Abstract
Locked nucleic acids (LNAs) are synthetic nucleic acid analogs that bind to complementary target molecules (DNA, RNA or LNA) with very high affinity. At the same time, this binding affinity is decreased substantially when the hybrids thus formed contain even a single mismatched base pair. We have exploited these properties of LNA probes to develop a new method for single nucleotide polymorphism genotyping. In this method, very short (hexamer or heptamer) LNA probes are labeled with either rhodamine or hexachlorofluorescein (HEX), and their hybridization to target DNAs is followed by measuring the fluorescence polarization (FP) of the dyes. The formation of perfectly complementary double-stranded hybrids gives rise to significant FP increases, whereas the presence of single mismatches results in very small or no changes of this parameter. Multiplexing of the assay can be achieved by using differentially labeled wild-type and mutant specific probes in the same solution. The method is homogeneous, and because of the use of extremely short LNA probes, the generation of a universal set of genotyping reagents is possible.
INTRODUCTION
Numerous methods for single nucleotide polymorphism (SNP) genotyping have been described and have recently been reviewed (1,2). However, there is still continuing interest in the development of new approaches, especially those that would be suitable for high throughput genotyping experiments. With the completion of the Human Genome Project, it is anticipated that such projects will become of great importance in gene discovery and pharmacogenomics.
We have been exploring the possibility of using peptide nucleic acids (PNAs) as probes for such high throughput genotyping applications and have recently reported two fluorescence polarization (FP) based methods using short, fluorescein or rhodamine labeled PNA probes. In the first such method (3), we used fluorescein labeled PNA probes and detected their hybridization to target DNA molecules by FP in the presence of polylysine. Later, we discovered that the use of another fluorescent dye, rhodamine, does not require the presence of polylysine, because the hybridization of such probes resulted in very significant increases of the FP signal even in its absence (4). Unfortunately, there were several problems associated with the use of PNA probes. First, some of these were poorly soluble and precipitated upon storage. Second, the length of these probes had to be at least seven bases, because the lower affinity of shorter probes made the hybridization at room temperature unfavorable. Third, guanosine-rich, rhodamine-labeled probes showed very high intrinsic FP values, and these changed very little upon target hybridization. Because of these problems, we decided to evaluate another recently described class of nucleic acid analogs, locked nucleic acids (LNAs) (5). The main structural feature of these probes is the presence of an additional methylene bridge linking the 2′ hydroxyl group of an RNA monomer to the 4′ carbon of the ribose ring. The presence of this bridge ‘locks’ the sugar ring in one fixed conformation (3′-endo), which is the conformation observed for that ring in RNA or DNA hybrids. On the basis of NMR studies of LNA/DNA duplexes, it has been concluded that the main factor contributing to the extraordinary high stability of LNA-containing duplexes is a local change of the phosphate backbone geometry that favors a much higher degree of base stacking (6).
LNA probes have already been used for solid-phase single nucleotide genotyping (7,8). In these methods, the authors covalently attached LNA probes to the surface of microtiter plates and used them to capture PCR products containing sequences surrounding the Leiden V and the ApoB 100 R3500Q mutation. An ELISA format was used to detect the hybridization event, and the authors reported excellent specificity and sensitivity of the assays. For high-throughput genotyping applications, a homogeneous method requiring as few post-amplification steps as possible would be of great interest. To our knowledge, no hybridization-based homogeneous method for SNP genotyping using LNA probes has been reported to date. In this article we present the first such approach. It is based on our observations that the FP values of a fluorescent dye such as rhodamine or hexachlorofluorescein (HEX) atached to an LNA probe increase significantly upon the hybridization of the probe to a target DNA molecule. Due to the very high affinity of the LNA molecules, very short probes can be used. In this article, we demonstrate that hexamer and heptamer LNA probes hybridize with very high affinity to perfectly complementary targets, and at the same time show an extraordinary specificity, allowing the discrimination of targets that differ by a single base.
