Abstract
The real-time quantitative polymerase chain reaction (rtqPCR) has overcome the limitations of conventional, time-consuming quantitative PCR strategies and is maturing into a routine tool to quantify gene expression levels, following reverse transcription (RT) of mRNA into complementary DNA (cDNA). Expression profiling with single-cell resolution is highly desirable, in particular for complex tissues like the brain that contain a large variety of different cell types in close proximity. The patch-clamp technique allows selective harvesting of single-cell cytoplasm after recording of cellular activity. However, components of the cDNA reaction, in particular the reverse transcriptase itself, significantly inhibit subsequent rtqPCR amplification. Using undiluted single-cell cDNA reaction mix directly as template for rtqPCR, I observed that the amplification kinetics of rtqPCRs were dramatically altered in a non-systematic fashion. Here, I describe a simple and robust precipitation protocol suitable for purification of single-cell cDNA that completely removes inhibitory RT components without detectable loss of cDNA. This improved single-cell real-time RT–PCR protocol provides a powerful tool to quantify differential gene expression of individual cells and thus could complement global microarray-based expression profiling strategies.
INTRODUCTION
Gene expression is regulated at the level of individual cells, with different cell types or different developmental stages of the same cell expressing distinct sets of genes. Thus, analysis of the gene expression pattern of individual cells is a desirable goal. Laser-based microdissection techniques enable the isolation of identified single cells from fixed tissue (1,2), while the patch-clamp method enables the harvesting of mRNAs from a single living cell (3,4). The latter technique is of particular importance as it permits correlation of the functional properties of individual cells with their specific gene expression profile (5). Reverse transcription of mRNA followed by polymerase chain reaction (RT–PCR) is a reliable method of detecting gene expression. To analyze mRNA expression from single cells without loss of low abundance mRNAs, it is common practice to use all of the total single-cell complementary DNA (cDNA) reaction mixture as template for subsequent PCR (3). It is of increasing importance to detect not only qualitative but also quantitative differences in gene expression levels, and several approaches have been used to obtain quantitative data from single-cell RT–PCR experiments. These include the addition of known amounts of competitor mRNA (6,7), serial dilution of single-cell cDNA pools (8), and fluorescence-based real-time quantitative PCR (rtqPCR) (9). The latter has the advantage that it is easy to perform, highly reproducible, and, importantly, has the sensitivity to amplify and quantify even a single DNA template molecule (10–14). Thus, rtqPCR has become the method of choice for quantitative analysis of gene expression levels (14–17). However, I and others have observed that components of the undiluted RT reaction mixture considerably distort the subsequent PCR amplification reaction, presumably by inhibiting Taq-polymerase (9,18,19). As DNA quantification via rtqPCR depends critically on the presence of an unperturbed exponential phase of PCR amplification (15,16) this inhibition restrains the use of rtqPCR for quantitative single-cell gene expression profiling. To date, RT–rtqPCR analysis of single cells has therefore been limited to either diluted (2,9), or PCR pre-amplified (20) single-cell cDNA pools. Both these strategies limit sensitivity. Here, I describe a simple single-cell cDNA precipitation protocol that overcomes these inhibition problems and thus enables the full application of rtqPCR in combination with established single-cell RT protocols (3–5,21,22).
MATERIALS AND METHODS
Generation of control cDNA and DNA standard
For tissue RT–PCR, RNA was prepared from the midbrains of three 13-day-old C57Bl/6J mice using the Micro-FastTrack™ Kit (Invitrogen). RT was performed with 500 ng of poly(A)+ RNA overnight at 37°C in a total reaction volume of 10 µl, containing random hexamer primers (5 µM; Roche), dithiothreitol (DTT, 10 mM; Gibco BRL), the four deoxyribonucleotide triphosphates (dNTPs, 0.5 mM each; Pharmacia), 20 U of ribonuclease inhibitor (Promega) and 100 U of reverse transcriptase (Superscript™II; Gibco BRL). For T4 gene 32 protein experiments, T4gp32 (ChimerX) was diluted to a concentration of 3.2 µg/µl with sterile water. Freshly diluted T4gp32 (1.6 µg) was added to the single-cell reaction mixture directly before the reverse transcriptase was added. Single-cell cDNA was kept at –70°C until PCR amplification. cDNA was purified, diluted and quantified using a BioPhotometer (Eppendorf). For the calibration curve, a tyrosine hydroxylase (TH) cDNA fragment (accession no. M69200) was amplified using conventional PCR [forward primer (387 bp), CACCTG GAGTACTTTGTGCG; reverse primer (1525 bp), CCTGTG GGTGGTACCCTATG], purified (Qiaquick Gel Extraction and PCR Purification Kits; Qiagen) and quantified using a BioPhotometer (Eppendorf). The number of DNA molecules was calculated and the DNA was diluted accordingly in serial steps.
Harvesting of single-cell mRNA and cDNA synthesis
Single-cell mRNA was harvested from dopaminergic neurons in acute mouse brain slices as previously described (23). Briefly, for patch-clamp recording and cytoplasm harvest, patch-clamp capillaries (baked overnight at 220°C) were filled with 6 µl of RNase-free patch-clamp buffer (140 mM KCl, 5 mM HEPES, 5 mM EGTA, 3 mM MgCl2, pH 7.3; all chemicals were from Sigma-Aldrich). After electrophysiological recording of neuronal activity using the whole-cell configuration, the cytoplasm of the same cell was harvested via the patch-pipette, under visual control, without losing the gigaseal to prevent contamination with extracellular fluid. Subsequently, the pipette contents were expelled into a sterile 0.5 ml reaction tube (Biopure; Eppendorf), containing the RT reaction mixture. RNasin and reverse transcriptase were added immediately, and single-cell cDNA synthesis was performed as described above.
