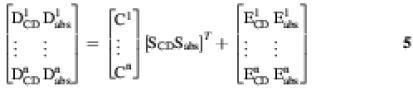Abstract
A successful application is reported of the multivariate curve resolution alternating least-squares method (MCR-ALS) for the analysis of nucleic acid melting and salt-induced transitions. Under conditions where several structures co-exist in a conformational equilibrium, MCR-ALS analysis of the UV and circular dichroism (CD) spectra at different temperatures, ionic strength and oligonucleotide concentration allows for the resolution of concentration profiles and pure spectra of the different species. The methodology is illustrated by the case of the cyclic oligonucleotide d<pTGCTCGCT>. The melting transition of this molecule at different oligonucleotide concentrations was studied at 0, 2 and 10 mM MgCl2 by UV and CD spectroscopy. In addition, salt titration experiments were carried out at 21.0 and 54.0°C. The MCR-ALS analysis indicates that three different conformations of this molecule co-exist in solution. In agreement with previous NMR studies, these conformations were assigned to a monomeric dumbbell-like structure, a dimeric four-stranded conformation and a disordered (random coil) structure. The MCR-ALS methodology allows for a detailed analysis of how this equilibrium is affected by temperature, salt and oligonucleotide concentration.
INTRODUCTION
The remarkable conformational flexibility of nucleic acids allows for the formation of a great variety of structures besides the canonic B-DNA duplex. Among these structures, those involving more than two strands have received considerable attention in the last decade (1–3).
There are two main reasons for the intense interest in such multistranded structures. First, there is more than circumstantial evidence for their biological role in replication, transcription and recombination (4,5). Second, several multi-stranded motifs are used as the basis of some therapeutic approaches. In the antigene strategy, triplex-forming oligonucleotides exhibit sequence-specific recognition of duplex DNA and can be used to modulate gene expression (6). A comprehensive review has recently appeared discussing the design of agents targeted toward a structure-specific molecular recognition of DNA triplexes or quadruplexes (7). The G-rich sequence of human telomeric DNA can adopt a G-tetraplex structure in vitro that blocks the telomerase-catalyzed telomer elongation (8). Since this enzyme is expressed in tumor cells but not in most somatic cells, it is a prime target for cancer therapy. Several small molecules that stabilize G-tetraplexes have shown themselves to be strong telomerase inhibitors and are promising antitumor drugs (9,10). On the other hand, the disruption of tetraplex forms by small molecules is a potential route to therapeutic intervention against triplet repeat diseases such as the fragile X syndrome (11). Finally, G-tetraplex-forming oligonucleotide aptamers are potent thrombin (12) or HIV-1 integrase (13) inhibitors.
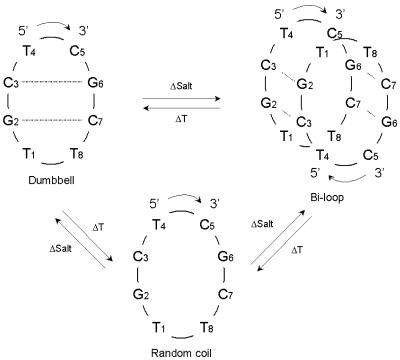
Numerous three- (14) and four-stranded (15) structures have been solved either by X-ray crystallography or by NMR spectroscopy. However, when the aim is only to assess the formation of such structures and to evaluate their stability under certain conditions, low resolution alternatives such as UV absorption spectroscopy or circular dichroism (CD) are the techniques of choice (16,17). These techniques have the advantage of requiring much smaller amounts of oligonucleotides and allowing easier performance of experimental measures. The drawback is that the interpretation of the experimental results may be difficult if more than two structurally different species are involved in an equilibrium and their occurrence strongly depends on experimental conditions such as ionic strength, type of cation, pH and temperature. A thorough review of energetic and structural considerations for low- and high-order nucleic acid systems can be found in the literature (18). Certain cyclic oligonucleotides, as shown by X-ray diffraction and NMR spectroscopy, can self-associate to form a new dimeric four-stranded structure, known as the bi-loop motif, containing G:C:G:C and A:T:A:T tetrads (19–21). The same tetrads, although differently arranged, have also very recently been described (22). In the solution NMR study of the cyclic octamer d<pTGCTCGCT> at low oligonucleotide concentration (<1 mM in water), we perceived the predomination of a dumbbell-like structure. But at higher oligonucleotide concentration, two molecules dimerize to form the quadruplex (20,21). When the temperature is increased, the two species evolve to a third one, the random coil structure.