MATERIALS AND METHODS
LNA probes and other reagents
All LNA probes used in this work were synthesized by Proligo, LLC (Boulder, CO). The sequences of all probes used in this work are shown in Table 1. All LNA probes were 5′ end-labeled with either rhodamine or HEX. The latter dye was attached to the 5′ end of the LNA probes using its commercially available phosphoramidite (Glen Research, Sterling, VA). Because of the instability of rhodamine to the concentrated ammonia treatment during the deprotection step, this dye was attached to 5′-amino modified LNA probes after their complete deprotection with concentrated ammonia. The reactive form of the rhodamine was an N-hydroxysuccinimidyl ester, available from Molecular Probes (Eugene, OR). All probes were purified by ion-exchange HPLC to a final purity of at least 90%. Stock solutions were prepared in water and stored frozen between experiments. The concentrations of the labeled probes were determined based on absorbance measurements at the absorbance maxima of the dyes. Synthetic DNA molecules were obtained from Oligos, Etc. (Wilsonville, OR).
Table 1. Melting temperatures and FP dynamic ranges of 5′-Rho LNAs.
| LNA name, sequence (5′ to 3′) | Perfect-match Tm (°C) | FP dynamic range (mP) | Mismatch Tm (°C) and position |
|---|---|---|---|
| LNA601, Rho-GTCGCC | 53 | 110 | <0, G/T center |
| LNA602, Rho-GTCACC | 50 | 140 | <0, A/C center |
| LNA603, Rho-CATGCC | 26 | 35 | ND |
| LNA604, Rho-TATGCC | 21 | 60 | ND |
| LNA605, Rho-TCTGAC | 26 | 60 | ND |
| LNA606, Rho-GCGGAG | 62 | 110 | 31, G/T center; <0, G/A center |
| LNA607, Rho-ACGGAG | 53 | 110 | 24, G/T center; <0, G/A center |
| LNA608, Rho-ATGGTG | 43 | 115 | 17, G/T center; <0, G/A center |
| LNA609, Rho-AAGTAG | 25 | 90 | ND |
| LNA610, Rho-TAGTAT | 19 (300 nM) | 85 (300 nM) | ND |
| LNA701, Rho-CCCGCCC | >85 (250 nM) | 55 (250 nM) | 72, G/T center |
| LNA702, Rho-TATTTTA | 10 (300 nM) | 35 (300 nM) | ND |
| LNA709, Rho-TAAAATA | 15 | 85 | ND |
| LNA713, Rho-AAGTAGT | 34 | 130 | ND |
| LNA714, Rho-TAGTATT | 18 | 110 | ND |
| LNA715, Rho-TAGTATG | 32 | 120 | <0, T/G center |
| LNA807, Rho-TAGTATTT | 28 | 130 | <0, T/G center |
Unless otherwise indicated, probe and target concentrations were 50 nM; ND, not determined; <0, no LNA/DNA duplex was detected even at the lowest temperatures employed (4°C).
Hybridization experiments
Hybridization experiments using synthetic DNA molecules were generally carried out in a buffer containing 50 mM HEPES pH 7.5, 100 mM NaCl. Room-temperature hybridizations and melting curve measurements were performed in a fluorescence spectrophotometer (Fluoromax-2 or Fluorolog-3, JY Horiba, Edison, NJ) equipped with polarizers and temperature bath.
PCR and SNP genotyping
Thermostable DNA polymerase (Taq) was procured from Qiagen (Valencia, CA) and used in the manufacturer-supplied buffer. PCR primers FS (5′ AGT CAA GGA CAC CGA GGA A, phosphorothioated at the first five bases) and R (5′ GCT TCT TAC AGT GCT GGA TGT) were synthesized by Oligos Etc. The incorporation of one partially phosphorothioated primer into the double-stranded PCR product allowed the selective enzymatic hydrolysis of the opposite strand by T7 gene 6 exonuclease (9). The exonuclease was from USB Corporation (Cleveland, OH).
