Abstract
The Dnmt3L protein belongs to the Dnmt3 family of DNA methyltransferases by virtue of its sequence homology in the plant homeodomain (PHD)-like motif. Dnmt3L is essential for the establishment of maternal genomic imprints and, given its lack of key methyltransferase motifs, is more likely to act as a regulator of methylation rather than as an enzyme that methylates DNA. Here, we show that Dnmt3L, like Dnmt3a and Dnmt3b, interacts both in vitro and in vivo with the histone deacetylase HDAC1. Consistent with the binding to a deacetylase, Dnmt3L purifies histone deacetylase activity from nuclear extracts. We find that Dnmt3L can repress transcription and that this repression is dependent on HDAC1 and is relieved by treatment with the HDAC inhibitor trichostatin A. Binding of Dnmt3L to HDAC1 as well as its repressive function require the PHD-like motif. Our results indicate that Dnmt3L plays a role in transcriptional regulation and that recruitment of the HDAC repressive machinery is a shared and conserved feature of the Dnmt3 family. The fact that, despite the absence of a methyltransferase domain, Dnmt3L retains the capacity to contact deacetylase further substantiates the notion that the Dnmts can repress transcription independently of their methylating activities.
INTRODUCTION
Methylation of cytosines within the CpG dinucleotide is essential for mammalian development and is associated with gene silencing (1,2). Transcriptional repression by DNA methylation plays a key role in several biological processes such as X-chromosome inactivation, genomic imprinting, the suppression of parasitic DNA sequences and tissue-specific gene expression (3–5). It is also becoming increasingly clear that alterations in DNA methylation contribute to tumorigenesis (6).
Methylation of CpG sites is established by the DNA methyltransferases—the Dnmts. The first isolated DNA methyltransferase, Dnmt1, has a high affinity for hemimethylated DNA substrates and is targeted to replication foci (7,8). Given these features, the primary function of Dnmt1 is thought to be its ability to function as a ‘maintenance’ methyltransferase, restoring the pre-existing methylation patterns during DNA replication. It is possible, however, that Dnmt1 also possesses de novo methyltransferase functions (9). Disruption of Dnmt1 in mice results in embryonic lethality (10). Besides Dnmt1, another set of DNA methyltransferases has recently been discovered, namely Dnmt3a and Dnmt3b, which are the founders of the Dnmt3 family of methyltransferases (11). As with Dnmt1, they can be divided at the sequence level into two domains, a C-terminal catalytic domain responsible for the methylation of DNA, and an N-terminal regulatory domain. The N-terminal non-catalytic portions of Dnmt3a and Dnmt3b show no strong sequence similarity to each other apart from a cysteine-rich motif, termed the plant homeodomain (PHD), present in many chromatin-associated proteins (12). In particular, the cysteine-rich region of Dnmt3a and Dnmt3b is most closely related to the imperfect PHD, or PHD-like, motif found in ATRX, a putative ATPase of the SNF2 family (13). Mice that lack Dnmt3a or Dnmt3b have been generated and reveal that these proteins are essential for de novo methylation and mammalian development (14). From this study as well as others (15), Dnmt3a and Dnmt3b have clearly been established as de novo methyltransferases. However, the possibility that they also act as ‘maintenance’ methyltransferases cannot be excluded (16).
Recent advances have begun to shed light on the mechanisms by which the Dnmts repress transcription. They can recruit the histone deacetylase (HDAC) repressive machinery, which removes acetyl groups from histones resulting in gene silencing (17–21). The N-terminal non-catalytic portion of the Dnmts mediates the association with HDAC and, intriguingly, the Dnmt enzymatic activity was found to be dispensable for transcriptional silencing (18,21). These observations suggest the attractive possibility that the Dnmts may have additional roles in the cell beyond their ability to methylate CpG dinucleotides. Recently, Dnmt3L, a novel isolated gene, has been placed in the Dnmt3 family (22) by virtue of its strong sequence similarity in the PHD-like fingers (see Fig. 1A). In contrast to the other Dnmts, Dnmt3L lacks most of the C-terminal catalytic domain and is therefore almost certainly devoid of intrinsic DNA methyltransferase activity. The inactivation of Dnmt3L in mice has been reported recently and it was found that Dnmt3L is required for the establishment of maternal methylation imprints during oogenesis (23). Given the lack of methyltransferase domain in Dnmt3L, this study suggests that Dnmt3L is likely to act as a regulator of imprint establishment rather than as an enzyme that methylates DNA.
Figure 1.