Preliminary UV and CD studies of this cyclic octamer were undertaken to assess the occurrence of these species under different conditions (water or different salt concentrations, temperatures). The studies were also designed to evaluate, at the dilution conditions used (5 µM), the thermodynamic stability of the structured dumbbell and bi-loop species (23). The differences perceived between the UV and CD spectra recorded under different conditions made it difficult to ascertain whether the spectrum of a unique species was being observed or that of an equilibrium mixture of two or three species. By way of example, in the UV melting profile in salt medium, an initial decrease in chromicity was followed by a final increase in the UV absorption. The anomalous shape of the curve, with a partially inverted denaturation profile (24) suggested that what was being recorded was the simultaneous melting of the two structured species into the random coil. The problem therefore, we assumed, was to describe how these different equilibria are reached, and also to achieve accurate speciation of the system with regard to temperature, salt and oligonucleotide concentration changes.
Traditionally, conformational equilibria changes have been monitored by UV or CD spectroscopies using a single wavelength (univariate data analysis). This approach has clear drawbacks, the most important being the assessment of the number of different conformations present in an experiment when no selective wavelengths are available and there is a lack of appropiate reference spectra. These difficulties can be overcome by multiple wavelength spectroscopic monitoring of these conformational transitions and the application of appropriate multivariate data analysis methods. One such method is the recently proposed multivariate curve resolution method based on alternating least squares or MCR-ALS (25,26 and references therein) and on factor analysis techniques (27).
This paper describes the application of MCR-ALS to the study of the conformational transitions of d<pTGCTCGCT>. The goal is the assessment and resolution of all the possible conformations, including intermediate states, and the estimation of their pure spectra and concentration profiles when temperature and salt concentration are changed. A certain challenge is involved in checking whether MCR-ALS results could describe the equilibria between the dimeric bi-loop and monomeric dumbbell structures in salt media and their denaturation into random coil structures. Hence, several experimental approaches using different initial conditions were used. First, melting experiments were run at different salt and oligomer concentrations to monitor their influence in conformational changes. Second, salt-induced conformation transitions were studied, carrying out salt titration experiments at two different temperatures.
MATERIALS AND METHODS
Sample preparation
Cyclic octamer d<pTGCTCGCT> was synthesized as reported (28) and purified by standard liquid chromatography. Oligonucleotide sodium salt solutions were prepared in Ultrapure water (Millipore). Appropriate volumes of buffer [200 mM piperazine-N,N′-bis(2-ethanensulfonic acid), PIPES, disodium salt, pH 7.0] and stock salt solutions (200 mM MgCl2 and 2 M NaCl) were added in salt medium experiments. Oligonucleotide concentration (referred to the monomer) ranged from 5 to 70 µM, and was determined by absorbance measurements at 260 nm at room temperature considering a micromolar extinction coefficient of 64.5 OD/(µmol·ml) in water (29). Samples for melting studies were heated at 90°C for 5 min and allowed to renaturalize, cooling slowly until room temperature. Oligonucleotide samples were kept at 4°C until their use.
CD and UV absorption experiments
CD and UV molecular absorption spectra (240–330 nm) were recorded on a Jasco J-720 spectropolarimeter equipped with Neslab RET-110 temperature control unit. Instrumental control, data acquisition and analysis were carried out using personal computers. The work parameters during the registration of the spectra were bandwidth of 1 nm, sensitivity of 10 mdeg, scan speed of 20 nm/min, response of 8 s, step resolution of 1 nm and two averaged spectra. For all measurements, a Hellma quartz cell (path length of 1.0 cm and volume of 750 µl) was used.