RESULTS
Detection of LNA/DNA hybridization by FP
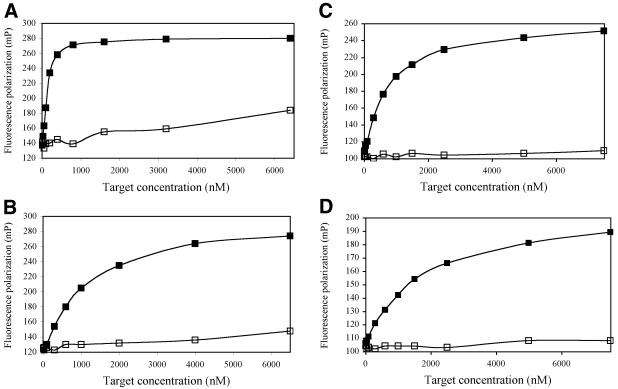
We have previously reported the successful use of FP to detect the hybridization of rhodamine-labeled PNA probes to DNA targets (4). We have now extended these observations to include fluorescently labeled LNA probes. The two fluorescent dyes that we have found to give the best FP responses upon hybridization were rhodamine and HEX. Since LNA molecules have been reported to form exceedingly stable duplexes with complementary target nucleic acids, we decided to test the hybridization behaviour of very short, dye-labeled probes. Thus, a series of dye-labeled hexamer and heptamer probes were obtained from Proligo, LLC. Figure 1 shows the results obtained when fixed concentrations of 50 nM of four different, rhodamine labeled LNA hexamers were titrated with increasing concentrations of synthetic target DNA molecules at room temperature. The targets were either perfectly complementary to the LNA probes, or contained a single nucleotide substitution that upon hybridization would give rise to a single nucleotide mismatch within the LNA/DNA duplex. A 50 nM probe concentration was chosen because this is within the same order of magnitude as the yield of a typical PCR amplification reaction. As expected, LNA probes with higher GC content (Fig. 1, LNA 601) were saturated at lower DNA target concentrations than probes that were more AT-rich.
Figure 1.
Room temperature hybridizations of 5′ rhodamine-labeled LNAs. LNA probes (50 nM in 50 mM HEPES, pH 7.5, 100 mM NaCl) were mixed thoroughly with the respective amounts of DNA targets and the steady-level FP was recorded in a fluorometer. Shown are the titration curves for (A) LNA 601, 5′ Rho-GTCGCC; (B) LNA 602, 5′ Rho-GTCACC; (C) LNA 603, 5′ Rho-CATGCC; and (D) LNA 604, 5′ Rho-TATGCC with their perfect-match (filled squares) and single-base mismatch (empty squares) DNA targets.
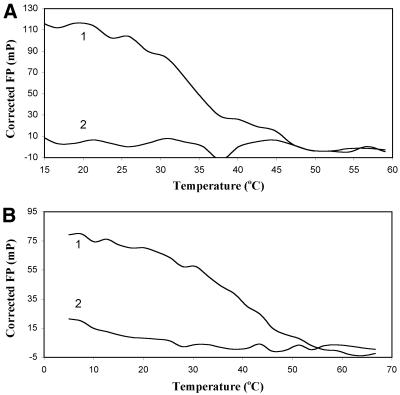
In order to evaluate the stability of the LNA/DNA duplexes, we performed melting curve measurements by recording the FP as a function of temperature for free LNA probes or mixtures of LNA probes and DNA target. Figure 2 shows the melting behavior of two probes, labeled with rhodamine and HEX, respectively. LNA 715 (5′ Rho-TAGTATG, Fig. 2A) is an AT-rich heptamer which nevertheless possesses sufficient binding affinity for its perfect-match target at 50 nM, combined with perfect discrimination of center T/G mismatch (Fig. 2A, curve 2). LNA 607H (5′ Hex-ACGGAG, Fig. 2B) is a GC-rich Hex-labeled hexamer which also presents high melting temperature (Tm) and excellent mismatch discrimination (G/T mismatch, Fig. 2B, curve 2). During the course of our studies, we noticed that although HEX-labeled LNA probes exhibited ample FP dynamic range, their values were generally lower compared with those of rhodamine-LNAs. On the other hand, FP detection of hybridization of LNAs labeled with fluorescein was practically impossible due to the very small (10–15 mP) changes in polarization (data not shown).