Precipitation of single-cell cDNA
All chemicals were molecular biology grade and certificated to be RNase/DNA free. One microgram of glycogen (Ambion), 250 ng of polyC RNA (Amersham Pharmacia), 250 ng of polydC DNA (Amersham Pharmacia) and a 1/10 vol of 2 M sodium acetate pH 4.0 were added to the single-cell cDNA reactions, or to 10 µl of diluted cDNA (generated from brain tissue) in water. cDNA was precipitated overnight with 3.5 vol of ethanol (100%; Sigma) at –20°C. After centrifugation for 60 min at 4°C (13 000 g) the supernatant was discarded, 100 µl of 75% ethanol was added and after a second centrifugation (13 000 g at 4°C) for 15 min, the supernatant was removed. The cDNA pellet was dried in a thermal heating block (Eppendorf) at 45°C until all ethanol had evaporated, and then dissolved in 10 µl of sterile water (Eppendorf). In order to completely resolubilize the cDNA, it was incubated for 60 min at 45°C prior to quantitative real-time PCR.
TaqMan quantitative real-time PCR and data analysis
rtqPCR was performed as recently described (13) using the GeneAmp 5700 instrument (Applied Biosystems). The TaqMan primer/hybridization probe real-time PCR approach uses a fluorescence resonance energy transfer probe as reporter system (24). Hybridization primer/probe assay specific for real-time PCR detection of TH (accession no. M69200) was optimized according to the recommended criteria using the Abiprism Primer express software (Applied Biosystems) and the 2× TaqMan hybridization-probe Mastermix (Applied Biosystems). The TaqMan Mastermix contains uracil-N-glycosylase (UNG). RT and PCR were performed with the same batches of enzymes, random hexamers and dNTPs for all cells analyzed. All pipetting steps were carried out in a flow hood (HERAEUS), with the same set of pipettes by the same person. Control DNA or purified single-cell cDNA (each in volumes of 10 µl) was used as a template in a 50 µl PCR reaction in 1× TaqMan Mastermix in the presence of 300 nM of the forward primer (TH-F1151, 5′-GAATGGGGAGCTGAAGGCTTA-3′), 300 nM of the reverse primer (TH-R1260, CTGCTGTGTCTGGTCAAAGG), and 125 nM of the specific probe (TH-probe1193, CTATGGAGAGCTCCTGCACTCCCTGTCA). The hybridization probe was 3′ labeled with 6-carboxytetramethylrhodamine as quencher and 5′-labeled 6-carboxyfluoroscein as reporter dye. Real-time PCR was performed in a GeneAmp 5700 thermocycler (PCR program: 2 min at 50°C, 10 min at 95°C, 50 cycles; 15 s at 95°C and 1 min at 60°C) and analyzed with GeneAmp 5700, Excel and IGOR (Wavemetrics) software. After defining a baseline (normalized background fluorescence of cycles 6–15) in a linear plot of relative fluorescence (Rf) against PCR cycle number, quantification of the initial template molecules was performed. The PCR cycle, where the increasing relative fluorescence (Rf) crossed a manually set detection threshold (Ct) was defined in a logarithmic plot of Rf values against PCR cycle numbers (Ct at Rf = 0.09 for all analyzed data). Calibration curves were generated in IGOR by plotting Ct values against respective numbers of DNA template molecules, cDNA concentrations or cDNA dilution factors. Overall efficiencies (E) of PCR were calculated from the slopes of the standard curves according to the E = 10(–1/slope) for serial dilution in steps of 10 [log(10) scale] or E = 2(–1/slope) for serial dilution in steps of 2 [log(2) scale]. E = 2 reflects a doubling of DNA in each PCR cycle over all dilution steps. Errors are given as standard deviations (SDs) of the means. Significance was defined according to P-values gained from either two-tailed t-test analysis, or for single-cell ΔCt comparisons, one-tailed t-test analysis.
RESULTS
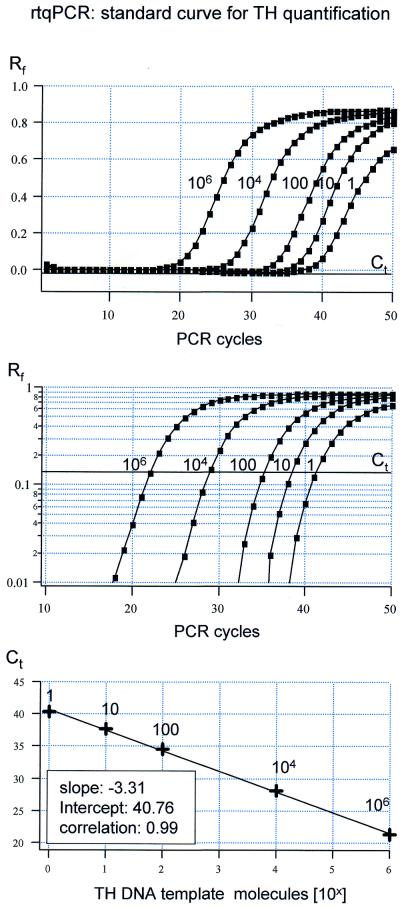
A TaqMan primer/probe assay was designed and optimized to detect TH, a marker gene expressed in monoaminergic neurons. Figure 1 shows a representative result for an experiment in which different amounts of TH DNA (from 1 up to 106 molecules) were used as templates for rtqPCR. Relative fluorescence levels are plotted against PCR cycle numbers on both a linear scale (Fig. 1, top) and a semi-logarithmic scale (Fig. 1, middle). Figure 1 illustrates that the PCR amplification kinetics are similar over the whole range of TH DNA template molecules. This is a precondition for the determination of Ct values and the generation of reliable calibration curves. The standard curve derived by plotting Ct values against DNA template molecule numbers has a calculated slope of –3.31, reflecting an overall PCR efficiency of 2.0 (mean slope for n = 5 experiments: –3.35 ± 0.09, E = 1.99 ± 0.04). This slope indicates a near perfect doubling of amplification products per cycle during the exponential phase of the PCR [the theoretical slope for an ideal PCR amplification is –1 / log(10)2 = –3.32].