Melting experiments were carried out measuring UV absorption and CD spectra at 3°C increments, using a temperature rate of 20°C/h. Each sample was initially stabilized at the starting temperature for 15 min. Three representative melting experiments at 0, 2 and 10 mM MgCl2 concentration (in experiments 1, 2 and 3, respectively) and 100 mM NaCl were selected to study the effect of temperature on equilibria between oligonucleotide conformations. In order to study and remove possible background contributions, melting experiments were also performed on samples without oligonucleotide. Background contributions were especially relevant in melting UV spectra because PIPES buffer UV absorption changes with temperature.
Salt titration experiments were carried out measuring UV absorption and CD spectra at each MgCl2 and NaCl concentration until no changes were observed in two consecutive spectra. These experiments were performed at 54°C (experiment 4) and 21°C (experiment 5) and MgCl2 concentrations ranging from 0 to 7.5 mM and 0 to10 mM, respectively. The first spectrum measured at each titration experiment was the one corresponding to oligonucleotide in buffer solution. The next recorded spectra corresponded to successive additions of stock salt solution (MgCl2 and NaCl). Salt experiments were carried out at 21 and 54°C because preliminary results showed that at these two temperatures the system was rather complex and difficult to be resolved when only melting experiments were analyzed (see Results and Discussion).
Nucleation equilibria between ordered conformations were studied in melting experiments at 5, 10, 20, 50 and 70 mM oligomer concentrations (in experiments 6, 7, 8, 9 and 10, respectively) in pure water and in salt media (10 mM MgCl2, 100 mM NaCl).
Multivariate curve resolution
Experimental CD and UV absorption data were analyzed using the MCR-ALS procedure previously described (25,26,30,31). This procedure is used to estimate the concentration profiles and pure spectra for each spectroscopically active conformation present in melting and salt titration experiments. MCR-ALS procedure is based on factor analysis methods (27) and can be used to analyze spectroscopic data obtained in biochemical or biophysical process monitoring. All MCR-ALS calculations were performed using in-house MATLAB (version 6; The Mathworks Inc, Natick, MA) routines (codes are freely available at www.ub.es/gesq/ mcr/mcr.htm). Only a short description of the MCR-ALS procedure is given here.
Spectra recorded in one experiment are collected in a table or matrix D (Nr × Nw), Nr rows being the Nr spectra recorded at successive temperature or salt concentration values, and Nw columns being the number of wavelengths measured in every spectrum. Mathematically, the goal of MCR-ALS is the recovery of the concentration profiles matrix C and of the pure spectra matrix ST (the superscript T means the transpose of matrix S). By considering the multiwavelength extension of Lambert-Beer’s law (in matrix form),
D = C ST + E1
E is the matrix of residual CD signal or absorbance not explained by the model. E should be close to experimental error. The first step of the data analysis procedure is the estimation of the number of spectroscopically distinct conformations present in a particular experiment. This number of conformations (Ns) is estimated by rank analysis or singular value decomposition (SVD) of each data matrix D (32). In this estimation, it is assumed that singular values associated with experimental noise are significantly lower than those associated with systematic chemical data variance. Therefore, it is assumed that they can easily be distinguished from the plot of their magnitudes. When doubts about the number of conformations occur, MCR-ALS analysis is repeated for the different possibilities and the results finally evaluated in terms of data fitting and of quality of MCR-ALS-resolved profiles.
Equation 1 is solved iteratively by an ALS algorithm that calculates concentration C and pure spectra ST matrices optimally fitting experimental data matrix D using the proposed number of Ns conformations. This iterative optimization requires initial estimations either of C or ST, which can be obtained from evolving factor analysis (EFA) (33) or from pure variable detection methods (34). During the ALS optimization, several constraints were applied including non-negativity for concentration profiles C and for UV absorbance spectra profiles ST (not applied in case of CD spectra profiles), unimodality for concentration profiles C, and closure also for concentration profiles C. See previous works for a more detailed explanation of the ALS iterative optimization procedure (25,26,30,31). Convergence is usually achieved in few iterations and the final value of lack of fit is defined as
where dij are the absorbance data of spectrum i and wavelength j, and dij* are the MCR-ALS-recalculated data using the specified number of conformations Ns.