Figure 2.
Melting curve detection by FP. The experiment was performed as described in the Materials and Methods. Shown are (A) LNA 715, 5′ Rho-TAGTATG; and (B) LNA 607H, 5′ Hex-ACGGAG at 50 nM concentration in the presence of 50 nM perfect-match (curve 1) or mismatch (curve 2) targets. FP correction was performed by subtraction of the free-probe FP at each temperature.
Affinity and GC-content of LNA probes
In order to assess the possibility of genotyping a large number of SNPs with extremely short LNA probes, we undertook a Tm study of a series of rhodamine-labeled short (6–7 nt) LNAs. The sequences of those probes were designed such that one could obtain a comprehensive picture of the affinities of such probes across the whole GC-content range. Table 1 shows the results for 17 different probes. In addition to the Tm values for perfectly matched hybrids, the table lists the FP dynamic ranges obtained during the melting experiments as well as results on mismatch discrimination. The mismatch positions were at or close to the center of the probes, as we expected this to have the most destabilizing effect on the hybrids. A quick survey of the GC-rich sequences (for example, LNA 601, 602, 606, 607 and 701) indicates that they possess enough affinity to be employed for SNP genotyping as hexamers. LNA hexamers with only 2 GCs (LNA 609) had marginal affinity which, however, increased sufficiently once an extra base was added (LNA 713). On the other hand, LNAs with only a single G or C required a total length of an octamer. Further studies would be needed to establish more precise and predictive rules for Tms of LNA/DNA hybrids as a function of the LNA sequence. As seen in Table 1, the FP dynamic ranges were variable but generally adequate (55–140 mP), with only few exceptions (e.g. LNA 603, 35 mP).
SNP genotyping of PCR products
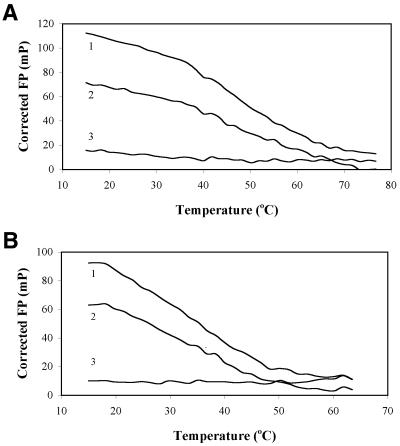
To demonstrate the possibility of single nucleotide genotyping of PCR products obtained from genomic DNA using rhodamine labeled hexamer LNA probes, we amplified a 107 bp sequence from genomic DNA samples using PCR primers FS and R. This product contains an SNP, a C to T transition, and the genomic DNA samples used in the experiment had been genotyped previously. PCR reactions were performed with four different templates: (i) no-template control (ntc); (ii) homozygous C human genomic DNA sample; (iii) heterozygous C/T human genomic DNA sample; and (iv) homozygous T human genomic DNA sample. One of the PCR primers used in the amplification, FS, contained five phosphorothioate bonds at its 5′ end. These rendered the corresponding strand of the double-stranded PCR product resistant to treatment with the T7 gene 6 exonuclease, which was added to the PCR mixture after the amplification to hydrolyze the opposite strand and generate a single stranded product (9). We had previously established that the LNA probes are completely resistant to this exonuclease, both in the single- and double-stranded (LNA/DNA hybrid) form (data not shown). To generate LNA–DNA melting curves by FP, PCR reaction products were first digested for 10 min with T7 gene 6 exonuclease, and then mixed with 50 nM probe in fluorometer cuvette. Duplex melting was effected by slow temperature increase in a water bath. The two LNA probes exhibited excellent discrimination, displaying typical melting behavior only where there was perfect match between probe and genotyping sample (Fig. 3).