Figure 1.
Real-time fluorescent RT–PCR standard curve for TH cDNA quantification. Top, sensitivity of the real-time fluorescent RT–PCR protocol for TH. Relative fluorescence intensities (Rf) were plotted against PCR cycle numbers for five different calculated numbers of TH dsDNA template molecules (ranging from 1 to 1 000 000 molecules, as indicated). Middle, same data as in the top panel are plotted on a logarithmic Rf scale, for better illustration of the exponential phase of the PCR. Slopes of the exponential amplification phases were highly reproducible and independent of template concentration. Bold line (Ct) indicates the fluorescence threshold of amplification detection at Rf = 0.09, manually set within the exponential PCR phase. Bottom, standard curve for TH cDNA quantification derived from data shown in the top and middle panels. Ct value is defined as the cycle number where the relative fluorescence crosses the set threshold of amplification detection. Mean Ct values for different numbers of TH template molecules are plotted against their respective numbers of template molecules on a logarithmic scale. The linear regression fit (r = 0.999) was highly reproducible (mean slope = –3.31 ± 0.09 SD, n = 3) and defined the intercept at PCR cycle = 40.76 for a single TH dsDNA molecule.
When undiluted single-cell cDNA was used as PCR template, the results were dramatically different from those obtained using purified DNA (Fig. 1), or cDNA in water (see Fig. 4A). The rtqPCR amplification kinetics varied from cell to cell in an unpredictable fashion, making the definition of the cycle threshold for detection problematic (Fig. 2A). These results appear to rule out reliable rtqPCR quantification of cDNA molecules using undiluted single-cell cDNA reaction mixtures as PCR templates.
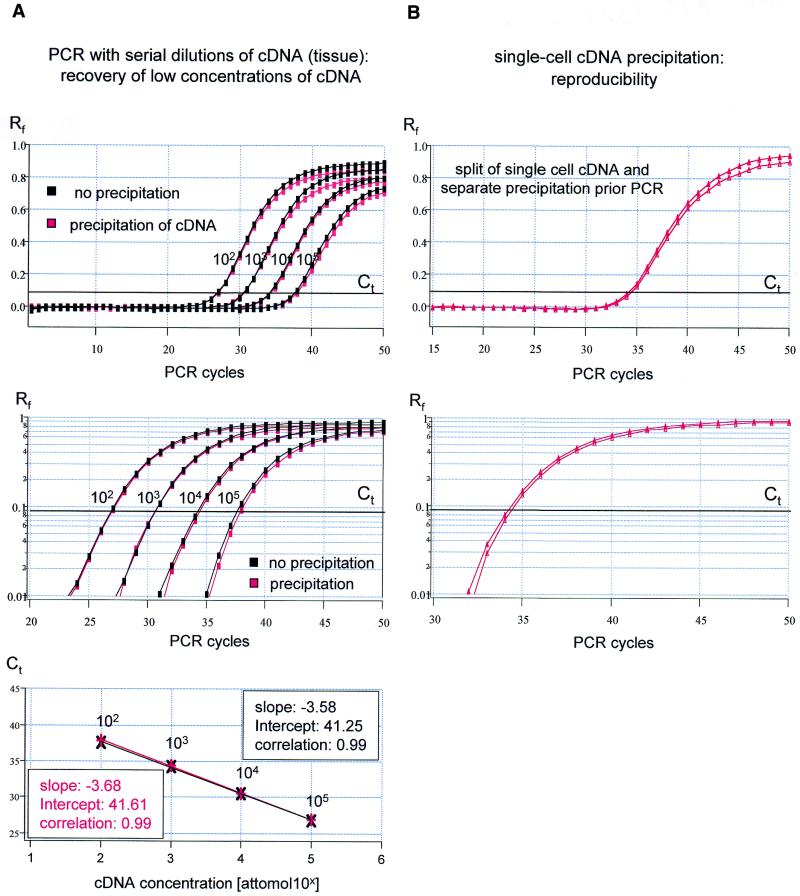
Figure 4.
No loss of cDNA occurs during the precipitation procedure. (A) rtqPCR of TH with serial dilutions (steps of 10) of cDNA in water, generated from brain tissue, as templates. Red traces, precipitation of cDNA prior to PCR; black traces, no precipitation of cDNA prior to PCR. Linear and logarithmic amplification plots reveal almost identical amplification kinetics (top/middle) over the full range of tested cDNA concentrations. The respective calibration curves (bottom) had very similar slopes and they were similar to the standard curves shown in Figure 1. (B) Reproducibility of rtqPCR following single-cell cDNA precipitation. Amplification plots for TH and Ct values of cDNA generated from a single cell, split into two reactions prior to precipitation and rtqPCR, are almost identical.
