Abstract
Formation of hairpin or tetraplex structures of the FMR1 gene d(CGG)n sequence triggers its expansion, setting off fragile X syndrome. In searching for proteins that destabilize d(CGG)n secondary structures we purified from rat liver quadruplex telomeric DNA binding protein 42 (qTBP42) that disrupts G′2 bimolecular tetraplex d(CGG)n while paradoxically stabilizing the G′2 structure of the telomeric sequence d(TTAGGG)n. Based on peptide sequence homology of qTBP42 and mouse CArG-box binding factor A (CBF-A), we provide direct evidence that recombinant CBF-A protein is physically and immunochemically indistinguishable from qTBP42 and that it too destabilizes G′2 d(CGG)n while stabilizing G′2 d(TTAGGG)n. We inquired whether CBF-A employs the same or different domains to differentially interact with G′2 d(CGG)n and G′2 d(TTAGGG)n. Mutant CBF-A proteins that lack each or combinations of its five conserved motifs: RNP11, RNP12, RNP21, RNP22 and ATP/GTP-binding box were tested for their G′2 d(CGG)n destabilization and G′2 d(TTAGGG)n stabilization activities. We find that either RNP11 or the ATP/GTP motifs are necessary and sufficient for G′2 d(CGG)n destabilization whereas RNP21 suppresses destabilization by either one of these two motifs. Neither RNP11 nor the ATP/GTP motif are required for G′2 d(TTAGGG)n stabilization. Hence, CBF-A employs different domains to destabilize G′2 d(CGG)n or stabilize G′2 d(TTAGGG)n.
INTRODUCTION
Runs of adjacent guanine residues in DNA are capable of self-association to form four-stranded structures termed DNA tetraplexes or quadruplexes. At the core of these DNA secondary structures are Hoogsteen hydrogen-bonded and cation-coordinated stacked guanine quartets (reviewed in 1,2). Three major classes of tetrahelical DNA are defined by the stoichiometry and orientation of the DNA strands: G′4 unimolecular tetraplexes, G′2 bimolecular tetraplexes, and G4 four-molecular tetraplexes. These types of tetrahelical DNA are further differentiated into sub-groups by parameters such as the glycosidic torsion angles and molecular geometry of the tetrahelix, the nucleotide sequence of non-guanine spacer stretches and their structure, inclusion of bases other than guanine in tetrad structures and the nature of the coordinating cation (1,2).
Although DNA tetraplexes are readily formed in vitro under physiological-like conditions, their existence in vivo still awaits direct demonstration. However, some indirect lines of evidence suggest that tetrahelical DNA might be present in living cells and contribute to diverse physiological and pathological processes. First, biologically important guanine-rich DNA regions fold into tetraplex structures under physiological-like conditions in vitro. It was argued that in vivo formation of tetraplex structures by such sequences might be necessary for the execution of their proposed biological roles. For instance, transient generation of tetraplex structures by the pairing of guanine runs at intra-chromosomal loci was suggested to mediate meiotic pairing of the homolog chromosome (3). Likewise, folding of the telomeric G-strand into tetraplex formations was proposed to contribute to the regulation of telomere extension (4). Also, tetrahelical parallel structures of guanine-rich stretches in regions upstream to genes such as c-myc (5) and insulin (6) were implicated in the regulation of their transcription. Lastly, formation of tetraplex structures by a d(CGG) trinucleotide repeat in the FMR-1 gene was suggested to prompt polymerase pausing and slippage and expansion of the repeat sequence that leads to silencing of FMR-1 and sets off fragile X syndrome (7). A second argument for the existence of tetraplex DNA structures in vivo is the presence of numerous cellular proteins that interact with tetraplex DNA. Proteins isolated from diverse organisms bind preferentially, and at a relatively high affinity, various types of tetraplex DNA (8–18). Other proteins were shown to selectively process tetraplex DNA or to modulate its structure. These are nucleases, identified in fission yeast (19,20), mouse (21) and human cells (22), that hydrolyze DNA (19,22) and RNA (21) next to tetraplex domains. Other proteins alter the equilibrium between single-stranded and tetraplex structures of guanine-rich DNA. The β-subunit of an Oxytricha telomere-binding protein promotes the formation of a tetraplex structure of telomeric DNA (23,24). Also, several mammalian proteins tightly bind to tetraplex DNA and increase its stability (14,16,25). Lastly, yeast and human helicases of the RecQ family were shown to preferentially unwind tetraplex structures of diverse guanine-rich sequences (26–29).
In searching for mammalian proteins that interact with tetraplex DNA we identified in rat hepatocytes a protein, designated quadruplex telomeric DNA binding protein 42 (qTBP42), that bound tightly (Kd = 3.7–14.6 nM) single-stranded and G′4 unimolecular and G′2 bimolecular tetraplex forms of the telomeric sequence d(TTAGGG)n and a G4 four-molecular quadruplex structure of an immunoglobulin switch region sequence (14). The association of qTBP42 with tetraplex telomeric DNA structures increased their resistance to heat denaturation and diminished their digestion by micrococcal nuclease (14). Conversely, without detectably binding to it, qTBP42 efficiently destabilized G′2 tetraplex d(CGG)n disrupting this tetrahelix into its constituent single strands (30). Amino acid sequences of qTBP42 peptides (15) are fully homologous to segments of the CArG-box binding factor A (CBF-A), a heterogeneous nuclear ribonucleoprotein-related protein originally identified as a muscle-specific transcriptional repressor (31). More recent data suggest that CBF-A might also be involved in transcriptional and post-transcriptional regulation of the expression of diverse genes (32–36).
Here we show that mouse recombinant CBF-A is physically and immunologically indistinguishable from qTBP42 and that similarly to qTBP42, CBF-A also contrastingly stabilizes tetraplex telomeric DNA while destabilizing tetraplex d(CGG)n. In undertaking to identify domains in CBF-A that mediate G′2 d(CGG)n destabilization or tetraplex telomeric DNA stabilization, we conducted a systematic study of the activities of truncated and deleted CBF-A mutant proteins. We report the identification of distinct domains in CBF-A that prompt or inhibit the destabilization of G′2 d(CGG)n. These regions are dispensable for the binding and stabilization of tetraplex telomeric DNA.
MATERIALS AND METHODS
Oligonucleotides
Deoxyoligonucleotides, 5′-tail TeR2, 5′-d[TAGACATG(TT AGGG)2TTA]-3′ and 3′-tail d(CGG)7, 5′-d[(CGG)7CGTG GACTC]-3′, were synthesized by Operon Technologies and purified by electrophoresis in 8 M urea, 14% polyacrylamide (acrylamide/bisacrylamide 19:1) denaturing gel (27).
Preparation of tetraplex forms of DNA oligomers
Gel-purified single-stranded 5′-tail TeR2 or 3′-tail d(CGG)7 were 5′-end labeled with 32P in a bacteriophage T4 polynucleotide kinase-catalyzed reaction (37). Generation of bimolecular tetraplex G′2 structures of the oligomers and their isolation by non-denaturing gel electrophoresis were performed as we described previously (30). The isolated tetraplex DNA was stored at –20°C in TE buffer, 100 mM KCl until used. Due to spontaneous dissociation of the tetraplex structures into their constituent single strands, tetraplex forms constituted 80–90% and 50–70%, respectively, of the total gel-purified 5′-tail TeR2 and 3′-tail d(CGG)7 DNA. Bimolecular stoichiometry of the tetraplexes was verified as detailed elsewhere (30). As previously demonstrated for the Hoogsteen hydrogen-bonded tetraplexes of these guanine-rich tracts (7,38–41), both G′2 5′-tail TeR2 and G′2 3′-tail d(CGG)7 DNA structures resisted methylation by dimethylsulfate.
