Abstract
CREB-binding protein (CBP) is a multifunctional cofactor implicated in many intracellular signal transduction pathways. We aimed to investigate the involvement of CBP in the cAMP response element-binding protein (CREB)-mediated pathway. The point mutation Tyr658Ala in the CREB-binding domain (CBD) was shown to abolish the binding activity of CBP to phospho-CREB, the activated form of CREB. By using a mutant Cre/loxP recombination system, this point mutation was aimed to be generated in the mouse genome in a tissue- and time-specific manner. A targeting construct in which CBD exon 5 and inverted exon 5* containing the point mutation flanked by two mutant loxP sites (lox66 and lox71) oriented in a head-to-head position was generated. When Cre recombinase is present, the DNA flanked by the two mutant loxP sites is inverted, forming one loxP and one double mutated loxP site. As the double mutated loxP site shows low affinity for Cre recombinase, the favorable reaction leads to a product where the mutated exon 5* is placed into the position to be correctly transcribed and spliced. Inversion was observed to be complete in both bacteria and mouse embryonic stem cells. Our results indicate that this Cre- mediated inversion method is a valuable tool to introduce point mutations in the mouse genome in a regulatable manner.
INTRODUCTION
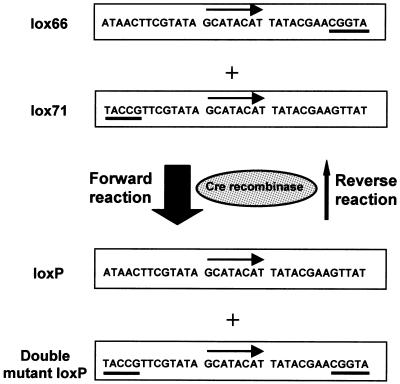
Tissue- and time-restricted genetic modifications in mice can be generated using the Cre/loxP recombination system (1). Cre is a 38 kDa site-specific DNA recombinase isolated from bacteriophage P1. loxP (locus of crossover in P1) was originally found in the phage P1 genome and is 34 bp in length, consisting of two 13 bp inverted repeats flanking an 8 bp non-palindromic core sequence that determines the polarity of the loxP site (2). Cre-mediated recombination between two directly repeated loxP sites results in an irreversible excision of the intervening sequence, while recombination between two loxP sites that are positioned in a head-to-head orientation leads to an inversion, which takes place continuously as long as Cre recombinase is present. Recently, various mutant loxP sites were generated and characterized (3), of which lox66 and lox71 are particularly interesting. After the first Cre-mediated recombination, one wild-type loxP site and one double mutant loxP site are formed; the latter, however, displays a very low affinity for Cre recombinase. Consequently, Cre-mediated recombination in the mutated Cre/loxP system has a favorable forward reaction equilibrium, as depicted in Figure 1. This mutant Cre recombination system was successfully tested for targeted integration into the mouse genome (4). However, the efficiency of inversion mediated by this system has not yet been evaluated in the context of the mouse genome.
Figure 1.
Schematic representation of the mutated Cre/loxP system. Nucleotide sequences of loxP and its mutated derivatives are listed; mutated sequences are underlined. Arrows indicate non-palindromic core sequence. Cre-mediated recombination between lox66 and lox71 sites generates one wild-type and one double mutant loxP site. Since the double mutant loxP site exhibits much reduced binding affinity for Cre recombinase, Cre- mediated inversion using the mutated Cre/loxP system prefers the forward reaction as indicated.
In the present study, we tested whether, in principle, this system could be used to introduce point mutations into CREB-binding protein (CBP) in a tissue- and time-restricted manner using the mutant Cre/loxP recombination system described above. CBP is a transcriptional cofactor implicated in many different intracellular signal transduction pathways (5). To dissect CBP function specifically in the cAMP response element-binding protein (CREB)-mediated pathway, we aimed to introduce a point mutation (tyrosine to alanine at amino acid residue 658 of mouse CBP, CBPTyr658Ala), which was reported to abolish the binding activity of CBP to phosphorylated and hence activated CREB (6), presumably leaving intact the activities of the other signaling pathways where CBP plays crucial roles.