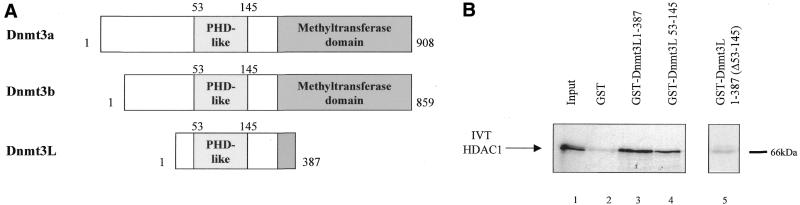
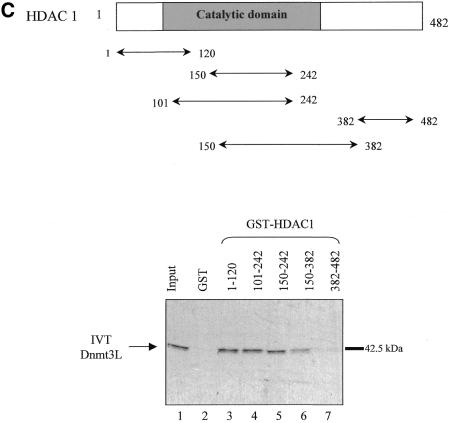
. Dnmt3L interacts with HDAC1 in vitro. (A) Schematic representation of the Dnmt3 family members. The conserved N-terminal cysteine-rich region, the PHD-like motif, characteristic of the Dnmt3 family is shown. The C-terminal domain is the catalytic methyltransferase domain present in Dnmt3a and Dnmt3b whereas the key catalytic motifs are absent in Dnmt3L. Numbers indicate amino acid residues. (B) Dnmt3L binds the histone deacetylase HDAC1 in GST pull-down experiments. Full-length HDAC1 was IVT and 35S-radiolabeled, incubated with equivalent amounts of GST (lane 2), GST fusion proteins of Dnmt3L 1–387 (full-length; lane 3), residues 53–145 (containing the PHD-like motif; lane 4) or Dnmt3L 1–387 Δ53–145 (a mutated version that lacks the PHD-like motif; lane 5). GST 1–387 Δ53–145 (lane 5) was tested in a separate experiment than the other constructs (lanes 1–4). Molecular weight in kDa is indicated on the right. The bound IVT HDAC1 is indicated by an arrow on the left. Lane 1, 35S-radiolabeled HDAC1 input (10%). (C) Dnmt3L binds specific regions of HDAC1 in vitro. The upper panel is a schematic representation of the HDAC1 deacetylase with its catalytic domain depicted by a grey box. The indicated GST–HDAC1 fusions were tested in GST pull-down experiments using IVT full-length Dnmt3L (lower panel, lanes 3–7). Lane 1, 35S-radiolabeled Dnmt3L input (10%).
In the present work, we have addressed the mechanisms by which Dnmt3L functions. Given that the PHD-like motif of Dnmt3a and Dnmt3b mediates the association with HDAC (20,21) and that this motif is conserved in Dnmt3L, we examined whether Dnmt3L can function as a transcriptional repressor through the recruitment of histone deacetylase. We now show, both in vitro and in vivo, that Dnmt3L binds the histone deacetylase HDAC1 and associates with histone deacetylase activity. We find that Dnmt3L can repress transcription and that this repression is dependent on HDAC1 and is sensitive to trichostatin A (TSA) treatment. Finally, we show that the association of Dnmt3L with HDAC1 and its repressive activity occur through the PHD-like motif. These data indicate that transcriptional silencing via the HDAC machinery is a shared and conserved feature within the Dnmt3 family. Given the lack of methyltransferase domain in Dnmt3L, this work also further supports the notion that the Dnmts possess functions other than the ability to methylate DNA.
MATERIALS AND METHODS
Constructs
For the GAL4 fusion experiments, we cloned full-length Dnmt3L (residues 1–387) or the PHD-like motif (residues 53–145), or Dnmt3L lacking the PHD-like motif (residues 1–387 Δ53–145) into the pcDNA3.1 GAL4 vector using PCR. pcDNA3.1GAL4 has the GAL4 DNA-binding domain (DBD) (residues 1–147) under the control of a CMV promoter. The pGEX plasmids with various domains of HDAC1 were described previously (21). We cloned the PHD-like motif (residues 53–145) of Dnmt3L or the full-length (residues 1–387) or PHD-like domain-deleted Dnmt3L (residues 1–387 Δ53–145) into the pGEX4T1 vector (Pharmacia) by PCR using appropriate sets of primers. pcDNA3HDAC1-F, pING 14A-HDAC1, pGEX-Rb (379–928) and the reporter construct 4XGAL4-TK-Luc have been described previously (24,25). We verified all constructs by DNA sequencing. pcDNA3.1Dnmt3L was a kind gift of U. Aapola (Tampere, Finland).
Glutathione S-transferase (GST) fusion proteins, in vitro translations and pull-down assays
We expressed GST and GST fusion proteins in Escherichia coli TOP 10F (Invitrogen) and proteins from crude bacterial lysates were purified using glutathione–Sepharose 4B (Pharmacia) according to the manufacturer’s instructions. In vitro transcription/translation was performed using the TNT system (Promega). GST pull-down experiments were performed essentially as described previously (26).
Cell culture, transfections and luciferase assays
Monolayer cultures of U2OS and 293 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum at 37°C in 5% CO2 atmosphere.
For transfection assays, 24 h after plating, the exponentially growing cells were transfected with OptiMEM (Life Technologies) and polyethylene imine (Euromedex). All transfections were carried out with the same total amount of DNA (1 µg). Four hours after transfection, the cells were washed once in phosphate buffered saline and recovered with fresh medium. The cells were incubated for an additional 24 h, in either the presence or the absence of TSA (200 nM; Waco Bioproducts). After 24 h of incubation, the cells were lysed in 200 µl of hypotonic IPH buffer (27). Supernatants were clarified by centrifugation. Luciferase assays were performed with the Promega Luciferase Assay System according to the manufacturer’s instructions. Transfection efficiencies were normalized using a cotransfected β-galactosidase plasmid.