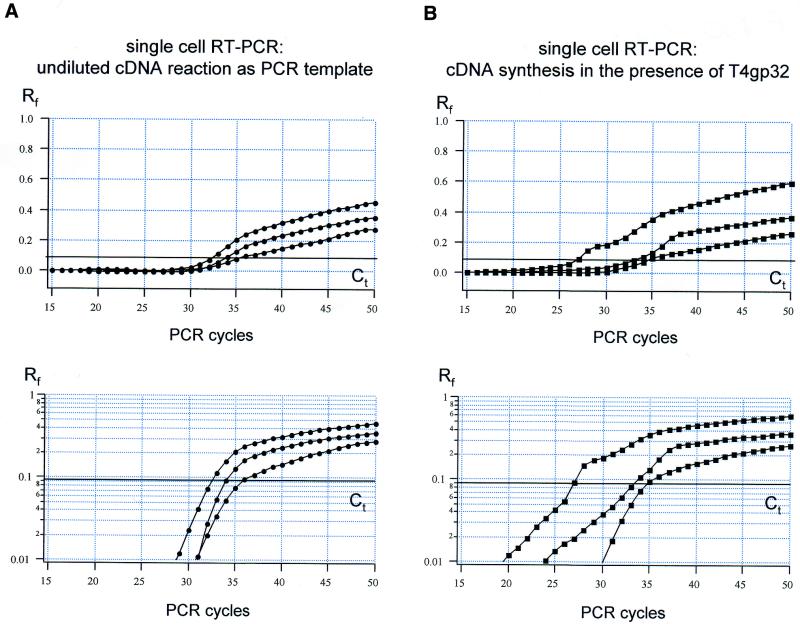
Figure 2.
Real-time PCR using undiluted single-cell cDNA as template. (A) Amplification plots for TH rtqPCR for three individual cells utilizing undiluted cDNA reactions as templates. Relative fluorescence intensities (Rf) were plotted against PCR cycle numbers on a linear (top) and logarithmic (bottom) scale, illustrating inhomogeneous amplification kinetics between different single cells and compared with the standard (compare Fig. 1). (B) Similar experiment as shown in (A). However, single-cell cDNA synthesis was performed in the presence of the T4 gene 32 protein.
To identify the RT reaction component(s) that distort the rtqPCR amplification kinetics, the inhibitory effect of each of the individual RT reaction components was determined. These included DTT, RT buffer (containing random hexamer primers and dNTPs), the patch-clamp buffer, reverse transcriptase and ribonuclease inhibitor. Real-time PCR was performed with identical amounts of template cDNA (1 fM), generated from brain tissue, in either water or in the presence of an individual RT reaction component. The Ct value for cDNA in water was used as a reference to identify significant inhibition of cDNA-synthesis reaction components. The following Ct values were obtained: Ct = 34.44 ± 0.18 (n = 6) for cDNA in water; Ct = 34.31 ± 0.33 (n = 4) for cDNA in patch-clamp buffer; Ct = 34.44 ± 0.16 (n = 4) for cDNA in 2 U/µl RNasin; Ct = 34.29 ± 0.18 (n = 4) for cDNA in RT buffer; Ct = 35.03 ± 0.23 for cDNA in DTT (n = 4); Ct = 50.00 ± 0.0 (n = 6) for cDNA in reverse transcriptase. A two-paired t-test demonstrated that the TaqMan rtqPCR amplification was significantly compromised only in the presence of 10 U/µl reverse transcriptase (P << 0.000005) and to a much lesser degree in the presence of 10 mM DDT (P = 0.002). Interestingly, the inhibitory effect of the reverse transcriptase was higher in these control experiments than in single-cell RT–PCR experiments (Fig. 2), resulting in complete rtqPCR inhibition (Ct = 50 after 50 PCR cycles).
It has been reported that adding of the T4 gene 32 protein (T4gp32) to the RT reaction mixture, prior to RT, could remove the inhibitory effects of reverse transcriptase on subsequent PCR amplification (18,25). However, as illustrated in Figure 2B, addition of 1.6 µg of T4gp32 prior to single-cell cDNA synthesis did not improve the real-time RT–PCR amplification kinetics.