MATERIALS AND METHODS
Plasmids
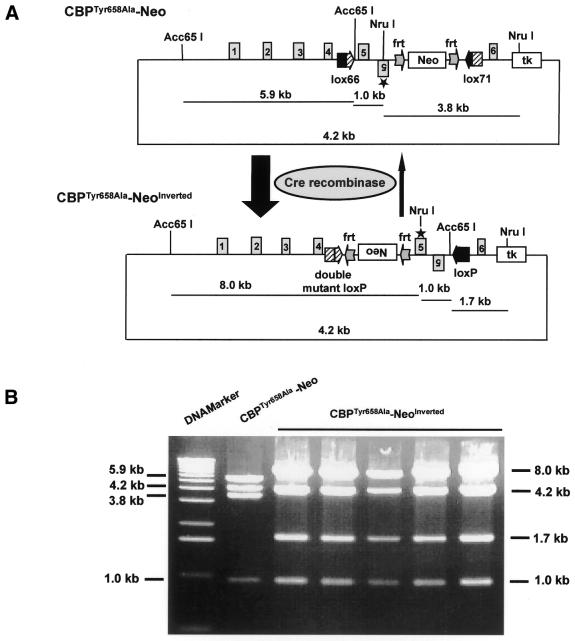
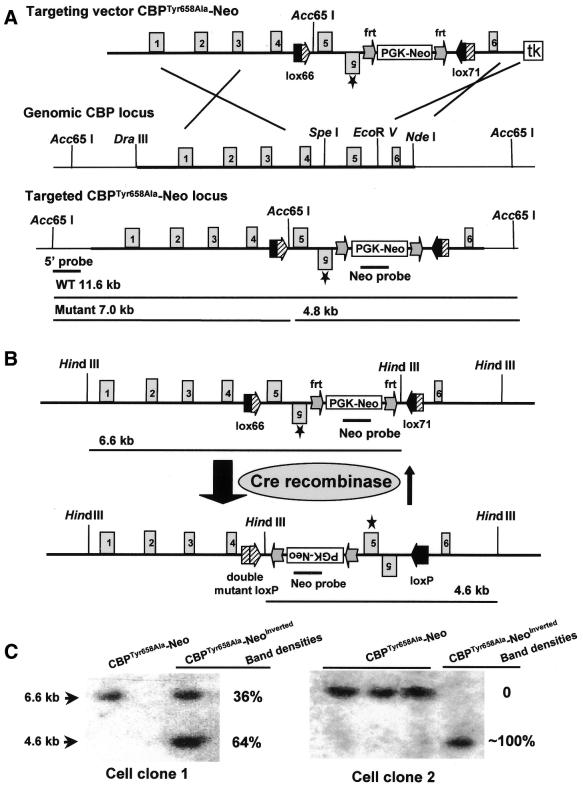
The mutant loxP sites, lox66 and lox71, were synthesized by PCR with two pairs of partly overlapping primers, Z24 and Z25, and Z26 and Z27, respectively. The following primers were used: Z24, 5′-gcc gaa gct tct cgt gat aac ttc gat tag cat aca tta tac gaa cgg t; Z25, 5′-acc gga att ccc gga cta ccg ttc gta taa tgt atg ct; Z26, 5′-gcc gaa gct tct cgt gta ccg ttc gta tag cat aca t; Z27, 5′-acc gga att ccc gga cat aac ttc gta taa tgt atg cta tac gaa cgg t. HindIII and EcoRI sites (underlined sequences) were introduced concomitantly and used to clone lox66 and lox71 into plasmid vector pBSIIKS(–) (Stratagene), respectively. The correct clones were confirmed by restriction digestion and sequencing. The plasmid CBPTyr658Ala–Neo (Fig. 2A) was the final targeting construct, in which the region containing exon 5 of the CREB-binding domain (CBD), an inverted exon 5 with the point mutation (tyrosine to alanine) at amino acid residue 658 and the positive selection marker PGK–Neo (phosphoglycerate kinase–neomycin phosphotransferase) were flanked by lox66 and lox71 sites in a head-to-head orientation. The negative selection marker tk (thymidine kinase) was put at the 3′ end of the targeting construct. The lox66 site was inserted into CBD intron 4 through a SpeI site. Sequences containing the inverted exon 5*, the PGK–Neo cassette flanked by two frt sites and the lox71 site were inserted into intron 5 through the EcoRV site. DraIII and NdeI sites were used for the margins of the left and right homology arms, respectively (Fig. 3A). The mutant exon 5* was generated by PCR with primers Z20 (5′-ttt gta ttt tc*g* c*ga ttt tct ctg ct), Z21 (5′-gca gag aaa atc g*c*g* aaa ata caa aaa ga), Z22 (5′-tag ctg tct cca gac act cca gaa) and Z23 (5′-aaa gac act tgc cag cac acc ctt). Mutations were introduced by the nucleotides marked with asterisks, generating a new NruI site. First, two PCRs with λ phage DNA containing the mouse genomic CBD region of CBP and primers Z22 and Z20, and Z21 and Z23, were performed. Both products were used as templates for a second PCR with primers Z22 and Z23. The product of this PCR was subcloned into plasmid pBSIIKS(–). The correct clone was confirmed by restriction digestion and sequencing. Primers Z33 (5′-ggt cca gct ggc aaa cta aat gat ggg ctc tca) and Z34 (5′-aac tcg agt tga tat caa aga gca cta agc aaa caa gct), containing PvuII, XhoI and EcoRV sites (underlined sequences), were used to shorten the mutant exon 5* clone and in subsequent cloning steps, as longer inverted repeat sequences could not be cloned.