Plasmids
A pGEX-A1 plasmid harboring mouse hnRNP A1 cDNA was the generous gift of Dr Benoit Chabot (Universitè de Sherbrooke, Canada). Mouse CBF-A cDNA in a pCDL-SR-CBFA plasmid was kindly contributed by Dr T. Miwa (Osaka University, Japan). Using primers with EcoRI ends, the CBF-A cDNA insert was amplified by polymerase chain reaction (PCR) using Pow DNA polymerase (Roche) and the product cDNA was cloned into pGEX-2T.
Generation of deletion, substitution and truncation mutations in CBF-A
Deletion or substitution mutations in the CBF-A cDNA insert were generated according to the Quickchange site-directed mutagenesis protocol (Stratagene) by extending over pGEX-2T DNA oligonucleotide primers that flank the deleted sequence or that contain a desired base substitution, respectively. To produce 5′- or 3′-truncation mutations, oligonucleotide primers positioned at the desired ends of the CBF-A cDNA sequence were extended by PCR using Pow polymerase. The 5′-end primers with EcoRI termini had a d(ATG) methionine-encoding start triplet whereas the EcoRI-ended 3′-primers had a d(TAG) termination triplet. Wild-type or mutant plasmid DNA was electroporated into Escherichia coli XL-1-Blue cells (Eppendorf electroporator 2510), the DNA was purified and mutations were validated by direct nucleotide sequencing of the CBF-A encoding cDNA tract.
Expression and purification of wild-type and mutant CBF-A proteins
Plasmid DNA harboring wild-type or mutant CBF-A cDNA was transformed by CaCl2 into E.coli BL21(DE3)pLysS cells. The cells were grown at 37°C for 2–3 h to A600 ∼0.6 and expression of glutathione-S-transferase (GST)-fused protein was induced by the addition of 100 µM IPTG for an additional 3–4 h. The induced cells were harvested, washed once with ice-cold STE buffer (100 mM NaCl, 1 mM EDTA, 10 mM Tris–HCl buffer, pH 8.0) and suspended at 4°C in 1:50 original volume of TED buffer (10 mM EDTA, 1 mM DTT, 2.5 mM PMSF, 10 µg/ml aprotinin, 10 µg/ml leupeptin, 50 mM Tris–HCl buffer, pH 8.0). All the subsequent steps were conducted at 4°C. The cells were disrupted by ultrasonic disintegration (Heat Systems XL sonicator) and the resulting extract was centrifuged at 27 000 g for 35 min. The supernatant fraction (10 ml) was adsorbed onto a 1.0 ml column of glutathione–agarose (Sigma) and after a wash with 10 ml of buffer T (1 mM EDTA, 50 mM Tris–HCl buffer, pH 8.0), the GST–CBF-A fusion protein was eluted from the column by 5 mM of freshly prepared reduced glutathione in buffer T. Following overnight dialysis of the protein solution against buffer D (0.5 mM DTT, 1 mM EDTA, 20% glycerol, 25 mM Tris–HCl buffer, pH 8.0), the GST protein tag was cleaved by incubating the fusion protein at 16°C for 18 h with 1 U bovine thrombin (Pharmacia) per 100 µg of protein. Proteins were resolved by SDS–PAGE to verify complete cleavage of GST and to establish that CBF-A and GST constituted more than 95% of the protein mass in the digest.
Purification of qTBP42
qTBP42 was purified to near homogeneity from extracts of rat hepatocytes by successive steps of column chromatography as previously described (14). The presence of stabilizing soy bean trypsin inhibitor (STI) protein in the highly purified fractions of qTBP42 prevented determination of its protein concentration and the amount of qTBP42 was thus calibrated by its 5′-tail G′2 TeR2 binding activity (14).
Assay of G′2 3′-tail d(CGG)7 destabilization
Measurement of G′2 3′-tail d(CGG)7 destabilization by wild-type or mutant CBF-A or by qTBP42 was conducted by incubating at 37°C for 15 min, specified amounts of the purified proteins in 10 µl reaction mixtures that contained 150–300 fmol of G′2 5′-32P 3′-tail d(CGG)7, 10 mM KCl and buffer D. The tetraplex destabilization reaction was terminated by adding to the mixtures 1% SDS to a final concentration of 0.3%. Intact and unwound G′2 3′-tail d(CGG)7 were resolved from one another by electrophoresis at 4°C and 200–250 V in a non-denaturing 10% polyacrylamide gel in 0.5× TBE buffer, 10 mM KCl, until the bromophenol blue tracking dye migrated 7–7.5 cm into the gel. The proportion of unwound G′2 3′-tail d(CGG)7 was quantified by phosphorimaging analysis of the dried gel.
Assays for G′2 5′-tail TeR2 DNA binding
G′2 5′-tail TeR2 binding to purified qTBP42 or to wild-type or mutant CBF-A proteins was conducted at 4°C for 20 min in 10 µl reaction mixtures that contained specified amounts of the respective protein, 150–300 fmol of 5′-32P G′2 5′-tail TeR2 DNA, and 10 mM KCl in buffer D. Protein–DNA complexes were resolved from unbound DNA by electrophoresis of the directly loaded reaction mixtures through a mobility shift non-denaturing 9% polyacrylamide gel in 0.5× TBE buffer (1.2 mM EDTA in 0.54 Tris-borate buffer, pH 8.3) containing 10 mM KCl. Electrophoresis at 4°C and 200–250 V was terminated after the bromophenol blue tracking dye migrated 7–7.5 cm into the gel. The formation of a protein–G′2 5′-tail TeR2 DNA complex was quantified by phosphorimaging of the dried gel. Some CBF-A mutant proteins failed to form complexes with labeled tetraplex TeR2 DNA that were detectable by mobility shift analysis.
Immunochemical identification of qTBP42 and CBF-A
Polyclonal antiserum against purified recombinant wild-type CBF-A protein was raised in a rabbit by Sigma (Israel). Anti-CBF-A antibodies were adsorbed to protein A/G-agarose (Santa Cruz) by incubating overnight at 4°C and under rotation, 50 µl of anti-CBF-A antiserum with 20 µl of a 50% slurry of protein A/G-agarose thrice pre-washed by buffer D, in a final volume of 1 ml of buffer D. Control mixtures contained 50 µl of either pre-immune serum or buffer D. The protein-coated A/G-agarose was washed three times to remove unabsorbed immunoglobulins and 3.9 µg of either purified CBF-A or an equivalent activity of TBP42 in 100 µl of buffer D were added to the mixtures. Immune adsorption was conducted under rotation at 4°C for 90 min, the protein A/G-agarose beads were removed by centrifugation and aliquots of the supernatant fraction were assayed for binding of 5′-32P-labeled G′2 5′-tail TeR2 DNA as described above. Immunochemical identification of GST-fused recombinant CBF-A was conducted by the same immunoprecipitation procedure except that anti-GST antibody (Santa Cruz) was employed in place of the anti-CBF-A antiserum.
RESULTS
CBF-A destabilizes G′2 d(CGG)n and binds G′2 telomeric DNA
We reported previously that rat liver qTBP42 protein binds tightly and stabilizes tetraplex forms of the telomeric sequence d(TTAGGG)n (14) while it paradoxically destabilizes bimolecular tetraplex structures of the fragile X syndrome expanded sequence d(CGG)n (30). Amino acid sequences of qTBP42 peptides are fully homologous to fragments of the hnRNP-related CArG-box binding protein CBF-A (15). We inquired whether, similarly to qTBP42, CBF-A also interacts contrastingly with tetraplex forms of d(CGG)n and d(TTAGGG)n. Mouse CBF-A cDNA (31), whose length and nucleotide sequence are identical to rat CBF-A cDNA (33), was expressed in E.coli cells. The recombinant CBF-A protein was purified and assayed for its ability to destabilize or bind and stabilize G′2 bimolecular tetraplex structures of 3′-tail d(CGG)7 and telomeric 5′-tail TeR2 DNA.