Figure 3.
SNP genotyping by FP melting curve measurement. The experiment was performed as described in Results. Shown are the melting curves when PCR reactions were analyzed with (A) LNA 601, 5′ Rho-GTCGCC; and (B) LNA 602, 5′ Rho-GTCACC. In both (A) and (B), curves denote the following: 1, homozygous perfect-match DNA; 2, heterozygous DNA; and 3, homozygous DNA of opposite genotype.
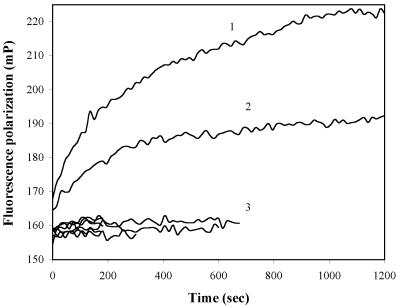
In addition to performing the genotyping by melting curve analysis, we tested the possibility of SNP detection by real-time FP measurement. Thus, the rhodamine-labeled probes 601 and 602 were added to separate aliquots of the PCR product, and the FP signals of the dye were recorded over 20 min. The result obtained from the analysis with probe LNA 601 is shown in Figure 4. We observed no changes in the FP signals upon addition of the double-stranded PCR products, in the absence of exonuclease. Then, following the addition of T7 gene 6 exonuclease to a final concentration of 2 U/µl, an increase of the FP was observed when the generated single-stranded PCR products were completely complementary to the LNA probe, but no changes were seen with the PCR product of the opposite genotype, containing a single nucleotide mismatch with the probe used. The heterozygous sample gave an intermediate increase of the FP signal, as expected.
Figure 4.
Real-time SNP genotyping by FP. The experiment was performed as described in Results. Shown are the FP time-courses when PCR reactions were analyzed with LNA 601, 5′ Rho-GTCGCC. Only the homozygous (curve 1) and heterozygous (curve 2) DNA samples show an FP increase upon exonuclease digestion. All other traces (curve cluster 3: no-template control, opposite genotype, no exonuclease) remain flat.
DISCUSSION
LNAs are a new class of synthetic DNA/RNA analogs. These molecules display a very high affinity for complementary nucleic acid targets, both RNA and DNA. At the same time, their sensitivity for mismatch discrimination makes them uniquely suited for hybridization-based SNP genotyping. These two properties could allow the generation of an essentially complete set of genotyping probes, applicable to the typing of the majority of SNPs. The size of this universal library will be determined by the length of the probes: 1024 for a library of all possible pentamers; 4096 for all possible hexamers; but 16 384 for all possible heptamers. The former two appear reasonable, but the latter would be prohibitively expensive and hard to manage. Ideally, all the probes of such universal genotyping probe libraries will hybridize at or close to room temperature, and provide excellent mismatch discrimination. The relatively low affinities of DNA probes and of all other nucleic acid probes available previously, including PNAs, are not high enough to allow the generation of such libraries of short probes fulfilling these criteria.