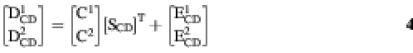
Concentration profiles C and pure spectra ST resolved for each conformation in the analysis of individual data matrices may differ from the true ones because of possible unresolved underlying factor analysis ambiguities (25,27). This means that concentration profiles and pure spectra may be only a solution within a band of feasible solutions that are bounded by the constraints applied in the calculation. Some of these ambiguity problems can be more easily solved by means of the simultaneous MCR-ALS analysis of several data matrices obtained under different experimental conditions (25). When a chemical system is monitored using two spectroscopies (e.g. CD and UV absorption), a row-wise augmented data matrix can be built up from the individual data matrices corresponding to each spectroscopy, Dabs and DCD, respectively. The dimensions of the new row-wise augmented matrix will be (Nr) × (Nwabs + NwCD), and the new linear model used for MCR-ALS analysis is
[DCDDabs] = [C][SCDSabs]T + [ECDEabs]3
Solving equation 3 for C and [SCDSabs] helps to resolve the species of the system, especially if their spectra are not well defined in one of the two spectroscopies simultaneously analyzed. For instance, in the study of the evolution of conformational equilibria, CD spectra are richer and with more features than UV absorption spectra. Therefore, the simultaneous analysis of both would allow a better resolution of UV absorption spectra.
In some other cases, one of the finally resolved components corresponds in fact to a mixture of two or more conformations due to rotational ambiguity problems (25) or to rank deficiency problems (35). In all these cases, and also to allow an improved resolution of the system, the simultaneous analysis of different independent experiments is usually very useful and powerful. For instance the model used for MCR-ALS simultaneous analysis of two different melting experiments at different salt concentrations monitored both by CD spectroscopy, giving experimental data matrices D1CD and D2CD, respectively, is described by the equation
In this case, better conditions for species resolution will be achieved when the two experiments are analyzed together compared with when each experiment is analyzed individually (see Results). We also used this kind of analysis in the present work for modeling UV absorption changes of background (PIPES salt) at low and high temperatures and at low and high salt concentrations.
Finally, a still more involved data analysis approach can be proposed for data acquired where different spectroscopies are applied simultaneously to multiple experiments. The linear model for n experiments studied by CD and UV absorption spectroscopies is described by equation
This kind of simultaneous analysis is the more powerful one and allows improvement of the resolution of very complex data structures, combining the profits of both approaches previously described. MCR-ALS analysis of row- and/or column-wise augmented data matrices has been shown to give more reliable solutions, eventually removing totally rotational ambiguities and rank deficiency problems. See previous references for a more thorough discussion of these topics (25,26).
Thermodynamic data analysis
Calculation of thermodynamic data for the equilibria between conformations is traditionally carried out by conversion of experimental absorbances at single wavelength versus temperature profiles into molar oligomer fraction versus temperature curves. However, this conversion is highly dependent on base line drifts and on the assumption that absorbance and CD signal changes at any temperature are directly proportional to the reaction extent at that particular temperature (36,37). Here, thermodynamic parameters were directly estimated instead from MCR-ALS-resolved concentration profiles. This more powerful multivariate approach eliminates base line problems and takes advantage of the improved resolution of concentration profiles. It allows a better estimation of melting temperatures and of the thermodynamic parameters because of signal averaging improvement and fewer error propagation problems. First, Tm values were estimated from the intersection of MCR-ALS-estimated concentration profiles of the different conformations resolved in melting experiments. Mean error deviation of these Tm values was estimated to be approximately ±0.7°C from two replicated independent melting experiments of each system. This mean error deviation value agrees with estimations in previous works (31). Equilibrium constants at the different temperatures were also directly evaluated from the ratio between concentration values of the two conformations resolved at the same temperature by MCR-ALS. Then, free energy ΔG values for the corresponding melting equilibrium between the two different conformations were roughly estimated from temperature dependence of equilibrium constants. In Table 1 more details about thermodynamic evaluations are given. Thermodynamic values are given here to corroborate the reliability of the resolved concentration profiles from the proposed MCR-ALS method. Additional work is pursued to validate the proposed method as a general method for the evaluation of thermodynamic parameters
Table 1. Melting and dimerization thermodynamic data.