Immunoprecipitations and western blot analysis
293 cells were transiently transfected in culture dishes (10 cm diameter) with 3 µg of each expression vector. Cells were harvested 24 h post-transfection, lysed in 300 µl of IPH lysis buffer (27) at 4°C for 30 min and debris removed by centrifugation. Immunoprecipitations and western blotting were then carried out as described previously (17). Anti-GAL4 (5C1; Santa Cruz) and anti-Flag (M2; Sigma) antibodies were used.
GST pull-down and immunoprecipitation of histone deacetylase activity from cell extracts
Equivalent amounts of GST and GST fusion proteins bound to glutathione–Sepharose beads were added to 50 µl of HeLa nuclear extracts (Computer Cell Culture Center, Belgium) in 300 µl of IPH buffer and incubated at 4°C for 2 h. The beads were washed four times in IPH buffer and assayed for histone deacetylase activity. We performed HDAC assays essentially as described previously (24,28) in a volume of 100 µl of IPH buffer containing 250 000 c.p.m. of tritium-labeled acetylated H4 peptide. For HDAC assays from transfected cells, 293 were transiently transfected as described above with an equal amount of expression plasmids (14 µg for a plate of 14 cm diameter). Cell extracts were immmunoprecipitated with 2 µg of anti-GAL4 antibody (5C1; Santa Cruz) and incubated with 50 µl of HeLa nuclear extracts. After six washes with IPH buffer, the immune complexes were tested for histone deacetylase activity.
RESULTS
Dnmt3L interacts with the histone deacetylase HDAC1 using its PHD-like motif
The de novo methyltransferases Dnmt3a and Dnmt3b have recently been reported to associate with the histone deacetylase HDAC1 using their conserved PHD-like motif (21). Given that Dnmt3L shows strong homology with the PHD-like motif of Dnmt3a and Dnmt3b (Fig. 1A), we set out to establish whether Dnmt3L associates with HDAC1. To test this possibility, we first performed in vitro GST pull-down assays using full-length Dnmt3L (residues 1–387) fused to GST and incubated with in vitro translated (IVT) 35S-labeled HDAC1. As shown in Figure 1B, GST-Dnmt3L 1–387 pulled down radiolabeled HDAC1 (lane 3) whereas GST alone did not (lane 2). As depicted in Figure 1B (lane 5), a mutated version of full-length Dnmt3L in which its PHD-like motif was deleted (GST-Dnmt3L 1–387 Δ53–145) failed to bind with HDAC1. Consistent with this observation, a GST Dnmt3L fusion that contains only the PHD-like motif (GST-Dnmt3L 53–145) pulled down HDAC1 (Fig. 1B, lane 4). Thus, the conserved PHD-like motif of Dnmt3L (residues 53–145) is required for the association with radiolabeled HDAC1.
We next wished to determine which part of HDAC1 mediated the observed in vitro association with Dnmt3L. To this end, various fragments of HDAC1 were fused to GST (Fig. 1C) and incubated with IVT full-length Dnmt3L. Figure 1C shows that two non-overlapping N-terminal regions of HDAC1 (residues 1–120 and 150–242) mediate the binding with Dnmt3L.
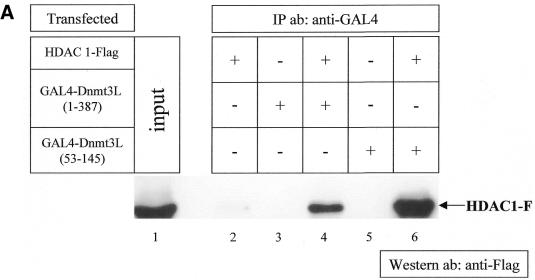
To further validate the interaction between Dnmt3L and HDAC1, we performed co-immunoprecipitation analyses from mammalian 293 cells co-transfected with GAL4-tagged full-length Dnmt3L (GAL4-Dnmt3L 1–387) and Flag-tagged full-length HDAC1 (HDAC1-F). The lysed transfected cells were immunoprecipitated with anti-GAL4 antibody and western blotted with anti-Flag antibody. Figure 2A shows that Dnmt3L interacted with HDAC1 (lane 4) whereas no precipitate was detected after transfection of either HDAC1-F or GAL4-Dnmt3L 1–387 alone (lanes 2 and 3, respectively). Similar co-immunoprecipitation experiments were performed using GAL4-tagged Dnmt3L 53–145, which contains the PHD-like motif, and Flag-tagged HDAC1. As shown in Figure 2A (lane 6), Dnmt3L 53–145 co-immunoprecipitated specifically with HDAC1-F. This is consistent with the finding presented in Figure 1B showing that residues 53–145 mediate the interaction with IVT HDAC1. Taken together, these data indicate that Dnmt3L specifically interacts with HDAC1 both in vitro and in vivo, and that this interaction requires the conserved PHD-like motif of Dnmt3L.
Figure 2.