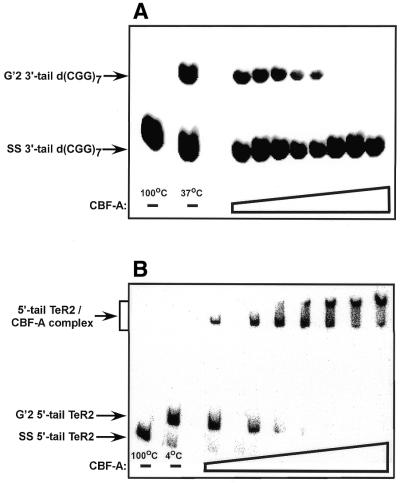
The results presented in Figure 1A show that, similarly to qTBP42 (30), recombinant mouse CBF-A destabilizes G′2 3′-tail d(CGG)7 and that the extent of destabilization is proportional to the amount of protein added. Destabilization of this tetraplex DNA substrate is undetectable in control reaction mixtures that contain GST protein in place of CBF-A (data not shown). Similarly to qTBP42, CBF-A destabilizes G′2 3′-tail d(CGG)7 at equal rates with or without the presence of ATP or Mg2+ ions (data not shown). Furthermore, purified recombinant CBF-A does not appear to contain bound ATP that contributes to its tetraplex DNA disruption activity. CBF-A pre-treated by apyrase (potato ATPase) or incubated in the presence of the ATP-consuming deoxyglucose-hexokinase system maintains 80 and 113%, respectively, of the G′2 3′-tail d(CGG)7 activity of control protein. Replicate titrations of the CBF-A tetraplex DNA destabilizing activity indicate that resolution of 50% of the tetraplex DNA substrate is attained at a 24–38-fold molar excess of protein over G′2 3′-tail d(CGG)7. As previously reported for qTBP42 (30), CBF-A destabilizes to a comparable extent G′2 structures of d(CGG)7, 3′-tail d(CGG)7, 5′-tail d(CGG)7 or 3′,5′-tail d(CGG)7 (data not shown). In contrast, with d(CGG)n tetrahelices, incubation of CBF-A with a tetraplex structure of telomeric DNA, G′2 5′-tail TeR2, under conditions of G′2 d(CGG)n destabilization, results in protein–DNA complex formation with no detectable tetraplex destabilization. As seen in Figure 1B, CBF-A and G′2 5′-tail TeR2 DNA form complexes whose amount is proportional to the amount of protein added. No complex is detected in control mixtures that contain GST protein in place of recombinant CBF-A (data not shown). Notably, larger sized complexes are generated in the presence of excess CBF-A protein. A similar association with G′2 telomeric DNA was reported for qTBP42 and the analysis indicated that the larger-sized complexes have a higher molecular ratio of protein to DNA (14,30). No binding of blunt-ended or tailed G′2 d(CGG)n substrates by CBF-A was detectable under these incubation conditions (data not shown). Multiple titrations of the complex formation between CBF-A and G′2 5′-tail TeR2 indicate that an 18–27-fold molar excess of protein over DNA is required to bind 50% of the telomeric tetraplex DNA.
Figure 1.
CBF-A destabilizes bimolecular tetraplex G′2 3′-tail d(CGG)n and binds bimolecular tetraplex G′2 5′-tail TeR2. (A) Destabilization of G′2 3′-tail d(CGG)7 by CBF-A. Increasing amounts of purified recombinant CBF-A, 5–2000 ng were incubated at 37°C for 15 min with 5′-32P G′2 3′-tail d(CGG)7 under standard tetraplex DNA destabilization assay conditions (see Materials and Methods). Control G′2 3′-tail d(CGG)7 with no added protein was either boiled for 10 min to denature the tetraplex DNA structure or was incubated at 37°C under standard tetraplex DNA destabilization conditions to reveal the initial amount of unwound G′2 3′-tail d(CGG)7. The reaction mixtures were cooled to 4°C and 1% SDS was added to a final concentration of 0.3% to denature DNA-bound protein. Intact and destabilized G′2 3′-tail d(CGG)7 were resolved by electrophoresis at 4°C in 10% polyacrylamide gels in 0.5× TBE buffer, 10 mM KCl. A phosphorimage of the electropherogram is shown. (B) Binding of 5′-tail TeR2 DNA by CBF-A. Increasing amounts of CBF-A, 5–600 ng, were incubated at 4°C for 20 min with 5′-32P G′2 5′-tail TeR2 DNA under standard tetraplex DNA-binding conditions (see Materials and Methods). Control G′2 5′-tail TeR2 DNA without added protein was either boiled for 10 min to denature the tetraplex DNA structure or was incubated at 4°C under standard tetraplex DNA-binding conditions to reveal the initial amount of unbound G′2 5′-tail TeR2 DNA. A phosphorimage of a 9% polyacrylamide, 0.5× TBE buffer, 10 mM KCl gel is shown.
A heat-resistant CBF-A stabilizes bound G′2 5′-tail TeR2 DNA
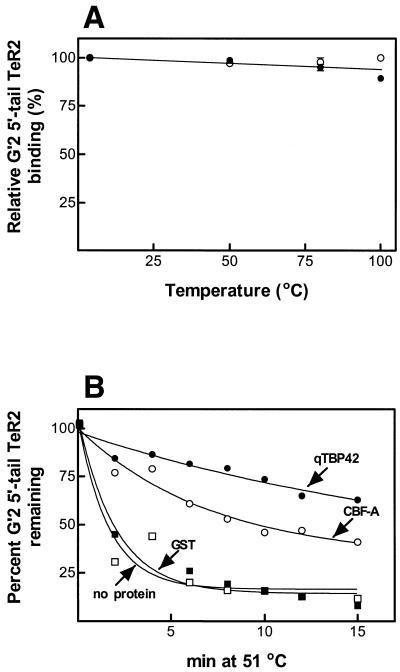
qTBP42 is a heat-stable protein whose association with tetraplex telomeric DNA raises the melting temperature of the bound DNA (14). The results presented in Figure 2A show that CBF-A and qTBP42 are similarly heat resistant, both maintaining nearly full DNA-binding activity after being incubated for 10 min at 100°C. To examine whether similarly to qTBP42, CBF-A protects tetraplex telomeric DNA against heat denaturation, complexes of G′2 5′-tail TeR2 DNA with each protein were incubated at 51°C, the protein residue was removed by SDS and the amounts of remaining tetraplex structure were determined by electrophoresis. As seen in Figure 2B, relative to free tetraplex DNA or to DNA heated in the presence of excess GST protein, the denaturation rate of DNA bound by CBF-A or qTBP42 is diminished. Since the amount of the added qTBP42 could not be determined directly (see Materials and Methods), the heated mixtures contained different amounts of CBF-A or qTBP42G′2. This difference in protein to DNA ratio is the probable cause for the somewhat greater degree of heat protection of the tetraplex DNA afforded by qTBP42 (Fig. 2B). Two lines of evidence indicate that CBF-A is directly responsible for the observed thermal stabilization of G′2 5′-tail TeR2 DNA. First, the heat stability of tetraplex telomeric DNA is not increased in the presence of carbonic anhydrase, bovine serum albumin, thrombin protease or ovalbumin added at 10-fold greater molecular excess than CBF-A (data not shown). Secondly, removal of GST-fused CBF-A by immune precipitation with anti-GST specifically obliterates the thermal protection of the DNA tetraplex (data not shown).
Figure 2.