| Coligoa (µM) | Dumbbell melting equilibrium (in water) | Bi-loop melting equilibrium (in salt medium) | |
|---|---|---|---|
| Tm (°C)b | ΔG (kJ/mol)c | Tm (°C)d | |
| 5 | 48.0 | –5.7 | 54.8 |
| 10 | 47.8 | –5.0 | 57.0 |
| 20 | 47.6 | –4.9 | 60.6 |
| 50 | –e | –e | 63.4 |
| 70 | 48.4 | –4.7 | 64.7 |
| –5 (1)f | |||
aTotal concentration of oligonucleotide in melting experiments in water and salt media (10 mM MgCl2).
bMelting temperatures obtained from crossing points of the resolved concentration profiles of the two conformations (dumbbell to random coil) involved in melting equilibrium in water. A standard deviation value of ± 0.7°C was calculated.
cFree energy values for the melting equilibrium in water calculated from the temperature dependence of the equilibrium constant estimated where the concentrations of the conformations in equilibrium have significant concentrations (i.e. >15%).
dMelting temperatures obtained from crossing points of the resolved concentration profiles of the two conformations (bi-loop to random coil) involved in melting equilibrium in salt.
eNo experimental estimation of Tm was done in this case.
fEstimated value of free energy for the melting equilibrium (dumbbell to random coil) in water from the average of the corresponding free energy values obtained at 0, 10, 20 and 70 µM concentration of the oligonucleotide. Standard deviations of these values is also given in parentheses.
RESULTS AND DISCUSSION
Melting experiments in water medium
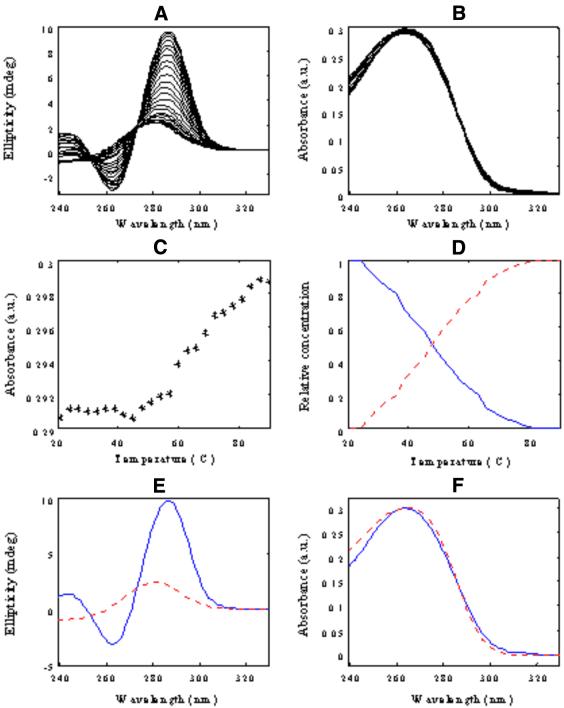
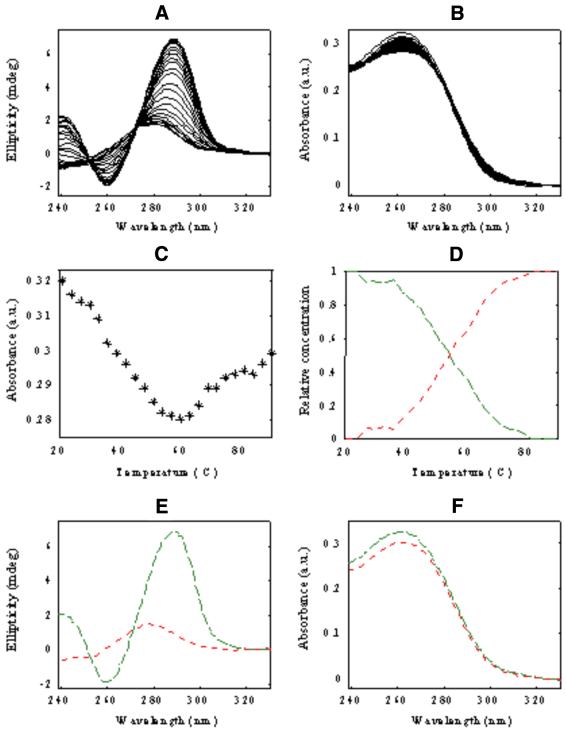
Thermal denaturation of d<pTGCTCGCT> was studied first in pure water, i.e., 0 M MgCl2 and 0 M NaCl (experiment 1). CD and UV absorbance spectra recorded in this experiment (Fig. 1A and B) were arranged in two data matrices [Dm,0CD] and [Dm,0UV], respectively. The superscripts refer to melting experiments and to no salt added (salt concentration is equal to zero) and the subscripts refer to the spectroscopic method used. The dimensions of both matrices were 24 rows (number of different temperatures/number of spectra) and 92 columns (number of wavelengths). CD bands at 21°C showed two maxima at 243.0 and 286.5 nm and a minimum at 263.0 nm. When temperature was raised, a gradual spectral variation was observed. The CD spectrum for the denatured conformation at the highest temperature showed the lowest intensity signal and only one maximum at 278 nm. Changes