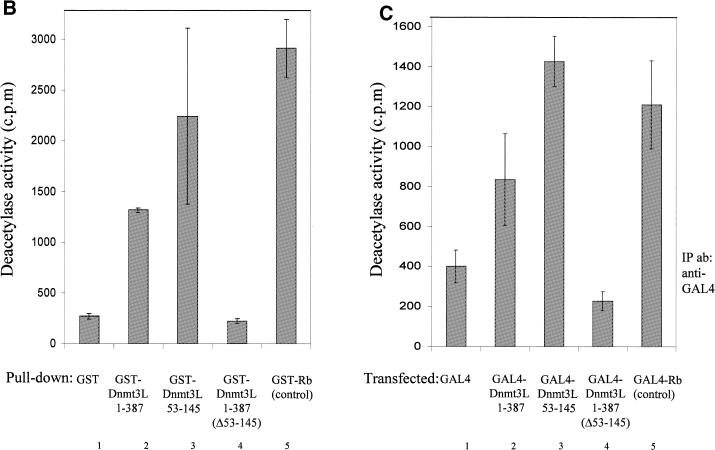
. Dnmt3L co-immunoprecipitates with HDAC1 and purifies HDAC activity in vitro and in vivo using its PHD-like finger. (A) 293 cells were transiently transfected as indicated (+) with 3 µg of either pcDNA3GAL4 Dnmt3L 1–387 or pcDNA3GAL4 Dnmt3L 53–145 (encompassing the PHD-like motif), together with 3 µg of pcDNA HDAC1-F (expressing Flag-tagged HDAC1). Whole cell extracts were then precipitated with anti-GAL4 antibody (5C1) and the presence of HDAC1-F in the immunoprecipitates was visualized by western blot analysis using anti-Flag antibody (M2). HDAC1-F is indicated by an arrow on the right. (B) Dnmt3L associates with deacetylase activity in vitro. Equivalent amounts of GST (lane 1) or GST fusion proteins (lanes 2–5) were incubated with HeLa nuclear extracts. After washing, the complexes were tested for histone deacetylase activity. Histone deacetylase activity is given as radioactivity (c.p.m.) released from an 3H-labeled acetylated histone H4 peptide. GST–Rb fusion protein (lane 5) was used as a positive control for the assay. The results shown are the average of at least two independent experiments with error bars displaying standard deviations. (C) Dnmt3L purifies deacetylase activity from transfected cells. Cells were transiently transfected with equal amounts of the indicated GAL4-tagged constructs. Whole cell extracts were immunoprecipited with anti-GAL4 antibody. After washing, the immune complexes were tested for histone deacetylase activity. Histone deacetylase activity is given as radioactivity (c.p.m.) released from an 3H-labeled acetylated histone H4 peptide. We used Gal4–Rb fusion protein as a positive control for the assay.
Dnmt3L associates with HDAC activity
The association of Dnmt3L with HDAC1 led us to expect that Dnmt3L would be associated with histone deacetylase activity. To test this, we determined whether a full-length GST fusion protein of Dnmt3L (residues 1–387) could purify HDAC activity from HeLa nuclear extracts. Histone deacetylase activity was assayed by liquid scintillation counting and measured as [3H]-acetate released from an acetylated H4 peptide. As shown in Figure 2B (lane 2), GST-Dnmt3L 1–387 precipitates deacetylase activity from HeLa nuclear extracts, whereas GST alone gave only background activity (lane 1). We used GST fused to the retinoblastoma protein (Rb), a protein known to associate with deacetylase activity (24,29,30), as a positive control (Fig. 2B, lane 5). As the PHD-like domain of Dnmt3L mediates the interaction with HDAC1 (Figs 1 and 2A), we next asked whether this domain was required for the association with HDAC activity. Figure 2B (lane 4) indicates that GST-Dnmt3L lacking the PHD-like motif (GST-Dnmt3L 1–387 Δ53–145) resulted in only background activity. In agreement with the requirement of the PHD-like finger for associated HDAC activity, GST-Dnmt3L 53–145 purified significant histone deacetylase activity from HeLa nuclear extracts (Fig. 2B, lane 3).
The association of Dnmt3L with deacetylase activity could also be observed in vivo. We transiently transfected GAL4-Dnmt3L 1–387 in 293 cells, followed by immunoprecipitation with anti-GAL4 antibody. Immunocomplexes were assayed for histone deacetylase activity. As shown in Figure 2C (lane 2), GAL4-Dnmt3L 1–387 immunocomplexes contained significant deacetylase activity compared with precipitates obtained from a control transfection with GAL4 alone (lane 1). GAL4 Rb, which was used as a positive control for the assay, precipitated HDAC activity (Fig. 2C, lane 5). Consistent with the finding that the PHD-like motif of Dnmt3L is required in vitro to associate with deacetylase activity (Fig. 2C), we find that exogenous expression of GAL4-Dnmt3L 53–145 precipitated significant amounts of histone deacetylase activity (Fig. 2C, lane 3), whereas GAL4-Dnmt3L 1–387 Δ53–145 (that is deleted of the PHD-like motif) gave only background activity (Fig. 2C, lane 4).
