J., Dai,H., Phillips,J.W., Linsley,P.S., Stoughton,R., Scherer,S. and Boguski,M.S. (2001) Experimental annotation of the human genome using microarray technology. Nature, 409, 922–927. [DOI] [PubMed] [Google Scholar]
- 37.Andrews J., Bouffard,G.G., Cheadle,C., Lu,J., Becker,K.G. and Oliver,B. (2000) Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogaster testis. Genome Res., 10, 2030–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kapranov P., Cawley,S.E., Drenkow,J., Bekiranov,S., Strausberg,R.L., Fodor,S.P. and Gingeras,T.R. (2002) Large-scale transcriptional activity in chromosomes 21 and 22. Science, 296, 916–919. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.