CBF-A and qTBP42 are heat-stable proteins that slow down denaturation of associated G′2 5′-tail TeR2 DNA. (A) Heat stability of CBF-A and qTBP42. Purified CBF-A at 100 µg/ml or qTBP42 protein to which bovine serum albumin was added to the same concentration, were incubated, each in duplicate, at temperatures ranging from 4 to 100°C for 10 min in a final volume of 10 µl of buffer D. The samples were rapidly cooled to 4°C and aliquots were assayed under standard conditions for the binding of 90 fmol of 5′-32P G′2 5′-tail TeR2 DNA. Protein–DNA complexes were resolved by electrophoresis at 4°C in 8% polyacrylamide gels, 0.5× TBE buffer, 10 mM KCl. Amounts of protein-bound 5′-32P G′2 5′-tail TeR2 DNA were quantified by phosphorimaging. Relative binding of 100% represents association of CBF-A or qTBP42 with 85.5–89 fmol of tetraplex DNA. Closed circles, qTBP42; open circles, CBF-A. (B) Binding of CBF-A and qTBP42 slows down the denaturation of G′2 5′-tail TeR2 DNA. DNA, 26 fmol of 5′-32P G′2 5′-tail TeR2, was incubated at 4°C for 20 min in 10 µl of buffer D, 10 mM KCl reaction mixtures that contained no protein or 167 pmol GST, 13 pmol of CBF-A, or qTBP42 (amount not determined, see Materials and Methods). The mixtures were transferred to 51°C for the indicated periods of time, rapidly cooled to 4°C and protein was removed from the DNA by adding 1% SDS to a final concentration of 0.3%. Intact and denatured G′2 5′-tail TeR2 DNA were resolved from one another by electrophoresis at 4°C in 10% polyacrylamide gels, 0.5× TBE buffer, 10 mM KCl. Amounts of G′2 and single-stranded DNA were quantified by phosphorimaging. A value of 100% G′2 5′-tail TeR2 DNA represents 57% G′2 out of the total 5′-tail TeR2 DNA in samples that were kept at 4°C. Closed circles, qTBP42; open circles, CBF-A; open boxes, GST; closed boxes, no protein.
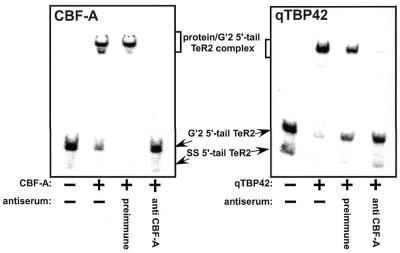
Anti-CBF-A antiserum recognizes qTBP42
To confirm synonymy of qTBP42 and CBF-A we inquired whether anti-mouse CBF-A rabbit antibodies bind rat qTBP42. The results presented in Figure 3 demonstrate that whereas pre-immune rabbit serum precipitates neither CBF-A nor qTBP42, both proteins are equally bound by the anti-CBF-A antiserum. The cross-antigenicity of qTBP42 and CBF-A, as well as their sequence homology, similar heat stability and comparable abilities to destabilize G′2 d(CGG)n and to bind and stabilize G′2 TeR2 DNA, establish their identity. It is highly likely, therefore, that rat qTBP42 and mouse CBF-A mediate their differential interaction with tetraplex d(CGG)n and d(TTAGGG)n through the same protein domains.
Figure 3.
Anti-CBF-A antiserum recognizes TBP42. Purified CBF-A or qTBP42 proteins were incubated at 4°C for 90 min with rabbit anti-CBF-A antibodies adsorbed to protein A/G-agarose (see Materials and Methods). Control mixtures contained pre-immune rabbit serum adsorbed to protein A/G-agarose or protein A/G-agarose with no adsorbed protein. The protein A/G-agarose beads were removed by centrifugation and aliquots of the supernatant fractions were assayed for binding of 5′-32P G′2 5′-tail TeR2 DNA as described in Materials and Methods. Phosphorimages of protein-G′2 5′-tail TeR2 complexes resolved from unbound DNA by electrophoresis through non-denaturing 9% polyacrylamide mobility shift gels are shown.
The RNP11 and ATP/GTP-binding motifs of CBF-A are specifically required for tetraplex d(CGG)n destabilization
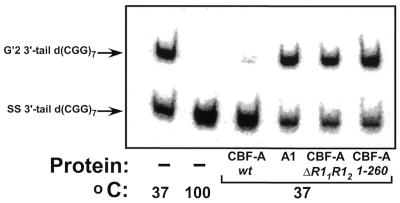
Mutant CBF-A proteins were expressed in E.coli cells that have truncated N- or C-terminal regions or that contain deletions in one or more of the evolutionary conserved and duplicated RNP1 or RNP2 boxes (42,43) or in the putative ATP/GTP-binding domain (42,44–46). The purified proteins were assayed for their capacity to destabilize G′2 3′-tail d(CGG)7. Figure 4 shows a typical analysis of G′2 3′-tail d(CGG)7 destabilization by wild-type and representative mutant CBF-A proteins. Whereas G′2 3′-tail d(CGG)7 is fully resolved by wild-type CBF-A, no destabilization is detectable in mixtures that contain equal amounts of mutant proteins with either deleted RNP11 and RNP12 boxes (mutant ΔR11R12) or truncated C-terminal tract that encompasses the ATP/GTP-binding motif (mutant 1–260). Notably, recombinant wild-type hnRNP A1 is also unable to unwind G′2 3′-tail d(CGG)7 (Fig. 4), indicating that the tetraplex d(CGG)n unwinding capacity of CBF-A is not shared by every hnRNP species.
Figure 4.
Destabilization of G′2 3′-tail d(CGG)7. Wild-type CBF-A or hnRNP A1 proteins, at 33 pmol each or 165 pmol each of R11R12 or 1–260 CBF-A mutant proteins were incubated at 37°C for 15 min with 0.16 pmol of 5′-32P G′2 3′-tail d(CGG)7 under standard tetraplex DNA destabilization assay conditions. Control samples that did not contain protein were similarly incubated at 37 or 100°C to reveal the initial amount of G′2 3′-tail d(CGG)7 and the position of displaced single-stranded DNA, respectively. The tetraplex unwinding reaction was terminated by the addition of 1% SDS to a final concentration of 0.3%, and tetraplex and single-stranded 3′-tail d(CGG)7 were resolved by electrophoresis through a 10% polyacrylamide gel, 0.5× TBE buffer, 10 mM KCl. A phosphorimage of the electropherogram is shown.
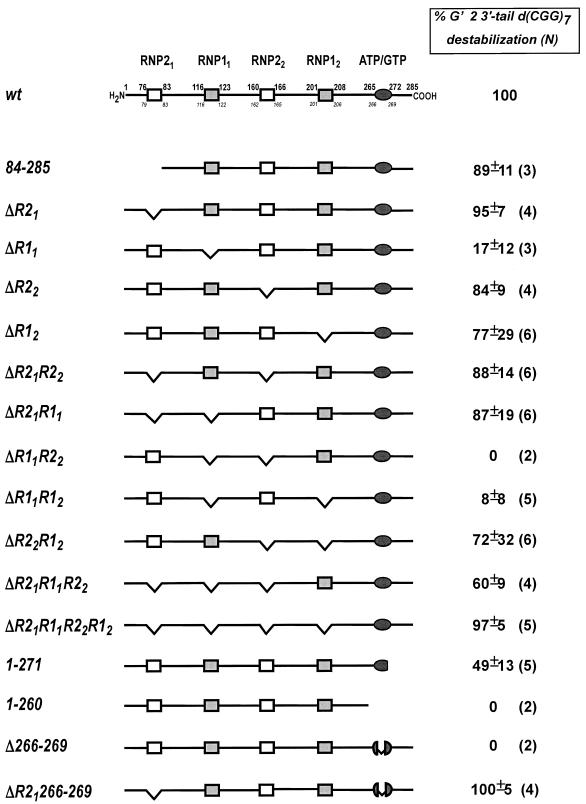
The schemes of all the CBF-A mutants examined and their relative G′2 3′-tail d(CGG)7 destabilization activities are shown in Figure 5. Our results indicate that the RNP11 box or the ATP/GTP-binding element are required for destabilization of G′2 3′-tail d(CGG)7. Deletion of residues 116–122 from the RNP11 motif (CBF-A mutant ΔR11) inactivates G′2 3′-tail d(CGG)7 destabilization, implying that this element is essential for the unwinding activity (Fig. 5). This proposition is supported by the similar loss of unwinding activity by CBF-A double mutants ΔR11R12 and ΔR11R22 that, in addition to RNP11, lack the RNP12 or RNP22 boxes which are dispensable for tetraplex destabilization (see below). Parallel data indicate that the destabilization activity is also dependent on the presence of an intact ATP/GTP-binding motif. The absence of this element in CBF-A Δ266–269 or in the truncation mutant 1–260 results in failure to destabilize G′2 3′-tail d(CGG)7 (Fig. 5). Interestingly, truncation that trims a single serine residue at the C-terminal end of the ATP/GTP motif (position 272; mutant 1–271) causes loss of ∼50% of the G′2 3′-tail d(CGG)7 destabilization activity (Fig. 5). Lastly, substituting a single tyrosine residue by lysine at position 267 within the ATP/GTP box (T267K mutation) also results in loss of destabilization activity (Fig. 6). Put together, these data substantiate the required role of the conserved ATP/GTP box in G′2 3′-tail d(CGG)7 disruption.