Dnmt3L actively represses transcription through the PHD-like motif
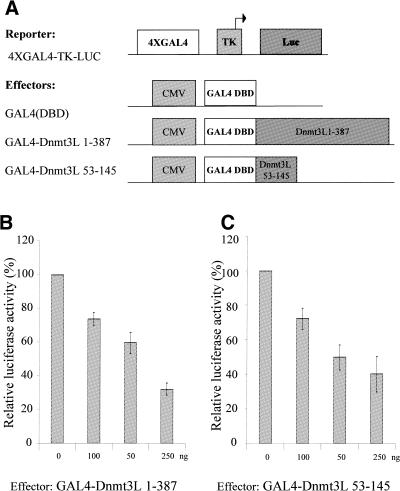
Dnmt3a and Dnmt3b have been shown to have repressive capacity using their PHD-like domain (20,21). Since the conserved PHD-like motif is present in Dnmt3L, we wished to determine whether Dnmt3L may have repressive potential and, if so, whether its PHD-like domain is responsible for the repression. To this end, we used GAL4-Dnmt3L 1–387, which contains full-length Dnmt3L fused to GAL4 DBD 1–147. Repressor activity was evaluated by transiently transfecting increasing amounts of the GAL4 fusion construct into U2OS osteosarcoma cells together with a reporter plasmid containing GAL4-binding sites upstream of the viral thymidine kinase promoter driving expression of a luciferase reporter gene (4XGAL4-TK-Luc; Fig. 3A). Fold repression was determined relative to the basal transcriptional activity of the reporter in the presence of GAL4(DBD) alone. Figure 4C shows that GAL4-Dnmt3L 1–387 can repress this promoter in a dose-dependent manner. This repression was specific, as a reporter lacking the GAL4 sites was not affected by GAL4-Dnmt3L 1–387 (data not shown). To address whether the PHD-like domain of Dnmt3L is responsible for the observed repression, we co-transfected the high basal reporter plasmid 4XGAL4-TK-Luc along with GAL4-Dnmt3L 53–145 containing the PHD-like motif. As presented in Figure 3C, GAL4-Dnmt3L 53–145 inhibited luciferase activity in a dose-dependent manner.
Figure 3.
. Dnmt3L represses transcription through its PHD-like motif when fused to the GAL4 DBD. (A) Schematic representation of the reporter and the effector constructs used. The reporter construct 4XGAL4-TK-Luc contains four copies of GAL4 sites upstream of the thymidine kinase promoter driving expression of the luciferase gene. The effectors used are also indicated and consist of GAL4 DBD alone [GAL4(DBD)], GAL4 DBD fused to either full-length Dnmt3L (residues 1–387) or Dnmt3L PHD (residues 53–145). (B and C) U2OS cells were transiently transfected with 250 ng of 4XGAL4-TK-luc reporter with increasing amounts (50–250 ng) of GAL4-Dnmt3L 1–387 (B) or GAL4-Dnmt3L 53–145 (C). Whole cell extracts were used in luciferase assays. The basal activity of the reporter is normalized to a value of 100%. Transfection efficiencies were normalized using β-galactosidase activity. The results are the average of at least four independent transfections done in duplicate with error bars displaying standard deviations.
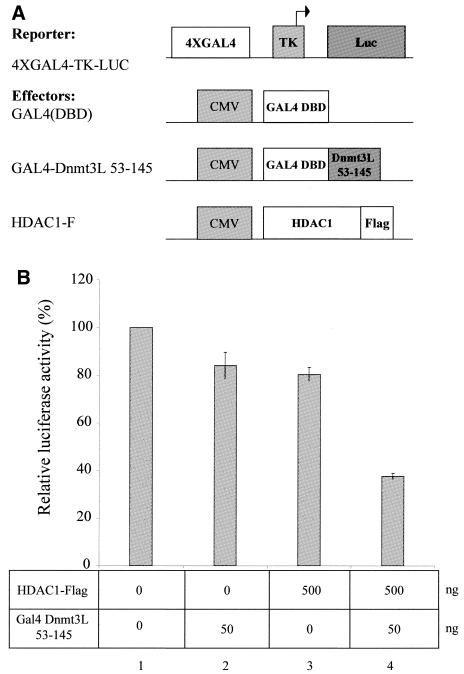
Figure 4.
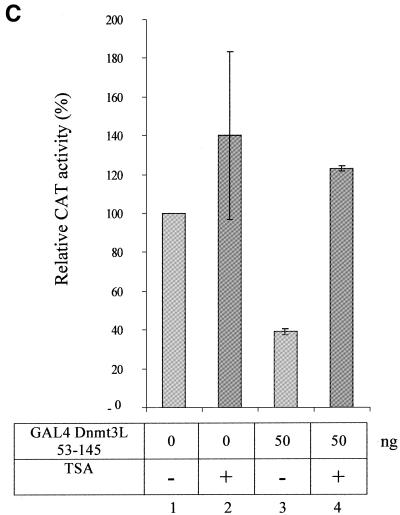
. Dnmt3L requires the deacetylase HDAC1 for transcriptional silencing and represses transcription in a TSA-sensitive manner. (A) Schematic representation of the reporter and the effector constructs used. The reporter construct 4XGAL4-TK-Luc contains four copies of GAL4 sites upstream of the thymidine kinase promoter driving expression of the luciferase gene. Also indicated are effector constructs expressing only the GAL4 DBD [GAL4(DBD)], the GAL4 DBD fused to the PHD of Dnmt3L (residues 53–145) and Flag-tagged full-length HDAC1 (HDAC1-F). (B) U2OS cells were transiently transfected with 250 ng of the 4XGAL4-TK-luc reporter and, as indicated, with 50 ng of GAL4 Dnmt3L 53–145 and/or 500 ng of HDAC1-F. Cells were then harvested and assayed for luciferase. The basal activity of the reporter is normalised to a value of 100%. Transfection efficiencies were normalized using β-galactosidase activity. The results shown are the average of at least two independent experiments with error bars displaying standard deviations. (C) Dnmt3L-mediated repression is sensitive to TSA. U2OS cells were transfected with 250 ng of the 5XGAL4-SV40-CAT reporter and 50 ng of GAL4 Dnmt3L 53–145. Sixteen hours after transfection, cells were treated (lanes 3 and 4) or not (lanes 1 and 2) with the HDAC inhibitor TSA (200 mM). The basal activity of the reporter is normalised to a value of 100%. Transfection efficiencies were normalized using β-galactosidase activity. The results shown are the average of at least two independent experiments with error bars displaying standard deviations.