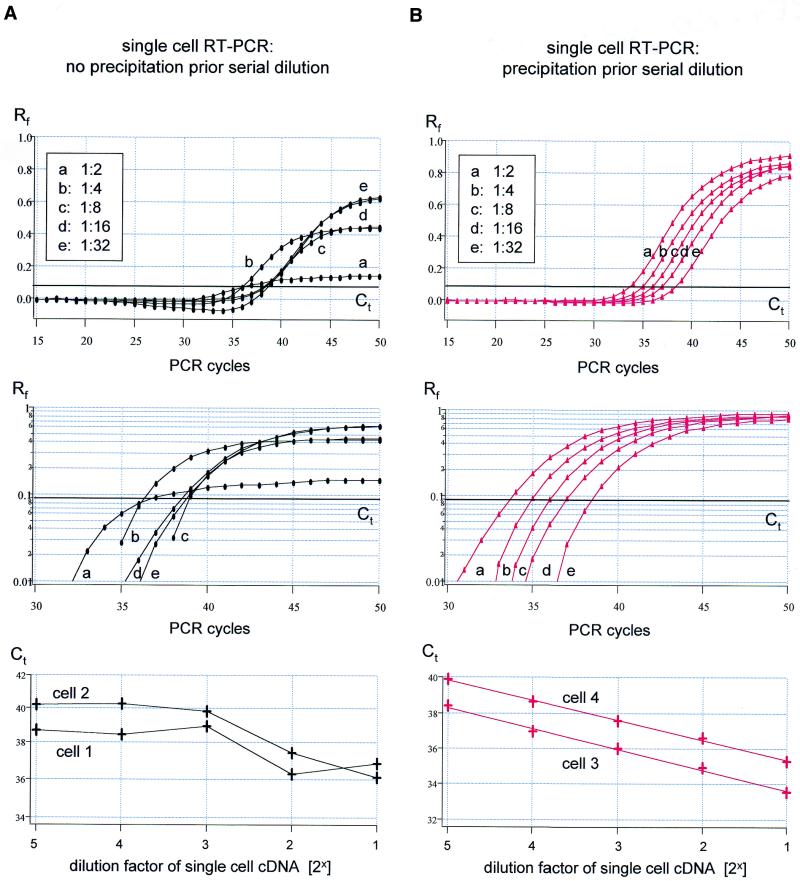
Figure 3 shows serial dilution of single-cell cDNA reaction mixtures. As illustrated, the rtqPCR amplification kinetics were not homogeneous across different cDNA dilutions, and varied both in the same cell (Fig. 3A, top and middle), and between individual cells (Fig. 3A, bottom). Thus, analysis of individual single-cell PCR calibration curves did not produce reliable values. The mean slope for the individual linear regressions was –3.45 ± 1.0 (n = 4), the high SD indicates the large scatter in the slopes of individual calibration curves. Figure 3B depicts the results of a similar set of experiments to those shown in Figure 3A, except that in this case the single-cell cDNAs were precipitated prior to serial dilution and rtqPCR. It is evident from the individual amplification plots that the kinetics of rtqPCR were independent of cDNA concentration both in the same cell (Fig. 3B, top and middle) and between different cells (Fig. 3B, bottom). The mean slope of the calibration curve was –1.03 ± 0.05 (n = 4) for the log(2) scale (ideal slope for serial dilution of 2 = 1.0), corresponding to –3.44 ± 0.16 on a log(10) scale and an efficiency of 1.96 ± 0.06. Importantly, neither the slope of the calibration curve nor the overall PCR efficiency was significantly different when precipitated single-cell cDNA or purified TH DNA was used as PCR template [P = 0.28 (slope) and P = 0.34 (E)].
Figure 3.
Real-time PCR of TH for cDNA derived from single cells. (A) Top/middle, amplification plots (linear/logarithmic y-axis) of TH rtqPCR with serial dilutions (steps of 2) of cDNA derived from a single cell as template (dilutions: a = 1:2, b = 1:4, c = 1:8, d = 1:16, e = 1:32, Ct at 0.09). Note the diverging slopes of individual amplification kinetics that also differs from those of controls (compare Figs 1 and 4A) preventing quantification of single-cell TH cDNA molecules. Bottom, linear plot of Ct values against dilution factor of cDNA derived from individual cells, further illustrating the unpredictability of altered amplification kinetics (data points for cell 1 correspond to amplification plots shown in the top and middle panel). (B) Similar experiment as shown in (A), but single-cell cDNA was precipitated and redissolved in water prior to rtqPCR. Top/middle, slopes of the exponential amplification phases (Ct = 0.09) were highly reproducible and independent of template concentration. Bottom, linear plot of Ct values against dilution factor of cDNA derived from individual cells, further illustrating the robustness and reproducibility of results (data points for cell 3 correspond to amplification plots shown in the top and middle panels).
I next tested whether the purification protocol leads to any loss of low concentration cDNA, which would bias quantification. Sets of serial dilutions of brain cDNA (10 µl each) were either used directly for rtqPCR or were precipitated and redissolved in 10 µl of water prior to rtqPCR. Figure 4A shows that the amplification plots and calibration curves obtained using these two protocols were almost identical, indicating that precipitation results in no significant loss of template cDNA over the full range of concentrations tested (102–105 aM). The mean values between the Ct value for non-precipitated and precipitated DNA (ΔCt) was –0.04 ± 0.15 (for n = 4 dilution steps). The slopes of the calibration curve obtained for non-precipitated or precipitated cDNA were –3.55 ± 0.17 (n = 4, E = 1.92 ± 0.06) and –3.62 ± 0.15 (n = 4, E = 1.89 ± 0.05), and were not significantly different [two-paired t-test: P = 0.54 (slope) and P = 0.52 (E)]. There was also no difference between the calibration curves generated from a single-cell cDNA and from brain tissue cDNA [P = 0.15 (slope) and P = 0.36 (E), respectively].