Figure 5.
CBF-A mutant proteins and their G′2 3′-tail d(CGG)7 destabilization activity. The conserved motifs marked in the scheme of wild-type CBF-A are: RNP21, MFVGGL; RNP11, SRGFGFIL; RNP22, IFVGGL; RNP12, RRGFVFIT; the putative ATP/GTP-binding domain, GSTNYGKS. The upper numbers in the wild-type protein scheme denote positions of residues at the boundaries of the conserved domains whereas the lower italicized numbers indicate the boundaries of segments deleted from each motif. Schemes show segments truncated or deleted in each mutant protein. The presented relative G′2 3′-tail d(CGG)7 destabilization activity of each mutant protein is an average of the indicated number of independent determinations.
Figure 6.
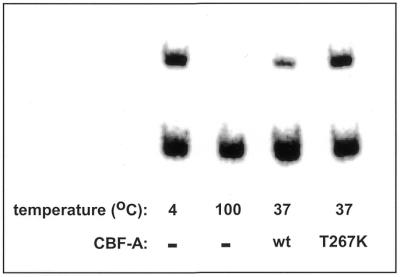
CBF-A mutant T267K fails to unwind G′2 3′-tail d(CGG)7. Wild-type or T267K variant CBF-A protein mutated in the ATP/GTP-binding motif at 48 pmol each, were incubated at 37°C for 15 min with 0.17 pmol of 5′-32P G′2 3′-tail d(CGG)7 under standard tetraplex DNA destabilization assay conditions. Incubation of control samples with no added protein, termination of the destabilization reaction and electrophoretic resolution of intact and destabilized G′2 3′-tail d(CGG)7 were conducted as detailed in the legend to Figure 4. Quantification of phosphorimages of four independent determinations reveal that wild-type and T267K CBF-A proteins destabilized 53 ± 3 and 4 ± 3%, respectively, of the tetraplex substrate.
The evidence summarized in Figure 5 indicates that in contrast to the essential RNP11 and ATP/GTP-binding motifs, the N-terminal sequence of CBF-A and its RNP21, RNP22 and RNP12 boxes are expandable for G′2 d(CGG)n destabilization activity. CBF-A is distinguished from other hnRNP homologs by its unique N-terminal acidic stretch of 75 amino acids (25,31). As seen in Figure 5, truncation of an 83 amino acid N-terminal tract (mutant 84–285) does not affect G′2 3′-tail d(CGG)7 destabilization, indicating the dispensability of this sequence for tetraplex disruption activity. Most of the G′2 3′-tail d(CGG)7 destabilization activity is also maintained by mutant proteins with residues deleted from the RNP21, RNP22 or RNP12 motifs as well as by proteins that contain double mutations; ΔR21R22 and ΔR22R12 (Fig. 5). These results demonstrate that RNP21, RNP22 and RNP12 are also superfluous for tetraplex d(CGG)n disruption.
The RNP21 box represses G′2 d(CGG)n destabilization by the RNP11 or ATP/GTP-binding motifs
Inspection of the activities of some of the compound CBF-A mutant proteins listed in Figure 5 reveals that the RNP21 motif inhibits tetraplex d(CGG)n disruption mediated by the RNP11 element or the ATP/GTP-binding box. Deletion of the RNP11 motif in CBF-A mutants ΔR11, ΔR11R22 or ΔR11R12, results in loss of G′2 3′-tail d(CGG)7 unwinding activity despite the presence of an intact ATP/GTP-binding motif (Fig. 5). Concordantly, the failure of CBF-A mutants Δ266–269 and 1–260 to destabilize G′2 3′-tail d(CGG)7 shows that tetraplex disruption is inactivated by the absence of an ATP/GTP-binding motif despite the presence of an undamaged RNP11 box. The vigorous destabilization activities of wild-type CBF-A and of the mutant proteins ΔR22, ΔR12 and ΔR2212 indicate, however, that tetraplex disruption is permitted in the presence of both the RNP11 and ATP/GTP-binding motifs (Fig. 5). At the same time, data show that removal of the RNP21 motif re-establishes G′2 d(CGG)n activity in mutant proteins that contain either an RNP11 motif but no ATP/GTP-binding element or an ATP/GTP box without RNP11. The nearly full disruption of G′2 3′-tail d(CGG)7 by CBF-A mutants ΔR21R11, ΔR21R11R22 or ΔR21R11R22R12 indicates that deletion of the RNP21 box restores destabilization activity to proteins that contain only an ATP/GTP-binding element without an RNP11 box (Fig. 5). Correspondingly, the full disruptive activity of CBF-A mutant protein ΔR21 indicates that deletion of the RNP21 motif confers tetraplex d(CGG)n destabilization activity to a mutant protein that has an RNP11 box but no ATP/GTP-binding motif (Fig. 5). Put together, these results indicate that in the absence of the RNP21 motif, RNP11 or the ATP/GTP-binding box are capable of mediating disruption of G′2 d(CGG)n independently of each other. Since the RNP21 box represses the activity of either one of these elements but is unable to simultaneously inhibit both, the presence of both the RNP11 and ATP/GTP-binding motifs secures destabilization activity despite the existence of an operational RNP21 inhibitory element. A tentative model that satisfies these observations is presented in the Discussion.
Binding and stabilization of G′2 TeR2 DNA do not require intact RNP11 motif or ATP/GTP-binding element
Parallel to possessing tetraplex d(CGG)n destabilizing capacity, CBF-A and qTBP42 are distinguished by their ability to bind and stabilize tetraplex telomeric DNA. We examined whether or not the RNP11 motif or the ATP/GTP-binding box that are required for G′2 d(CGG)n disruption, also mediate G′2 5′-tail TeR2 DNA binding and stabilization. The results summarized in Table 1 indicate that tetraplex telomeric DNA is bound to a similar extent by recombinant wild-type CBF-A or by the ΔR11 and 1–260 mutant proteins that lack these two domains. Likewise, wild-type CBF-A and the two mutant proteins protect to a similar extent G′2 5′-tail TeR2 against heat denaturation (Table 1). It appears, therefore, that the RNP11 or the ATP/GTP-binding motifs are not involved in the binding and stabilization of tetraplex telomeric DNA.
Table 1. Deletion of the RNP11 box or the ATP/GTP-binding motif does not affect binding or stabilization of G′2 5′-tail TeR2.
| CBF-A protein | % G′2 5′-tail TeR2 binding (N) | %G′2 5′-tail TeR2 stabilization (N) |
|---|---|---|
| wt | 100 | 100 |
| ΔR11 | 92.8 ± 3.1 (4) | 96.9 ± 24.4 (4) |
| 1–260 | 78.4 ± 1.1 (4) | 84.4 ± 11.0 (3) |
Wild-type (wt) or mutant CBF-A proteins were assayed for their ability to bind 5′-32P G′2 5′-tail TeR2 DNA as described in Materials and Methods. In parallel, the relative capacity of the proteins to stabilize 5′-32P G′2 5′-tail TeR2 DNA against denaturation for 7 min at 51°C was assayed as detailed in the legend to Figure 2B.