Dnmt3L-mediated repression is sensitive to TSA and it co-represses transcription together with HDAC1
Having shown that Dnmt3L can silence transcription using a minimal transferable repressor domain, we next wished to investigate the molecular mechanism governing this repression. HDAC activity has been shown to repress transcription (31). Since Dnmt3L associates with histone deacetylase activity and binds HDAC1 via its PHD-like motif (see above), we evaluated whether Dnmt3L 53–145 and HDAC1 may act together to silence transcription. To this end, we performed transient transfections in 293 cells using the reporter plasmid 4XGAL4-TK-Luc. As illustrated in Figure 4B, expression of a limiting amount of GAL4-Dnmt3L 53–145 (lane 2) or HDAC1 (lane 3) only slightly repressed transcription. In contrast, co-transfection of GAL4-Dnmt3L 53–145 along with HDAC1 provided a synergistic repressive effect on transcription (Fig. 4B, lane 4).
We next asked whether the repression mediated by GAL4-Dnmt3L could be relieved by treatment with the specific HDAC inhibitor TSA. As depicted in Figure 4C, the repressive effect observed with GAL4-Dnmt3L 53–145 on reporter activity was relieved by the addition of TSA. Collectively, these data indicate that Dnmt3L contains a transcriptional repressor domain, encompassing the PHD-like motif, and requires the histone deacetylase HDAC1 to exert this repressive effect.
DISCUSSION
Here, we report the first biochemical analysis of Dnmt3L, a third member of the Dnmt3 family. We show that Dnmt3L functions as a transcriptional repressor through its ability to associate with the histone deacetylase HDAC1. We find that Dnmt3L interacts both in vitro and in vivo with HDAC1 and, consistent with this, it associates with histone deacetylase activity. In addition, we demonstrate that Dnmt3L actively represses transcription and that this repression requires the recruitment of the histone deacetylase HDAC1 and is sensitive to the HDAC inhibitor TSA. We and others have recently reported a close connection between histone deacetylation and the two Dnmt3 family members Dnmt3a and Dnmt3b (20,21). Thus, the present study reveals that transcriptional repression through recruitment of histone deacetylation is a conserved and shared feature within the Dnmt3 family.
We find that the link between Dnmt3L and deacetylation is mediated by its conserved PHD-like motif. This cysteine-rich region shows high sequence similarity among all three Dnmt3 family members. In Dnmt3a and Dnmt3b, the PHD-like motif also functions as a transcriptional repression domain by recruiting histone deacetylase (20,21). Thus, together these data suggest that, within the Dnmt3 family, the conservation of the PHD-like fingers at the sequence level reflects a conservation at the functional level. It is worth noting that the PHD-like domain found in the Dnmt3 members most closely resembles a domain found in the ATRX gene that codes for a putative ATP-dependent chromatin-remodeling factor of the SNF2 family (13). Mutations in the ATRX gene are the cause of ATRX syndrome, an α-thalassemia X-linked mental retardation disease (32). It is interesting to note that the majority of mutations associated with ATRX syndrome are found in its PHD-like fingers (32), thus highlighting the biological significance of this domain.
As mentioned above, Dnmt3a and Dnmt3b recruit HDAC to silence gene expression through their PHD-like motif (20,21). Recent studies have demonstrated that Dnmt1 also binds to histone deacetylase through its non-catalytic N-terminal portion (17–19). Intriguingly, in both instances, the methyltransferase activity of the Dnmts seems to be dispensable for gene silencing. From these observations, a new concept is beginning to emerge concerning the functional roles of the Dnmts in that they are multi-faceted proteins with additional functions other than their capacity to methylate CpG dinucleotides. Dnmt3L lacks the key catalytic motifs characteristic of DNA methyltransferases and is not likely to function as an enzyme that methylates CpG sites. Dnmt3L, however, retains the PHD-like motif characteristic of the Dnmt3L family. Our finding that Dnmt3L can still recruit the HDAC repressive machinery, despite its lack of a methyltransferase domain, further supports the observation that in some instances the HDAC-associated functions of the Dnmts may be working independently of their methyltransferase activity.