in UV absorption spectra were not so dramatic as those for CD data. Variations in CD and UV absorption could also be observed by recording data ellipticity at 286 nm and UV absorption at 264 nm (univariate analysis, as shown in Fig. 1C). Melting profiles obtained in this way agreed with the presence of a single transition in the studied temperature range (Tm value ∼50°C).
Figure 1.
Melting of d<pTGCTCGCT> oligomer in pure water (experiment 1). (A) CD data [Dm,0CD], (B) UV molecular absorption once the blank contribution is removed [Dm,0UV], (C) absorbance versus temperature at 264 nm, (D) concentration profiles [Cm,0], (E) pure CD spectra [STCD] and (F) pure UV molecular absorption spectra [STUV]. Blue continuous line, monomeric dumbbell conformation; red dotted line, random coil conformation.
MCR-ALS was applied to check the validity of the conclusions derived from this single wavelength data analysis. Rank analysis by SVD of the whole CD spectra (data matrix [DCDm,0]) indicated the presence of two components (i.e., Ns = 2). Accordingly, an initial estimation of the concentration profiles was obtained by EFA and later optimized by ALS using the constraints of non-negativity, unimodality and closure for concentration profiles (25,26,30,31). A Tm value of 48.2°C was obtained from MCR-ALS-resolved concentration profiles, which was in agreement with the value obtained from single wavelength data analysis. The ALS lack of fit calculated for the model with two components was 3.0%, which was considered reasonably good, taking into account the higher noise levels of CD data. MCR-ALS was then applied to analyze UV molecular absorption data [Dm,0UV]. In this case, the resolved profiles were slightly worse than those described above in the analysis of CD data. Resolved concentration profiles did not show a smooth shape like those for CD data, which made it more difficult to estimate an accurate value for Tm. The reason for this difficulty was related to the small UV absorbance variation observed during the melting experiment, the variation being hidden by the experimental noise contribution. When CD and UV absorption data were simultaneously analyzed according to equation 5, the resolution of UV absorption data improved significantly. Figure 1D–F shows the resolved concentration profiles [Cm,0] and pure CD [STCD] and UV absorption spectra [STUV]. ALS lack of fit for the simultaneous analysis was 2.0%.