DISCUSSION
Tetrahelical structures of the FMR1 gene d(CGG)n tract have been suggested to promote polymerase slippage and expansion of the trinucleotide repeat sequence that silences FMR1 and sets off fragile X syndrome (7). Disruption of d(CGG)n tetraplexes could possibly lower the probability of expansion of this sequence. Proteins such as the Werner syndrome DNA helicase (27,47), qTBP42 (14,30) and uqTBP25 (16,30) were found to unwind G′2 bimolecular tetraplex forms of d(CGG)n. In this work, we undertook to unravel the protein structure basis for the G′2 d(CGG)n destabilization activity of qTBP42/CBF-A. The results described here have two major corollaries: First, the hnRNP-related rat protein qTBP42 (14,15,30) and the transcriptional regulator CBF-A (31–36) are indistinguishable. Secondly, two conserved domains of CBF-A, the RNP11 box and the putative ATP/GTP-binding fold mediate destabilization of tetraplex d(CGG)n whereas the RNP21 motif acts to suppress this activity.
Rat liver qTBP42 is distinguished by its tight binding and stabilization of tetraplex forms of the telomeric sequence d(TTAGGG)n (14,15,30) and by its contrasting capacity to destabilize G′2 tetraplex structures of d(CGG)n (30). Based on peptide sequence homology of qTBP42 and CBF-A, we inquired whether physical–chemical properties of recombinant mouse CBF-A and its differential interaction with G′2 tetraplex structures of d(TTAGGG)n and d(CGG)n resemble those of qTBP42. Our results show that, similarly to qTBP42, CBF-A also disrupts G′2 bimolecular d(CGG)n tetraplexes while binding G′2 d(TTAGGG)n (Fig. 1). Furthermore, CBF-A resembles qTBP42 in being a heat-stable protein that also slows down the heat denaturation of bound tetraplex telomeric DNA (Fig. 2). Lastly, the identity of the two proteins is corroborated by the specific precipitation of rat qTBP42 by anti-mouse CBF-A antibodies (Fig. 3).
We identified domains in CBF-A that mediate G′2 d(CGG)n destabilization by determining this activity in an array of CBF-A mutant proteins. Although CBF-A is distinguished from other hnRNP-related homologous proteins by its unique acidic N-terminal stretch of 75 amino acids (25,31), results show that this domain is not required for G′2 3′-tail d(CGG)7 destabilization as evidenced by the full unwinding activity of the N-terminal truncated mutant protein 83–285 (Fig. 5). The nearly full G′2 3′-tail d(CGG)7 disruption activity of CBF-A mutant proteins with deletions in the RNP21, RNP22 or RNP12 boxes or combinations thereof, indicates that these elements are also expendable for tetraplex destabilization (Fig. 5). In contrast, the RNP11 box and/or the ATP/GTP-binding elements are required for G′2 d(CGG)n disruption. CBF-A mutants that have residues deleted from their RNP11 motif alone or in combination with the dispensable RNP12 or RNP22 boxes lack unwinding activity (Fig. 5). Likewise, CBF-A mutant proteins with truncated or deleted ATP/GTP-binding motifs (Fig. 5), or with amino acid substitution within this domain (mutant T267K, Fig. 6) fail to disrupt G′2 d(CGG)n. Notably, the putative ATP/GTP-binding fold, solely assigned by amino acid sequence homology, has no known function (42,44–46). The participation of this element in tetraplex DNA destabilization is, to the best of our knowledge, its first identified activity. However, since disruption of G′2 d(CGG)n by CBF-A does not require nucleotide triphosphates (see Results), the involvement of this domain in tetraplex disruption is probably unrelated to its presumed binding of ATP or GTP.
Surprisingly, we find that the RNP21 motif acts to suppress tetraplex d(CGG)n destabilization. Analysis of multiple mutant proteins indicates that whereas the disruption of G′2 d(CGG)n by intact CBF-A requires the presence of both the RNP11 motif and an ATP/GTP-binding element, removal of the inhibitory RNP21 box allows mutant proteins that lack either RNP11 or ATP/GTP-binding box to conduct tetraplex unwinding by the single remaining element (Fig. 5).
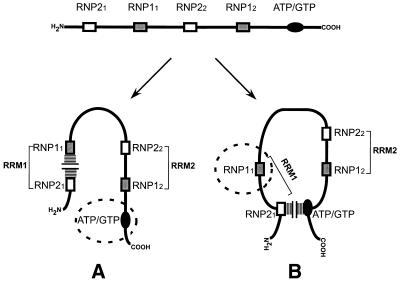
Which then is the domain in wild-type CBF-A that is active in G′2 d(CGG)n disruption and which is inactivated by the suppressor RNP21 box? Two alternative modes of destabilization of G′2 d(CGG)n by intact CBF-A protein are modeled in Figure 7. In one modality the RNP21 box blocks RNP11, leaving an uninhibited ATP/GTP-binding motif to conduct tetraplex destabilization (Fig. 7A). In an alternative mode RNP21 obstructs the ATP/GTP-binding element and allows the unblocked RNP11 to mediate disruption of G′2 d(CGG)n (Fig. 7B). A third possibility is that the two depicted conformations of CBF-A are in equilibrium and either the RNP11 motif or the ATP/GTP-binding box operates within any individual protein molecule to destabilize G′2 d(CGG)n. However, the known mode of interaction of hnRNP proteins with single-stranded DNA or RNA favors the view that the ATP/GTP-binding box is the primary domain responsible for tetraplex unwinding by wild-type CBF-A. Proteins of the hnRNP super family possess two RNA recognition motifs (RRMs). The crystal structure of hnRNPA1 reveals that RRM1, formed by the coupling of RNP21 to RNP11, and RRM2, consisting of RNP22 and RNP12, are held in close contact in an anti-parallel orientation (48,49). Several closely related hnRNPs, including CBF-A/qTBP42, bind tightly the single-stranded telomeric repeat d(TTAGGG)n (14,16,50–55). The crystal structure of a complex of d(TTAGGG)2 with a dimer of the N-terminal proteolytic fragment of hnRNPA1 (protein UP1) reveals that each of the two DNA strands is in contact with RRM1 of one UP1 monomer and with RRM2 of the other monomer (54). It appears, therefore, that when they interact with telomeric DNA, RNP21 and RNP11 are coupled together in RRM1. Conceivably, the interaction of CBF-A with G′2 d(CGG)n is also mediated by its RRM1 and RRM2 domains. If this is the case, the RNP11 box of the wild-type CBF-A is coupled within an RRM1 element to the RNP21 suppressor motif that renders it inactive while allowing the uninhibited ATP/GTP box to mediate disruption of tetraplex d(CGG)n (Fig. 7A). However, with the inhibitory RNP21 element deleted, no RRM1 is formed and the unsuppressed RNP11 takes over the function of unwinding G′2 d(CGG)n when the ATP/GTP-binding motif is absent (Fig. 5).
Figure 7.
Models of two alternative modes of tetraplex d(CGG)n destabilization by CBF-A. CBF-A, schematically presented in the upper diagram with its conserved RNP boxes and ATP/GTP-binding motif, might assume either one of the two illustrated alternative conformations that conduct G′2 d(CGG)n destabilization through either RNP11 or the ATP/GTP-binding motif. (A) G′2 d(CGG)n unwinding mediated by the ATP/GTP-binding motif. The RNP11 box is blocked by the inhibitory RNP21 element (horizontal hatches) whereas the ATP/GTP-binding motif (dashed circle) remains available to conduct G′2 d(CGG)n destabilization. (B) RNP11-mediated G′2 d(CGG)n unwinding. The ATP/GTP-binding motif is blocked by the inhibitory RNP21 element (vertical hatches) whereas the RNP11 box (dashed circle) remains available to conduct G′2 d(CGG)n destabilization. Details of the models are considered in the Discussion.