Figure 2.
Cre recombinase-mediated inversion in bacteria. (A) Targeting construct CBPTyr658Ala–Neo was transformed into competent DH5α bacteria expressing Cre recombinase (plasmid 705-Cre) and inversion was induced at 30°C. (B) Restriction pattern analysis after Acc65I/NruI double digestion of clones before and after transformation revealed complete and irreversible Cre recombinase-mediated inversion.
Figure 3.
Cre recombinase-mediated inversion in ES cells. (A) ES cell clones containing the targeted CBPTyr658Ala–Neo allele were generated and analyzed by Southern blotting. Relevant restriction sites used for construction of targeting vector are listed: SpeI and EcoRV sites for insertion of lox66 and lox71, respectively; DraIII and NdeI sites as the margins of the 5′ and 3′ homology arms; Acc65I sites for screening for correct homologous recombinants. The tk cassette was inserted into a SacII site located in the polylinker region of the vector, adjacent to the end of the 3′ homology arm. (B) Correctly recombined ES cell clones were electroporated with the Cre recombinase-expressing plasmid pCre–pac. (C) Southern blot analysis using the Neo probe showed a 6.6 kb band for the targeted CBP allele, a 4.6 kb band for the inverted targeted allele of CBP and both bands for an incomplete inversion. Two examples of inversion are shown. As evaluated by densitometric quantification of the autoradiography film, the inversion ratio was 64% (left) and 100% (right).
In the plasmid pCre–pac (7), the Cre recombinase gene was cloned into vector pUC9 containing the puromycin resistance gene (Pac) driven by the PGK promoter. Plasmid 705-Cre (8) was used to express Cre recombinase in Escherichia coli. The plasmid pSVaZ11-PGK–Neo contains the PGK–Neo cassette flanked by two frt sites. The plasmid pUSEFUL contains the tk gene.
Cre recombinase-mediated inversion in bacteria
Plasmid 705-Cre (containing the chloramphenicol resistance gene) was transformed into competent DH5α bacteria, which were in turn made transformation competent using polyethylene glycol. Plasmid CBPTyr658Ala–Neo (containing the ampicillin resistance gene) was then transformed into these competent Cre-DH5α bacteria. Colonies were inoculated in 2 ml of LB medium containing ampicillin (100 µg/ml) and chloramphenicol (25 µg/ml), and the cultures were incubated overnight with shaking at 30°C. Cre recombinase-mediated inversion was examined by restriction enzyme digestion of isolated DNA using Acc65I and NruI.