To evaluate the reproducibility of the precipitation method at the level of the single cell, cDNA reaction mixtures from individual cells were split after RT into two halves, and both halves were separately precipitated and used for comparative rtqPCR. Figure 4B illustrates the high reproducibility of the single-cell cDNA precipitation protocol. The Ct values of single-cell duplicates were very similar (mean ΔCt = 0.27 ± 0.16, n = 6) and the mean ΔCt was not significantly different from those of standard replicates, calculated to have 10 DNA molecules as PCR template (compare Fig. 1, mean ΔCt = 0.31 ± 0.13, n = 6, P = 0.32, one-tailed t-test). In contrast, if only one half of the single-cell cDNA reaction mixture was precipitated, and the other was not, the Ct values were very different from cell to cell (mean ΔCt = 1.73 ± 1.33, n = 6; the high SD indicates the large scatter of ΔCt values), and ΔCt values were significantly higher than those of standard replicates (P = 0.01, one-tailed t-test). These results further illustrate that the extent of PCR inhibition is not predictable using unpurified single-cell cDNA reaction mixtures as templates for rtqPCR. In contrast, the precipitated single-cell cDNA behaves like the purified TH DNA template during rtqPCR amplification.
Taken together, absolute quantification of single-cell cDNA molecules using real-time PCR is possible, without diluting the single-cell cDNA reaction mixture, providing a quantitative cDNA precipitation step is included in the protocol.
DISCUSSION
Quantitative analysis of gene expression at the level of individual cells is of particular relevance in tissues that contain a large variety of different cell types in close proximity, like the brain. Determination of the exact amount of a cDNA related to a specific gene using rtqPCR requires the definition of a cycle threshold for detection (Ct) in the exponential phase of the PCR amplification. For both relative and absolute quantification and comparison of gene expression between different samples, similar PCR amplification kinetics, especially in the exponential phase, are required for all samples analyzed. Relative quantification requires a housekeeping gene that shows constant expression levels between individual samples. The choice of such a gene is problematic at the tissue level (26) and is likely to be even more difficult at the single-cell level. We have previously shown that the classic housekeeping gene beta-actin display large variations in cDNA expression levels between individual dopaminergic neurons (13). This indicates that beta-actin, at least at the level of single dopaminergic neurons, is not suitable for relative quantification. In order to allow absolute quantification of cDNA molecule numbers, a standard curve is required for the respective gene, which is generated by using defined numbers of DNA molecules as rtqPCR templates (compare Fig. 1). In addition, the amplification kinetics and efficiencies of PCR reactions for sample DNA and for standard DNA need to be similar (15,17,27,28). In general, it is problematic to extrapolate from the number of cDNA molecules determined, to the absolute number of corresponding mRNA transcripts, as the efficiency of reverse transcriptase depends on the secondary structure, the specific RNA/protein complexation, and the absolute number of the respective individual target mRNA species (26,29,30).
I show here that the requirements for reliable rtqPCR quantification of TH cDNA molecules were clearly not met when undiluted single-cell cDNA reaction mixtures were used as templates; rtqPCR amplification kinetics were altered in a non-systematic fashion from cell to cell (compare Fig. 2). Serial dilution experiments of single-cell cDNA reaction mixtures revealed that the rtqPCR amplification kinetics were also inhomogeneous across different cDNA dilutions, not only for the same cell, but also between different individual cells (Fig. 3A). Inhibition of the PCR amplification reaction, and thus altered PCR amplification kinetics, has been found to be a major problem for quantitative PCR (18,27,31). Mathematical procedures to adjust altered PCR amplification kinetics are only valid if PCR kinetics are distorted in a systematic and thus predictable manner (19,28,32,33). Thus, mathematical procedures to correct for the different PCR amplification kinetics found for undiluted single-cells cDNA reaction mixtures are not applicable.
To find an experimental way to overcome rtqPCR inhibition when undiluted cDNA reaction mixtures are used as templates, I analyzed which component of the cDNA reaction mixture was responsible for PCR inhibition. I found that the presence of the reverse transcriptase itself in the rtqPCR reaction dramatically inhibited PCR amplification. PCR inhibition by reverse transcriptases has previously been described, and reported to be particularly significant at lower concentrations of template DNA (<105 molecules) (18).
It has been reported that the addition of the T4 gene 32 protein prior to RT increases the efficiency of reverse transcriptases and could overcome PCR inhibition (18). However, I found that addition of T4gp32 prior to single-cell cDNA synthesis did not result in improved amplification kinetics of subsequent rtqPCR (Fig. 2B). Heat inactivation of the reaction mix after cDNA synthesis, to destroy secondary structure and thus enzymatic activity, has previously been shown not to be preventing the inhibitory effects of reverse transcriptase on PCR (18). This is in accordance with my results, as all rtqPCR reactions were incubated for 10 min at 95°C prior to PCR amplification to inactivate the UNG enzyme of the TaqMan Mastermix.
Decreasing the amount of reverse transcriptase to reduce its inhibitory effect on PCR is expected to result in loss, or under-representation, of low abundance mRNAs from single cells because the efficiency of the cDNA synthesis is reduced at low mRNA concentrations (15). Dilution of the single-cell cDNA reaction (>one-eighth-fold, compare Fig. 2A) resulted in amplification kinetics that were very similar to those performed with either cDNA in water, or purified DNA as PCR template. However, dilution of single-cell cDNA will inevitably lead to loss of low abundance cDNAs and thus compromise the application of single-cell rtqPCR for the study of low abundance mRNAs.