Whereas motifs in CBF-A that contribute positively or negatively to the disruption of tetraplex d(CGG)n were pinpointed, CBF-A mutants lacking either RNP11 or the ATP/GTP-binding motif maintain full capacity to bind and stabilize tetraplex telomeric DNA. Hence, specific CBF-A domains that are responsible for the binding and stabilization of G′2 d(TTAGGG)n remain to be identified. Notably, analysis of co-crystals of UP1 and single-strand telomeric DNA reveals multiple contacts between the protein and the d(TTAGGG)2 sequence (55). Amino acids within and outside the RNP elements interact with bound DNA through hydrogen bonds and charge and van der Waals interactions (55). If a similar large number of amino acids in CBF-A interact with tetraplex telomeric DNA it is plausible that the binding and stabilization of the tetrahelical telomeric sequence by CBF-A can be impaired only by the removal of amino acids at multiple sites.
In the past we speculated that qTBP42 might act in vivo bi-functionally by stabilizing desirable tetrahelices, such as perhaps at telomere ends, and destabilizing unwanted tetrahelices such as G′2 d(CGG)n structures that might be generated during DNA transactions (30). Interestingly, additional hnRNP-related proteins were also found to interact differentially with tetraplex DNA structures. These are uqTBP25, which binds unimolecular and bimolecular tetraplexes of the telomeric sequence d(TTAGGG)n but destabilizes G′2 d(CGG)n (16,30) and hnRNP D, which binds tightly the telomeric tract and destabilizes its G–G paired structures (52). Since hnRNPs play major roles in RNA metabolism, it is highly likely that the observed interactions with tetraplex DNA reflect functions of hnRNPs in stabilizing or disrupting tetraplex structures in specific RNA molecules. Interestingly, recent results indicate that the FMR1 protein binds preferentially tetraplex-forming mRNA molecules, modulating their association with polysomes (56–58). A tempting conjecture is that by differentially modulating the stability of tetraplex domains in different mRNA molecules, proteins such as CBF-A, uqTBP25 or hnRNP D might regulate their nuclear– cytoplasm translocation or control their translation.
Acknowledgments
ACKNOWLEDGEMENTS
This study was supported by grants to M.F. from the U.S.–Israel Binational Science Foundation, The Israel Science Foundation, the Conquer Fragile X Foundation Inc., The Chief Scientist—Israel Ministry of Health, and the Fund for Promotion of Research in the Technion.
REFERENCES
- 1.Simonsson T. (2001) G-quadruplex DNA structures—variations on a theme. Biol. Chem., 382, 621–628. [DOI] [PubMed] [Google Scholar]
- 2.Arthanari H. and Bolton,P.H. (2001) Functional and dysfunctional roles of quadruplex DNA in cells. Chem. Biol., 8, 221–230. [DOI] [PubMed] [Google Scholar]
- 3.Sen D. and Gilbert,W. (1988) Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature, 334, 364–366. [DOI] [PubMed] [Google Scholar]
- 4.Henderson E. (1995) Telomere DNA structure. In Blackburn,E.H. and Greider,C.W., (eds), Telomeres. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, pp. 11–34.
- 5.Catasti P., Chen,X., Moyzis,R.K., Bradbury,E.M. and Gupta,G. (1996) Structure–function correlations of the insulin-linked polymorphic region. J. Mol. Biol., 264, 534–545. [DOI] [PubMed] [Google Scholar]
- 6.Simonsson T., Pecinka,P. and Kubista,M. (1998) DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res., 26, 1167–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fry M. and Loeb,L.A. (1994) The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl Acad. Sci. USA, 91, 4950–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walsh K. and Gualberto,A. (1992) MyoD binds to the guanine tetrad nucleic acid structure. J. Biol. Chem., 267, 13714–13718. [PubMed] [Google Scholar]
- 9.Pearson A.M., Rich,A. and Krieger,M. (1993) Polynucleotide binding to macrophage scavenger receptors depends on the formation of base-quartet-stabilized four-stranded helices. J. Biol. Chem., 268, 3546–3554. [PubMed] [Google Scholar]
- 10.Weisman-Shomer P. and Fry,M. (1993) QUAD, a protein from hepatocyte chromatin that binds selectively to guanine-rich quadruplex DNA. J. Biol. Chem., 268, 3306–3312. [PubMed] [Google Scholar]
- 11.Schierer T. and Henderson,E. (1994) A protein from Tetrahymena thermophila that specifically binds parallel-stranded G4-DNA. Biochemistry, 33, 2240–2246. [DOI] [PubMed] [Google Scholar]
- 12.Frantz J.D. and Gilbert,W. (1995) A yeast gene product, G4p2, with a specific affinity for quadruplex nucleic acids. J. Biol. Chem., 270, 9413–9419. [DOI] [PubMed] [Google Scholar]
- 13.Frantz J.D. and Gilbert,W. (1995) A novel yeast gene product, G4p1, with a specific affinity for quadruplex nucleic acids. J. Biol. Chem., 270, 20692–20697. [DOI] [PubMed] [Google Scholar]
- 14.Sarig G., Weisman-Shomer,P., Erlitzki,R. and Fry,M. (1997) Purification and characterization of qTBP42, a new single-stranded and quadruplex telomeric DNA-binding protein from rat hepatocytes. J. Biol. Chem., 272, 4474–4482. [DOI] [PubMed] [Google Scholar]
- 15.Sarig G., Weisman-Shomer,P. and Fry,M. (1997) Telomeric and tetraplex DNA binding properties of qTBP42: a homologue of the CArG box binding protein CBF-A. Biochem. Biophys. Res. Commun., 237, 617–623. [DOI] [PubMed] [Google Scholar]
- 16.Erlitzki R. and Fry,M. (1997) Sequence-specific binding protein of single-stranded and unimolecular quadruplex telomeric DNA from rat hepatocytes. J. Biol. Chem., 272, 15881–15890. [DOI] [PubMed] [Google Scholar]
- 17.Lu Q., Schierer,T., Kang,S.G. and Henderson,E. (1998) Purification, characterization and molecular cloning of TGP1, a novel G-DNA binding protein from Tetrahymena thermophila. Nucleic Acids Res., 26, 1613–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kee K., Niu,L. and Henderson,E. (1998) A Tetrahymena thermophila G4-DNA binding protein with dihydrolipoamide dehydrogenase activity. Biochemistry, 37, 4224–4234. [DOI] [PubMed] [Google Scholar]
- 19.Liu Z., Frantz,J.D., Gilbert,W. and Tye,B.K. (1993) Identification and characterization of a nuclease activity specific for G4 tetrastranded DNA. Proc. Natl Acad. Sci. USA, 90, 3157–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Z. and Gilbert,W. (1994) The yeast KEM1 gene encodes a nuclease specific for G4 tetraplex DNA: implication of in vivo functions for this novel DNA structure. Cell, 77, 1083–1092. [DOI] [PubMed] [Google Scholar]
- 21.Bashkirov V.I., Scherthan,H., Solinger,J.A., Buerstedde,J.M. and Heyer,W.D. (1997) A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol., 136, 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun H., Yabuki,A. and Maizels,N. (2001) A human nuclease specific for G4 DNA. Proc. Natl Acad. Sci. USA, 98, 12444–12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang G. and Cech,T.R. (1993) Characterization of a G-quartet formation reaction promoted by the beta-subunit of the Oxytricha telomere-binding protein. Biochemistry, 32, 11646–11657. [DOI] [PubMed] [Google Scholar]
- 24.Fang G. and Cech,T.R. (1993) The beta subunit of Oxytricha telomere-binding protein promotes G-quartet formation by telomeric DNA. Cell, 74, 875–885. [DOI] [PubMed] [Google Scholar]