Embryonic stem (ES) cell cultures
ES cells (clone E14) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, high glucose, plus sodium pyruvate) (Gibco, Germany) with 15% heat-inactivated fetal calf serum (Roche, Germany), 2 mM l-glutamine (Gibco), 1/12 000 (v/v) leukemia inhibitory factor (Gibco), 100 U/ml penicillin/100 µg/ml streptomycin (Gibco) and 50 µM β-mercaptoethanol (Gibco), incubated at 37°C in 5% CO2. Targeting construct CBPTyr658Ala–Neo was electroporated into ES cells and screened for homologous recombinant clones according to standard procedures. For Cre recombinase-mediated inversion in ES cells, two of the recombinant cell clones were expanded separately on Petri dishes containing mouse embryonic fibroblast feeder cells. Lastly, cells were harvested from 10 cm gelatinized dishes for electroporation. For each recombinant, 1 × 106 ES cells in 800 µl of phosphate-buffered saline were prepared, transferred into the electroporation cuvette (Gene Pulser cuvette, 0.4 cm electrode gap; Bio-Rad, USA) and mixed with 20 µg plasmid pCre–pac. Electroporation was performed at 240 V, 500 µF in a Bio-Rad gene pulser. After electroporation, the cuvette was put onto ice and incubated for 15 min. The cell suspension was then distributed evenly onto three 10 cm feeder dishes, fed with DMEM complete ES cell medium and incubated overnight at 37°C in 5% CO2. Cells were then fed daily with fresh complete ES cell medium plus 1 µg/ml puromycin for selection of Cre recombinase-transfected cells for 2 days, then fed daily with medium without puromycin until formation of colonies. Finally, 96 single colonies for each cell clone were picked and expanded on gelatinized 96-well plates for Southern blot analysis.
Southern blot analysis
Southern blots with the Neo probe were carried out according to standard procedures. The Neo probe was the 300 bp fragment from a XbaI digestion of plasmid pSVaZ11-PGK–Neo.
RESULTS
Generation of targeted CBP locus
To achieve the Cre recombinase-mediated mutation CBPTyr658Ala, we generated a targeting construct containing the wild-type exon 5 of the CBD and a mutated exon 5* (Tyr658Ala) in an inverted orientation. These sequences were flanked by lox66 and lox71 sites, which were positioned in a head-to-head orientation. The notion was that after removal of the Neo selection cassette, Cre recombinase-mediated inversion would place the mutated exon 5* into a position where it would be spliced properly, while the inverted wild-type exon 5 would be spliced out. After several cloning steps using mouse genomic DNA of the CBP locus isolated from mouse strain 129, the targeting construct CBPTyr658Ala–Neo was obtained. A schematic representation of the region encoding the CBD region of CBP and several restriction sites used for cloning are shown in Figures 2A and 3A. SpeI and EcoRV sites were used for insertion of the lox66 and lox71 sites, respectively. Inverted mutant exon 5* and the PGK–Neo cassette flanked by two frt sites were introduced concomitantly with the lox71 site. DraIII and NdeI sites formed the margins of the 5′ and 3′ homology arms, respectively. The tk cassette was inserted into a SacII site located in the polylinker region at the end of the 3′ homology arm (plasmid data not shown). Integrity of the targeting construct was confirmed by sequencing of all cloning junctions, recombination sites, exon 5 and mutant exon 5*.
Cre-mediated inversion in bacteria
We next tested the targeting construct CBPTyr658Ala–Neo in bacteria regarding Cre-mediated inversion through the lox66 and lox71 sites. For this, the targeting construct was transformed into bacteria expressing Cre recombinase. DNA was isolated from five transformed colonies and digested with Acc65I and NruI. All five DNA samples analyzed displayed a digestion pattern with four bands of 1.0, 1.7, 4.2 and 8.0 kb, while the digestion of CBPTyr658Ala–Neo before inversion showed bands of 1.0, 3.8, 4.2 and 5.9 kb (Fig. 2B). According to the restriction map of the targeting construct and its Cre-mediated inversion product (Fig. 2A), this result indicates that the sequence flanked by lox66 and lox71 sites was completely inverted and that the sequence after inversion containing one wild-type loxP and one double mutated loxP no longer appeared to be recognized by Cre recombinase (Fig. 2B).
Cre-mediated inversion in ES cells
To investigate the lox66 and lox71 sites regarding Cre-mediated inversion in the mouse genome, targeting construct CBPTyr658Ala–Neo was electroporated into ES cells. Several clones containing the correct homologous recombination event were characterized by Southern blot analysis (Fig. 3A and data not shown). Two of these clones were then transiently transfected with the plasmid pCre–pac, in order to express Cre recombinase and to induce concomitant inversion. Southern blot analysis of HindIII-digested DNA from these transfected ES cell clones was carried out using the Neo probe (Fig. 3B). The targeted CBPTyr658Ala–Neo allele gave one band of 6.6 kb. After transient transfection with Cre recombinase, some cell clones showed only one 4.6 kb band for the inversion product (Fig. 3C), indicating that inversion was complete and apparently irreversible. Other clones that were analyzed by densitometric evaluation of the autoradiography film revealed values of the inverted band (4.6 kb) lower than 100% (e.g. 64% in Fig. 3C), suggesting that incomplete inversions can also occur.