In our initial evaluation of the hybridization properties of LNA molecules we used probes containing eight to nine bases, but after observing their extraordinary high affinities for the target DNAs, we decided to explore the idea of a library of hexamer LNA probes for SNP typing. As the method for hybridization detection, we decided to explore an approach that we have previously successfully employed to detect the hybridization of PNA probes to DNA targets. This approach uses rhodamine-labeled probes and relies on measuring the FP changes of this dye upon probe hybridization. As shown in the present paper, this approach proved highly suitable for detecting the formation of LNA/DNA hybrid formation in a homogeneous solution. At the same time, the sensitivity of the LNA probes for single nucleotide mismatches allowed the discrimination of perfectly matched and mismatched targets with high efficiency. Application of LNAs in this assay was additionally facilitated by their nuclease stability and general ease of handling. Unlike PNAs, which occasionally tend to aggregate and have limited solubility, we encountered no such issues when working with LNA.
From our results, it appears that in theory it may be possible to generate a useful, universal set of genotyping reagents based on LNA probes. Based on our results, all probes containing at least three G or C would be hexamers, and probes containing two or one G or C would be heptamers or octamers. For multiplexing experiments, two identical libraries will be needed, one with all probes labeled with rhodamine and the other with all probes labeled with HEX. Sequences that will be especially problematic to genotype will be the ones that encompass palindromes because self-complementary LNAs are expected to have greater affinity to themselves than to their perfect-match DNA targets. In reality though, at the relatively high current prices of LNA oligomers, the synthesis of such universal reagent set is prohibitively expensive.
REFERENCES
- 1.Shi M. (2001) Enabling large-scale pharmacogenetic studies by high-throughput mutation detection and genotyping technologies. Clin. Chem., 47, 164–172. [PubMed] [Google Scholar]
- 2.Landegren U., Nilsson,M. and Kwok,P.-Y. (1998) Reading bits of genetic information: methods for single-nucleotide polymorphism analysis. Genome Res., 8, 769–776. [DOI] [PubMed] [Google Scholar]
- 3.Nikiforov T.T. and Jeong,S. (1999) Detection of hybrid formation between peptide nucleic acids and DNA by fluorescence polarization in the presence of polylysine. Anal. Biochem., 275, 248–253. [DOI] [PubMed] [Google Scholar]
- 4.Nikiforov T.T., Coffin,J., Jeong,S., Simeonov,A.M. and Bi,X. (2001) New applications of fluorescence polarization for enzyme assays and in genomics. Proceedings of SPIE, 4255, 94–105. [Google Scholar]
- 5.Koshkin A.A., Nielsen,P., Meldgaard,M., Rajwanshi,V.K., Singh,S.K. and Wengel,J. (1998) LNA (locked nucleic acid): an RNA mimic forming exceedingly stable LNA:LNA duplexes. J. Am. Chem. Soc., 120, 13252–13253. [Google Scholar]
- 6.Nielsen K.E., Singh,S.K., Wengel,J. and Jacobsen,J.P. (2000) Solution structure of an LNA hybridized to DNA: NMR study of the d(CT(L)GCT(L)T(L)CT(L)GC):d(GCAGAAGCAG) duplex containing four locked nucleotides. Bioconjugate Chem., 11, 228–238. [DOI] [PubMed] [Google Scholar]
- 7.Ørum H., Jakobsen,M.H., Koch,T., Vuust,J. and Borre,M. (1999) Detection of the factor V Leiden mutation by direct allele-specific hybridization of PCR amplicons to photoimmobilized locked nucleic acids. Clin. Chem., 45, 1898–1905. [PubMed] [Google Scholar]
- 8.Jacobsen N., Fenger,M., Bentzen,J., Rasmussen,S.L., Jakobsen,M.H., Fenstholt,J. and Skouv,J. (2002) Genotyping of the apolipoprotein B R3500Q mutation using immobilized locked nucleic capture probes. Clin. Chem., 48, 657–660. [PubMed] [Google Scholar]
- 9.Nikiforov T.T., Rendle,R.B., Kotewicz,M.L. and Rogers,Y.-H. (1994) The use of phosphorothioate primers and exonuclease hydrolysis for the preparation of single-stranded PCR products and their detection by solid-phase hybridization. PCR Methods Appl., 3, 285–291. [DOI] [PubMed] [Google Scholar]