Recently, Dnmt3L knock-out mice have been generated by two groups (23,33). They both found that Dnmt3L+/– embryos derived from homozygous mutant oocytes show a lack of maternal genomic imprints, causing genes that are normally methylated and maternally repressed to lack methylation marks and to be expressed on both alleles. This indicates that Dnmt3L is required for the establishment of maternal methylation imprints during oogenesis. As stated above, Dnmt3L is most certainly not acting as a DNA methyltransferase and it is therefore likely that it functions as a regulator of methylation at imprinted loci. Interestingly, Hata et al. (33) have recently shown that Dnmt3L interacts with the DNA methyltransferase Dnmt3a as well as Dnmt3b, and that the absence of Dnmt3a, like the lack of Dnmt3L, results in the loss of maternal imprints in mice. Thus, Dnmt3L may act as a co-factor for the Dnmt3a and Dnmt3b methyltransferases. Our finding that Dnmt3L is associated with HDAC activity could also be relevant to its role as a regulator of maternal imprints. It is indeed interesting to note that several imprinted genes that show loss of maternal-specific methylation imposed by Dnmt3L deficiency, such as Snrpn, have been reported to be normally regulated by histone deacetylation (34). The methylated and non-expressed maternal allele of the imprinted Snrpn gene is found to be underacetylated relative to the unmethylated paternal allele (34) and treatment of cells with the HDAC inhibitor TSA results in transcriptional reactivation of the silent maternal allele (35). Thus, it is tempting to speculate that the HDAC-associated function of Dnmt3L may contribute to its role as a regulator of methylation at imprinted genes. This would imply that histone deacetylation can influence DNA methylation patterns. This notion is supported by the observation in the fungus Neurospora crassa that HDAC inhibition by TSA can induce DNA demethylation (36). Taken together, a model could be envisaged in which Dnmt3L would not only act as a co-factor for the Dnmt3a and Dnmt3b methyltransferases but would also recruit HDAC activity to maternally imprinted genes. The methyltransferase and deacetylase activities recruited by Dnmt3L would then act together to potentiate the repressed state. The targeting of the Dnmt3L-containing enzymatic activities to the relevant loci is unlikely to occur through direct binding to DNA, as Dnmt3L does not contain any obvious DNA-binding domain. It is more likely that, as was shown for Dnmt1 and Dnmt3a (21,37), Dnmt3L is recruited to DNA by an as yet unidentified DNA-binding repressor.
In conclusion, our results further emphazise the close connection that exists between the Dnmts and histone deacetylation. All Dnmts identified so far have now been shown to silence gene expression by contacting histone deacetylases. The challenge for the future is to understand how a cell establishes and maintains a transcriptional repressive state through the use of multiple HDAC activities associated with the Dnmts.
Acknowledgments
ACKNOWLEDGEMENTS
We thank U. Aapola for the pcDNA3.1 Dnmt3L construct and K. Calame for the 4XGAL4-TK-luc reporter plasmid. R.D. was supported by the F.R.I.A., C.B. was funded by Télévie, W.A.B. was supported by the South African National Research Foundation (NRF) and F.F. was funded by the Fonds National de la Recherche Scientifique (‘Chargé de recherches du F.N.R.S.’). This work was funded by grants from the Association Belge contre le Cancer and F.N.R.S. and a grant from the ‘Action de Recherche Concertée de la Communauté Française de Belgique’.
REFERENCES
- 1.Jones P.A. and Takai,D. (2001) Cancer epigenetics comes of age. Science, 293, 1068–1070. [DOI] [PubMed] [Google Scholar]
- 2.Bird A. (2002) DNA methylation patterns and epigenetic memory. Genes Dev., 16, 6–21. [DOI] [PubMed] [Google Scholar]
- 3.Jaenisch R. (1997) DNA methylation and imprinting: why bother? Trends Genet., 13, 323–329. [DOI] [PubMed] [Google Scholar]
- 4.Surani M.A. (2001) Reprogramming of genome function through epigenetic inheritance. Nature, 414, 122–128. [DOI] [PubMed] [Google Scholar]
- 5.Bestor T.H. (2000) The DNA methyltransferases of mammals. Hum. Mol. Genet., 9, 2395–2402. [DOI] [PubMed] [Google Scholar]
- 6.Jones P.A. and Laird,P.W. (1999) Cancer epigenetics comes of age. Nature Genet., 21, 163–167. [DOI] [PubMed] [Google Scholar]
- 7.Leonhardt H., Page,A.W., Weier,H.U. and Bestor,T.H. (1992) A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell, 71, 865–873. [DOI] [PubMed] [Google Scholar]
- 8.Chuang L.S., Ian,H.I., Koh,T.W., Ng,H.H., Xu,G. and Li,B.F. (1997) Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science, 277, 1996–2000. [DOI] [PubMed] [Google Scholar]
- 9.Vertino P.M., Yen,R.W., Gao,J. and Baylin,S.B. (1996) De novo methylation of CpG island sequences in human fibroblasts overexpressing DNA (cytosine-5-)-methyltransferase. Mol. Cell. Biol., 16, 4555–4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li E., Bestor,T.H. and Jaenisch,R. (1992) Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell, 69, 915–926. [DOI] [PubMed] [Google Scholar]
- 11.Okano M., Xie,S. and Li,E. (1998) Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nature Genet., 19, 219–220. [DOI] [PubMed] [Google Scholar]