I describe here a simple ethanol precipitation protocol that enables quantitative precipitation and recovery of very low concentrated cDNA, and that removes all rtqPCR-inhibiting components. Ethanol precipitation was carried out in the presence of sodium acetate, in accordance with the original protocol (34). Precipitation of cDNA was carried out overnight at –20°C, as precipitation at –70°C has been found to inhibit complete DNA precipitation when ammonium acetate was used as source of monovalent cations (35,36). The addition of the carrier nucleic acids polyC and polydC allowed quantitative precipitation of low concentrated DNA without affecting subsequent rtqPCR, as illustrated in Figure 4A. Glycogen was added to increase the reproducibility of cDNA recovery. I did not test linear acrylamide as co-precipitants as there were no problems either with DNA or RNase contamination of glycogen or with re-dissolving of precipitated cDNA. Linear acrylamide has the advantage of being derived from a non-biological source, however, it contributes to absorbance at 260 and 280 nm (37).
Experiments using the same amount of highly diluted cDNA with or without precipitation prior to rtqPCR allowed direct monitoring of any cDNA loss due to the precipitation procedure and demonstrated that this did not happen (Fig. 4A). PCR amplification plots, PCR efficiencies, and the slopes of calibration curves generated from precipitated single-cell cDNA were very similar to those using purified TH DNA or brain cDNA as templates. Taken together, single-cell cDNA after precipitation fulfills all the requirements for absolute quantification of TH cDNA molecules at the level of individual cells.
The protocol enables rtqPCR in combination with established specific RT conditions for single-cell RNA, harvested after electrophysiological characterization (3–5,9,23). With the method described here we have quantified single-cell cDNA expression after electrophysiological characterization for six different genes with high, medium and low expression levels, and identified a high correlation between cDNA transcript numbers and respective functional protein levels in the individual neurons (13). Furthermore, this approach provides a reliable method for cell-specific validation of tissue-based DNA microarray-generated global gene expression-profiling data (38,39).
Acknowledgments
ACKNOWLEDGEMENTS
I thank Frances Ashcroft and Jochen Roeper for critically reading the manuscript. B.L. is supported by a Todd-Bird Junior Research Fellowship at New College, Oxford, and a Royal Society Dorothy Hodgin Research Fellowship.
REFERENCES
- 1.Schutze K., Posl,H. and Lahr,G. (1998) Laser micromanipulation systems as universal tools in cellular and molecular biology and in medicine. Cell Mol. Biol. (Noisy-le-grand), 44, 735–746. [PubMed] [Google Scholar]
- 2.Fink L., Seeger,W., Ermert,L., Hanze,J., Stahl,U., Grimminger,F., Kummer,W. and Bohle,R.M. (1998) Real-time quantitative RT–PCR after laser-assisted cell picking. Nature Med., 4, 1329–1333. [DOI] [PubMed] [Google Scholar]
- 3.Monyer H. and Lambolez,B. (1995) Molecular biology and physiology at the single-cell level. Curr. Opin. Neurobiol., 5, 382–387. [DOI] [PubMed] [Google Scholar]
- 4.Sucher N.J. and Deitcher,D.L. (1995) PCR and patch-clamp analysis of single neurons. Neuron, 14, 1095–1100. [DOI] [PubMed] [Google Scholar]
- 5.Lambolez B., Audinat,E., Bochet,P., Crepel,F. and Rossier,J. (1992) AMPA receptor subunits expressed by single Purkinje cells. Neuron, 9, 247–258. [DOI] [PubMed] [Google Scholar]
- 6.Tsuzuki K., Lambolez,B., Rossier,J. and Ozawa,S. (2001) Absolute quantification of AMPA receptor subunit mRNAs in single hippocampal neurons. J. Neurochem., 77, 1650–1659. [DOI] [PubMed] [Google Scholar]
- 7.Baro D.J., Levini,R.M., Kim,M.T., Willms,A.R., Lanning,C.C., Rodriguez,H.E. and Harris-Warrick,R.M. (1997) Quantitative single-cell-reverse transcription–PCR demonstrates that A-current magnitude varies as a linear function of shal gene expression in identified stomatogastric neurons. J. Neurosci., 17, 6597–6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tkatch T., Baranauskas,G. and Surmeier,D.J. (1998) Basal forebrain neurons adjacent to the globus pallidus co-express GABAergic and cholinergic marker mRNAs. Neuroreport, 9, 1935–1939. [DOI] [PubMed] [Google Scholar]
- 9.Tkatch T., Baranauskas,G. and Surmeier,D.J. (2000) Kv4.2 mRNA abundance and A-type K(+) current amplitude are linearly related in basal ganglia and basal forebrain neurons. J. Neurosci., 20, 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lockey C., Otto,E. and Long,Z. (1998) Real-time fluorescence detection of a single DNA molecule. Biotechniques, 24, 744–746. [DOI] [PubMed] [Google Scholar]
- 11.Steuerwald N., Cohen,J., Herrera,R.J. and Brenner,C.A. (1999) Analysis of gene expression in single oocytes and embryos by real-time rapid cycle fluorescence monitored RT–PCR. Mol. Hum. Reprod., 5, 1034–1039. [DOI] [PubMed] [Google Scholar]
- 12.Svanvik N., Stahlberg,A., Sehlstedt,U., Sjoback,R. and Kubista,M. (2000) Detection of PCR products in real time using light-up probes. Anal. Biochem., 287, 179–182. [DOI] [PubMed] [Google Scholar]