DISCUSSION
Our results showed that the inversion system using lox66 and lox71 sites represents a novel and very valuable tool to introduce conditional point mutations into transcription units of the genome. While the inversion was always complete and irreversible in bacteria, we observed different efficiencies of inversion patterns in ES cells, ranging from 64 to 100%. It could be that inversion is not as efficient in ES cells as in bacteria, in agreement with another report where the evaluation of inversion was performed using a luciferase assay in transient transfections of cell lines. Cre-mediated integration was shown to be 4- to 7-fold more efficient between lox66 and lox71 than between double mutant loxP and wild-type loxP (3). The variabilities we observed in our experiments using ES cells and inverting a genomic locus might be caused by the different levels of Cre recombinase expression and/or by the different duration of Cre recombinase action in the transiently transfected clones. At any rate, we were able to clearly show that it is possible to get up to 100% inversion using this method. It remains to be investigated how efficient inversion will be in the living mouse, by crossing the CBPTyr658Ala–Neo allele with Cre recombinase-expressing transgenic mice.
The method presented here is very appropriate to study CBP function, since the CBP gene is very sensitive to gene dose. Mice lacking only one CBP allele display quite strong phenotypical changes (9) and Rubinstein–Taybi syndrome patients, who have one defective CBP allele, also show many severe developmental defects (10). Thus, even if the inversion ratio might not reach 100% in all cells, inversion is very likely to be sufficient to introduce phenotypic changes and, thus, we would be able to analyze CBP function in the mouse by the generation of tissue-specific point mutations in CBP.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Drs B. Lüscher for plasmid pSVaZ11-PGK–Neo, F. Stewart for plasmid 705-Cre, A. Bradley for plasmid pUSEFUL and K. Pfeffer for E14 ES cells. This work was supported by a grant from the Volkswagen-Stiftung.
REFERENCES
- 1.Gu H., Marth,J.D., Orban,P.C., Mossmann,H. and Rajewsky,K. (1994) Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science, 165, 103–106. [DOI] [PubMed] [Google Scholar]
- 2.Sauer B. (1998) Inducible gene targeting in mice using the Cre/lox system. Methods, 14, 381–392. [DOI] [PubMed] [Google Scholar]
- 3.Albert H., Dale,E.C., Lee,E. and Ow,D.W. (1995) Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J., 7, 649–659. [DOI] [PubMed] [Google Scholar]
- 4.Araki K., Araki,M. and Yamamura,K. (1997) Targeted integration of DNA using mutant lox sites in embryonic stem cells. Nucleic Acids Res., 25, 868–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodman R.H. and Smolik,S. (2000) CBP/p300 in cell growth, transformation and development. Genes Dev., 14, 1553–1577. [PubMed] [Google Scholar]
- 6.Radhakrishnan I., Perez-Alvarado,G.C., Parker,D., Dyson,H.J., Montminy,M.R. and Wright,P.E. (1997) Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell, 91, 741–752. [DOI] [PubMed] [Google Scholar]
- 7.Taniguchi M., Sanbo,M., Watanabe,S., Naruse,I., Mishina,M. and Yagi,T. (1998) Efficient production of Cre-mediated site-directed recombinants through the utilization of the puromycin resistance gene, pac: a transient gene-integration marker for ES cells. Nucleic Acids Res., 26, 679–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y., Buchholz,F., Muyrers,J.P. and Stewart,A.F. (1998) A new logic for DNA engineering using recombination in Escherichia coli. Nature Genet., 20, 123–128. [DOI] [PubMed] [Google Scholar]
- 9.Kung A.L., Rebel,V.I., Bronson,R.T., Ch’ng,L.E., Sieff,C.A., Livingston,D.M. and Yao,T. (2000) Gene dose-dependent control of hematopoeisis and hematologic tumor suppresssion by CBP. Genes Dev., 14, 272–277. [PMC free article] [PubMed] [Google Scholar]
- 10.Petrij F., Giles,R.H., Dauwerse,H.G., Saris,J.J., Hennekam,R.C.M., Masuno,M., Tommerup,N., van Ommen,G.J.B., Goodman,R.H., Peters,D.J.M. and Breuning,M.H. (1995) Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature, 376, 348–351. [DOI] [PubMed] [Google Scholar]