- 25.Weisman-Shomer P. and Fry,M. (1994) Stabilization of tetrahelical DNA by the quadruplex DNA binding protein QUAD. Biochem. Biophys. Res. Commun., 205, 305–311. [DOI] [PubMed] [Google Scholar]
- 26.Sun H., Karow,J.K., Hickson,I.D. and Maizels,N. (1998) The Bloom’s syndrome helicase unwinds G4 DNA. J. Biol. Chem., 273, 27587–27592. [DOI] [PubMed] [Google Scholar]
- 27.Fry M. and Loeb,L.A. (1999) Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem., 274, 12797–12802. [DOI] [PubMed] [Google Scholar]
- 28.Sun H., Bennett,R.J. and Maizels,N. (1999) The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G-G paired DNAs. Nucleic Acids Res., 27, 1978–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohaghegh P., Karow,J.K., Brosh,R.M.,Jr, Bohr,V.A. and Hickson,I.D. (2001) The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res., 29, 2843–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weisman-Shomer P., Naot,Y. and Fry,M. (2000) Tetrahelical forms of the fragile X syndrome expanded sequence d(CGG)n are destabilized by two heterogeneous nuclear ribonucleoprotein-related telomeric DNA-binding proteins. J. Biol. Chem., 275, 2231–2238. [DOI] [PubMed] [Google Scholar]
- 31.Kamada S. and Miwa,T. (1992) A protein binding to CArG box motifs and to single-stranded DNA functions as a transcriptional repressor. Gene, 119, 229–236. [DOI] [PubMed] [Google Scholar]
- 32.Bemark M., Olsson,H., Heinegård,D. and Leanderson,T. (1998) Purification and characterization of a protein binding to the SP6 kappa promoter. A potential role for CArG-box binding factor-A in kappa transcription. J. Biol. Chem., 273, 18881–18890. [DOI] [PubMed] [Google Scholar]
- 33.Rushlow W.J., Rajakumar,N., Flumerfelt,B.A. and Naus,C.C.G. (1999) Characterization of CArG-binding protein A initially identified by differential display. Neuroscience, 94, 637–649. [DOI] [PubMed] [Google Scholar]
- 34.Rushlow W.J., Rajakumar,B., Flumerfelt,B.A., Naus,C.C.G. and Rajakumar,N. (2000) Changes in CArG-binding protein A expression levels following injection(s) of the D1-dopamine agonist SKF-82958 in the intact and 6-hydroxydopamine-lesioned rat. Neuroscience, 98, 69–78. [DOI] [PubMed] [Google Scholar]
- 35.Mikheev A.M., Mikheev,S.A., Zhang,Y., Aebersold,R. and Zarbl,H. (2000) CArG binding factor A (CBF-A) is involved in transcriptional regulation of the rat Ha-ras promoter. Nucleic Acids Res., 28, 3762–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inoue A., Omori,A., Ichinose,S., Takahashi,K.P., Kinoshita,Y. and Mita,S. (2001) S1 proteins C2 and D2 are novel hnRNPs similar to the transcriptional repressor, CArG box motif-binding factor A. Eur. J. Biochem., 268, 3654–3663. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J. and Russel,D.W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, Vol. 3, pp. A4.35–A4.36.
- 38.Henderson E., Hardin,C.C., Walk,S.K., Tinnoco,I.,Jr and Blackburn,E.H. (1987) Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine–guanine base pairs. Cell, 51, 899–908. [DOI] [PubMed] [Google Scholar]
- 39.Sundquist W.I. and Klug,A. (1989) Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature, 342, 825–829. [DOI] [PubMed] [Google Scholar]
- 40.Williamson J.R., Raghuraman,M.K. and Cech,T.R. (1989) Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell, 59, 871–880. [DOI] [PubMed] [Google Scholar]
- 41.Kettani A., Kumar,R.A. and Patel,D.J. (1995) Solution structure of a DNA quadruplex containing the fragile X syndrome triplet repeat. J. Mol. Biol., 254, 638–656. [DOI] [PubMed] [Google Scholar]
- 42.Dreyfuss G., Matunis,M.J., Pinol-Roma,S. and Burd,C.G. (1993) hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem., 62, 289–321. [DOI] [PubMed] [Google Scholar]
- 43.Weighardt F., Biamonti,G. and Riva,S. (1996) The roles of heterogeneous nuclear ribonucleoproteins (hnRNP) in RNA metabolism. Bioessays, 18, 747–756. [DOI] [PubMed] [Google Scholar]
- 44.Swanson M.S., Nakagawa,T.Y., LeVan,K. and Dreyfuss,G. (1987) Primary structure of human nuclear ribonucleoprotein particle C proteins: conservation of sequence and domain structures in heterogeneous nuclear RNA, mRNA and pre-rRNA-binding proteins. Mol. Cell. Biol., 7, 1731–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khan F.A., Jaiswal,A.K. and Szer,W. (1991) Cloning and sequence analysis of a human type A/B hnRNP protein. FEBS Lett., 290, 159–161. [DOI] [PubMed] [Google Scholar]
- 46.Cvekl A., McDermott,J.B. and Piatigorsky,J. (1995) cDNA encoding a chicken protein (CRP1) with homology to hnRNP type A/B. Biochim. Biophys. Acta, 1261, 290–292. [DOI] [PubMed] [Google Scholar]
- 47.Kamath-Loeb A.S., Loeb,L.A., Johansson,E., Burgers,P.M.J. and Fry,M. (2001) Interactions between the Werner syndrome helicase and DNA polymerase delta specifically facilitate copying of tetraplex and hairpin structures of the d(CGG)n trinucleotide repeat sequence. J. Biol. Chem., 276, 16439–16446. [DOI] [PubMed] [Google Scholar]
- 48.Shamoo Y., Krueger,U., Rice,L.M., Williams,K.R. and Steitz,T.A. (1997) Crystal structure of the two RNA binding domains of human hnRNP A1 at 1.75 Å resolution. Nature Struct. Biol., 4, 215–222. [DOI] [PubMed] [Google Scholar]
- 49.Xu R.-M., Jokhan,L., Cheng,X., Mayeda,A. and Krainer,A.R. (1997) Crystal structure of human UP1, the domain of hnRNP A1 that contains two RNA-recognition motifs. Structure, 5, 559–570. [DOI] [PubMed] [Google Scholar]
- 50.McKay S.J. and Cooke,H. (1992) hnRNP A2/B1 binds specifically to single stranded vertebrate telomeric repeat TTAGGGn. Nucleic Acids Res., 20, 6461–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishikawa F., Matunis,M.J., Dreyfuss,G. and Cech,T.R. (1993) Nuclear proteins that bind the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol. Cell. Biol., 13, 4301–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eversole A. and Maizels,N. (2000) In vitro properties of the conserved mammalian protein hnRNP D suggest a role in telomere maintenance. Mol. Cell. Biol., 20, 5425–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kamma H., Fujimoto,M., Fujiwara,M., Matsui,M., Horiguchi,H., Hamasaki,M. and Satoh,H. (2001) Interaction of hnRNP A2/B1 isoforms with telomeric ssDNA and the in vitro function. Biochem. Biophys. Res. Commun., 280, 625–630. [DOI] [PubMed] [Google Scholar]
- 54.Denisenko O. and Bomsztyk,K. (2002) Yeast hnRNP K-like genes are involved in regulation of the telomeric position effect and telomere length. Mol. Cell. Biol., 22, 286–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding J., Hayashi,M.K., Zhang,Y., Krainer,A.R. and Xu,R.-M. (1999) Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev., 13, 1102–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schaeffer C., Bardoni,B., Mandel,J.L., Ehresmann,B., Ehresmann,C. and Moine,H. (2001) The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J., 20, 4803–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brown V., Jin,P., Ceman,S., Darnell,J.C., O’Donnell,W.T., Tenenbaum,S.A., Jin,X., Feng,Y., Wilkinson,K.D., Keene,J.D., Darnell,R.B. and Warren,S.T. (2001) Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell, 107, 477–487. [DOI] [PubMed] [Google Scholar]
- 58.Darnell J.C., Jensen,K.B., Jin,P., Brown,V., Warren,S.T. and Darnell,R.B. (2001) Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell, 107, 489–499. [DOI] [PubMed] [Google Scholar]