- 13.Liss B., Franz,O., Sewing,S., Bruns,R., Neuhoff,H. and Roeper,J. (2001) Tuning pacemaker frequency of individual dopaminergic neurons by Kv4.3L and KChip3.1 transcription. EMBO J., 20, 5715–5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker N.J. (2002) A technique whose time has come. Science, 296, 557–559. [DOI] [PubMed] [Google Scholar]
- 15.Freeman W.M., Walker,S.J. and Vrana,K.E. (1999) Quantitative RT–PCR: pitfalls and potential. Biotechniques, 26, 112–122, 124–125. [DOI] [PubMed] [Google Scholar]
- 16.Bustin S.A. (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol., 25, 169–193. [DOI] [PubMed] [Google Scholar]
- 17.Medhurst A.D., Harrison,D.C., Read,S.J., Campbell,C.A., Robbins,M.J. and Pangalos,M.N. (2000) The use of TaqMan RT–PCR assays for semiquantitative analysis of gene expression in CNS tissues and disease models. J. Neurosci. Methods, 98, 9–20. [DOI] [PubMed] [Google Scholar]
- 18.Chandler D.P., Wagnon,C.A. and Bolton,H.,Jr (1998) Reverse transcriptase (RT) inhibition of PCR at low concentrations of template and its implications for quantitative RT–PCR. Appl. Environ. Microbiol., 64, 669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfaffl M.W. (2001) A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res., 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Taher A., Bashein,A., Nolan,T., Hollingsworth,M. and Brady,G. (2000) Global cDNA amplification combined with real-time RT–PCR: accurate quantification of multiple human potassium channel genes at the single cell level. Yeast, 17, 201–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song W.J., Tkatch,T., Baranauskas,G., Ichinohe,N., Kitai,S.T. and Surmeier,D.J. (1998) Somatodendritic depolarization-activated potassium currents in rat neostriatal cholinergic interneurons are predominantly of the A type and attributable to coexpression of Kv4.2 and Kv4.1 subunits. J. Neurosci., 18, 3124–3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sucher N.J., Deitcher,D.L., Baro,D.J., Warrick,R.M. and Guenther,E. (2000) Genes and channels: patch/voltage-clamp analysis and single-cell RT–PCR. Cell Tissue Res., 302, 295–307. [DOI] [PubMed] [Google Scholar]
- 23.Liss B., Bruns,R. and Roeper,J. (1999) Alternative sulfonylurea receptor expression defines metabolic sensitivity of K-ATP channels in dopaminergic midbrain neurons. EMBO J., 18, 833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lie Y.S. and Petropoulos,C.J. (1998) Advances in quantitative PCR technology: 5′ nuclease assays. Curr. Opin. Biotechnol., 9, 43–48. [DOI] [PubMed] [Google Scholar]
- 25.Villalva C., Touriol,C., Seurat,P., Trempat,P., Delsol,G. and Brousset,P. (2001) Increased yield of PCR products by addition of T4 gene 32 protein to the SMART PCR cDNA synthesis system. Biotechniques, 31, 81–83, 86. [DOI] [PubMed] [Google Scholar]
- 26.Bustin S.A. (2002) Quantification of mRNA using real-time RT–PCR: trends and problems. J. Mol. Endocrinol., 29, 23–40. [DOI] [PubMed] [Google Scholar]
- 27.Halford W.P. (1999) The essential prerequisites for quantitative RT–PCR. Nat. Biotechnol., 17, 835. [DOI] [PubMed] [Google Scholar]
- 28.Pfaffl M.W., Horgan,G.W. and Dempfle,L. (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res., 30, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerard G.F. and D’Alessio,J.M. (1993) Reverse transcriptase (EC 2.7.7.49). Methods Mol. Biol., 16, 73–93. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J. and Byrne,C.D. (1999) Differential priming of RNA templates during cDNA synthesis markedly affects both accuracy and reproducibility of quantitative competitive reverse-transcriptase PCR. Biochem. J., 337, 231–241. [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Soud W.A. and Radstrom,P. (2001) Purification and characterization of PCR-inhibitory components in blood cells. J. Clin. Microbiol., 39, 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vu H.L., Troubetzkoy,S., Nguyen,H.H., Russell,M.W. and Mestecky,J. (2000) A method for quantification of absolute amounts of nucleic acids by (RT)–PCR and a new mathematical model for data analysis. Nucleic Acids Res., 28, e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meijerink J., Mandigers,C., van de Locht,L., Tonnissen,E., Goodsaid,F. and Raemaekers,J. (2001) A novel method to compensate for different amplification efficiencies between patient DNA samples in quantitative real-time PCR. J. Mol. Diagn., 3, 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wallace D.M. (1987) Precipitation of nucleic acids. Methods Enzymol., 152, 41–48. [DOI] [PubMed] [Google Scholar]
- 35.Crouse J. and Amorese,D. (1987) Ethanol precipitation: ammonium acetate as an alternative to sodium acetate. Focus, 19, 13–16. [Google Scholar]
- 36.Zeugin J.A. and Hartley,J.L. (1985) Ethanol precipitation of DNA. Focus, 7, 1–2. [Google Scholar]
- 37.Gaillard C. and Strauss,F. (1990) Ethanol precipitation of DNA with linear polyacrylamide as carrier. Nucleic Acids Res., 18, 378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mills J.C., Roth,K.A., Cagan,R.L. and Gordon,J.I. (2001) DNA microarrays and beyond: completing the journey from tissue to cell. Nature Cell Biol., 3, E175–E178. [DOI] [PubMed] [Google Scholar]
- 39.Rajeevan M.S., Ranamukhaarachchi,D.G., Vernon,S.D. and Unger,E.R. (2001) Use of real-time quantitative PCR to validate the results of cDNA array and differential display PCR technologies. Methods, 25, 443–451. [DOI] [PubMed] [Google Scholar]