According to previous results based on NMR spectroscopy (20,21) carried out in the absence of salt, the two resolved components in Figure 1D–F can be assigned to the dumbbell and the random coil conformations, respectively (Scheme 1). From the resolved concentration profiles, a Tm value of 50.4°C was estimated. The two resolved pure CD and UV absorption spectra showed spectral features explaining experimental spectra at low and high temperatures.
Scheme 1. Conformations of d<pTGCTCGCT> and the equilibria between them when temperature, salt and oligonucleotide concentration are changed [according to Lane et al. (18) and Salisbury et al. (19)].
Melting experiments in salt media
In order to study the influence of salt media on the denaturation equilibria, two melting experiments (experiments 2 and 3) were carried out at 10 and 2 mM MgCl2, respectively, and 100 mM NaCl. CD and UV absorption spectra obtained at 10 mM MgCl2 (experiment 2) and arranged in data matrices [Dm,10CD] and [Dm,10UV], are given in Figure 2A and B. There were some spectral features that could not be explained satisfactorily using a single wavelength approach. Whereas the CD signal at 289 nm showed a continuous smooth decrease along the whole temperature range studied, UV absorbance at 263 nm decreased its value from 20 to ∼60°C and, finally, increased slightly from 60 to 90°C (Fig. 2C). Whereas information obtained from CD data seemed to indicate the presence of only two conformations, UV absorption data showed a behavior that cannot be explained by assuming the presence of only two conformations.
Figure 2.
Melting of d<pTGCTCGCT> oligomer in salt medium (10 mM PIPES pH 7, 10 mM MgCl2 and 100 mM NaCl; experiment 2). (A) CD data [Dm,10CD], (B) UV molecular absorption once the blank contribution is removed [Dm,10UV], (C) absorbance versus temperature at 263 nm, (D) concentration profiles [Cm,10], (E) pure CD spectra [STCD] and (F) pure UV molecular absorption spectra [STUV]. Green dashed line, dimeric bi-loop conformation; red dotted line, random coil conformation.
As in experiment 1, CD and UV absorption data from experiment 2 were analyzed using a row-wise data matrix arrangement, i.e., according to equation 5. Rank analysis of the row-wise matrix showed the presence of two components. Figure 2D–F shows the results obtained by MCR-ALS analysis. Lack of fit was 1.8%. The Tm value of 55.4°C agreed well with the value determined from univariate analysis of CD data. Resolved CD, [STCD] and UV, [STUV], spectra for the component at the highest temperature, 90°C, were very similar to the spectra previously obtained in pure water at the same temperature. Therefore, they were assigned to the same random coil conformation. The resolved pure CD spectrum for the component at 20°C showed band maxima shifted to 240.5 nm and 289.0 nm and a band minimum shifted to 259.5 nm. These values were not the same as those observed for the major conformation present at 20°C in water (experiment 1), which would indicate a different initial conformation at 10 mM MgCl2. According to previous studies, this initial conformation can be related to the bi-loop dimeric structure (Scheme 1; 23) formed by antiparallel association of two monomer molecules.
However, MCR-ALS results for experiment 2 (shown in Fig. 2D–F) did not explain satisfactorily the univariate melting profile previously observed. Since the absorptivity of the resolved UV absorption spectra for the initial ordered conformation is higher than the absorptivity of the resolved UV spectrum of the random coil conformation, the absorbance at any wavelength should always have decreased smoothly along the whole temperature range. However, the experimental spectra showed a clear increase of absorbance values at temperatures >60°C. This fact suggested that MCR-ALS results obtained from the analysis of experiment 2 according to equation 5, although mathematically acceptable (as reflected by the low lack of fit value obtained), could not entirely explain the experimental results.