- 12.Aasland R., Gibson,T.J. and Stewart,A.F. (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci., 20, 56–59. [DOI] [PubMed] [Google Scholar]
- 13.Gibbons R.J., Bachoo,S., Picketts,D.J., Aftimos,S., Asenbauer,B., Bergoffen,J., Berry,S.A., Dahl,N., Fryer,A., Keppler,K., Kurosawa,K., Levin,M.L., Masuno,M., Neri,G., Pierpont,M.E., Slaney,S.F. and Higgs,D.R. (1997) Mutations in transcriptional regulator ATRX establish the functional significance of a PHD-like domain. Nature Genet., 17, 146–148. [DOI] [PubMed] [Google Scholar]
- 14.Okano M., Bell,D.W., Haber,D.A. and Li,E. (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell, 99, 247–257. [DOI] [PubMed] [Google Scholar]
- 15.Hsieh C.L. (1999) In vivo activity of murine de novo methyltransferases, Dnmt3a and Dnmt3b. Mol. Cell. Biol., 19, 8211–8218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhee I., Jair,K.W., Yen,R.W., Lengauer,C., Herman,J.G., Kinzler,K.W., Vogelstein,B., Baylin,S.B. and Schuebel,K.E. (2000) CpG methylation is maintained in human cancer cells lacking DNMT1. Nature, 404, 1003–1007. [DOI] [PubMed] [Google Scholar]
- 17.Fuks F., Burgers,W.A., Brehm,A., Hughes-Davies,L. and Kouzarides,T. (2000) DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nature Genet., 24, 88–91. [DOI] [PubMed] [Google Scholar]
- 18.Robertson K.D., Ait-Si-Ali,S., Yokochi,T., Wade,P.A., Jones,P.L. and Wolffe,A.P. (2000) DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nature Genet., 25, 338–342. [DOI] [PubMed] [Google Scholar]
- 19.Rountree M.R., Bachman,K.E. and Baylin,S.B. (2000) DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nature Genet., 25, 269–277. [DOI] [PubMed] [Google Scholar]
- 20.Bachman K.E., Rountree,M.R. and Baylin,S.B. (2001) Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem., 276, 32282–32287. [DOI] [PubMed] [Google Scholar]
- 21.Fuks F., Burgers,W.A., Godin,N., Kasai,M. and Kouzarides,T. (2001) Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J., 20, 2536–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aapola U., Kawasaki,K., Scott,H.S., Ollila,J., Vihinen,M., Heino,M., Shintani,A., Minoshima,S., Krohn,K., Antonarakis,S.E., Shimizu,N., Kudoh,J. and Peterson,P. (2000) Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics, 65, 293–298. [DOI] [PubMed] [Google Scholar]
- 23.Bourc’his D., Xu,G.L., Lin,C.S., Bollman,B. and Bestor,T.H. (2001) Dnmt3L and the establishment of maternal genomic imprints. Science, 294, 2536–2539. [DOI] [PubMed] [Google Scholar]
- 24.Brehm A., Miska,E.A., McCance,D.J., Reid,J.L., Bannister,A.J. and Kouzarides,T. (1998) Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature, 391, 597–601. [DOI] [PubMed] [Google Scholar]
- 25.Yu J., Angelin-Duclos,C., Greenwood,J., Liao,J. and Calame,K. (2000) Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol. Cell. Biol., 20, 2592–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bannister A.J. and Kouzarides,T. (1995) CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J., 14, 4758–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bannister A.J. and Kouzarides,T. (1996) The CBP co-activator is a histone acetyltransferase. Nature, 384, 641–643. [DOI] [PubMed] [Google Scholar]
- 28.Taunton J., Hassig,C.A. and Schreiber,S.L. (1996) A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science, 272, 408–411. [DOI] [PubMed] [Google Scholar]
- 29.Magnaghi-Jaulin L., Groisman,R., Naguibneva,I., Robin,P., Lorain,S., Le Villain,J.P., Troalen,F., Trouche,D. and Harel-Bellan,A. (1998) Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature, 391, 601–605. [DOI] [PubMed] [Google Scholar]
- 30.Luo R.X., Postigo,A.A. and Dean,D.C. (1998) Rb interacts with histone deacetylase to repress transcription. Cell, 92, 463–473. [DOI] [PubMed] [Google Scholar]
- 31.Kouzarides T. (1999) Histone acetylases and deacetylases in cell proliferation. Curr. Opin. Genet. Dev., 9, 40–48. [DOI] [PubMed] [Google Scholar]
- 32.Gibbons R.J., Picketts,D.J., Villard,L. and Higgs,D.R. (1995) Mutations in a putative global transcriptional regulator cause X-linked mental retardation with α-thalassemia (A-TRX-syndrome). Cell, 80, 837–845. [DOI] [PubMed] [Google Scholar]
- 33.Hata K., Okano,M., Lei,H. and Li,E. (2002) Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development, 129, 1983–1993. [DOI] [PubMed] [Google Scholar]
- 34.Gregory R.I., Randall,T.E., Johnson,C.A., Khosla,S., Hatada,I., O’Neill,L.P., Turner,B.M. and Feil,R. (2001) DNA methylation is linked to deacetylation of histone H3, but not H4, on the imprinted genes Snrpn and U2af1-rs1. Mol. Cell. Biol., 21, 5426–5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshioka H., Shirayoshi,Y. and Oshimura,M. (2001) A novel in vitro system for analyzing parental allele-specific histone acetylation in genomic imprinting. J. Hum. Genet., 46, 626–632. [DOI] [PubMed] [Google Scholar]
- 36.Selker E.U. (1998) Trichostatin A causes selective loss of DNA methylation in Neurospora. Proc. Natl Acad. Sci. USA, 95, 9430–9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Croce L., Raker,V.A., Corsaro,M., Fazi,F., Fanelli,M., Faretta,M., Fuks,F., Lo Coco,F., Kouzarides,T., Nervi,C., Minucci,S. and Pelicci,P.G. (2002) Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science, 295, 1079–1082. [DOI] [PubMed] [Google Scholar]