MCR-ALS results for experiment 3 at 2 mM MgCl2 concentration (data not shown) were similar to those obtained for experiment 2 at 10 mM MgCl2 concentration. Rank analysis also showed the presence of only two components. MCR-ALS resolved concentration profiles were similar to those shown in Figure 2D, i.e., a major component at 20°C and another one at 90°C. As for experiment 2, resolved spectra for the major component at 90°C showed the same spectral features as those observed in water at higher temperatures. Therefore, this component was also related to the random coil conformation. Band positions of CD spectrum for the major component present at 20°C were different to those found in pure water, but also different to those found at 10 mM MgCl2. In fact, MCR-ALS-resolved CD spectra for the initial state at 2 mM MgCl2 showed two maxima at 242.0 and 288.0 nm, respectively, and a minimum at 261 nm. Therefore, these maxima and minimum values were located between those obtained in experiments 1 and 2. The CD band positions suggested that the initial state of the oligonucleotide at 2 mM MgCl2 could actually be a mixture of the conformations observed in pure water (dumbbell) and 10 mM MgCl2 (bi-loop). In other words, the MCR-ALS-resolved concentration profile for the first component would correspond to the sum of concentration profiles of, at least, two conformations. This problem is known in chemometrics as rank deficiency and several ways to circumvent it have been proposed (35). In the present paper, rank deficiency could be solved by the simultaneous analysis of several experiments at different initial conditions (see below).
Salt titration experiments at a fixed temperature were carried out and the experimental data analyzed by MCR-ALS. The aim of these experiments was to obtain a better understanding and resolution of the systems under study, taking into consideration previous proposals about the formation of dimeric bi-loop structures that predominate at high concentration salt media (Scheme 1) (20, 21) as well as the monomeric dumbbell structures that dominate in pure water media.
Salt titration experiments
The conformation found at low temperatures in pure water was also expected to be the main conformation at low temperatures in low salt concentrations. At high MgCl2 salt concentrations and low temperatures, the main conformation was also expected to be similar to that present at 10 mM MgCl2. MCR-ALS analysis of salt titration at 21°C (experiment 4, experimental data not shown) indicated the presence of two components. Concentration profiles showed a crossing point at 2 mM MgCl2 concentration, approximately. This value would suggest the presence, at the same initial conditions of melting experiment 3, of a mixture of ordered conformations. MCR-ALS analysis of salt titration at 54.0°C (experiment 5, experimental data not shown) gave poorer fitting results. Lack of fit value was now higher, near 7.0%, when analyzed according to equations 1 or 3, and it was possible to resolve (poorly) only two conformations. From previous results, it was apparent that at 54°C, and depending on salt concentration, the mixtures of three conformations can be present. A possible way to improve the resolution of the different conformations in this and other experiments is the simultaneous analysis of more than one experiment according to equations 4 and 5.
MCR-ALS simultaneous analysis of melting and salt titration experiments
Results of the simultaneous analysis of melting and salt titration experiments monitored using only one of the two spectroscopies (either CD or UV absorption, i.e., the model described in equation 4) are not given here for the sake of brevity. In any case, the results for the simultaneous analysis of CD and UV absorption data sets according to equation 5 were better. The analyzed data matrix consisted of the arrangement of 10 single data matrices corresponding to melting experiments at 0, 10 and 2 mM MgCl2 (experiments 1, 2 and 3) and salt titrations at 54 and 21°C (experiments 4 and 5). All of these were monitored by CD and UV molecular absorption. Additionally, background data matrices obtained at the same experimental conditions as the melting experiments but without changes in oligonucleotide concentration (UV absorption changes due to PIPES buffer) were also included in the simultaneous analysis. These background contributions were modeled appropriately with the inclusion of three additional components in the MCR-ALS model: one component is used to take into account temperature background changes in water; the other two components are used to take into account temperature background changes in salt media. Using this approach, lack of fit values and resolution of conformation profiles improved appreciably. Lack of fit for the global analysis was 6.1%, which was considered rather good taking into account the large number of independent experiments simultaneously analyzed and their variety. The results finally achieved are given in Figures 3 and 4. These results confirmed the presence of three independent oligonucleotide conformations, in agreement with what was proposed in Scheme 1. SVD analysis of augmented data matrices described by equations 4 and 5 already indicated a rank increase of the system due to the detection of these three conformations. This confirms that using the proposed approach of simultaneous analysis of appropriately designed independent experiments, the elimination of rank deficiency and rotational ambiguity problems is possible, in full agreement with previous results (25,26,30,31).
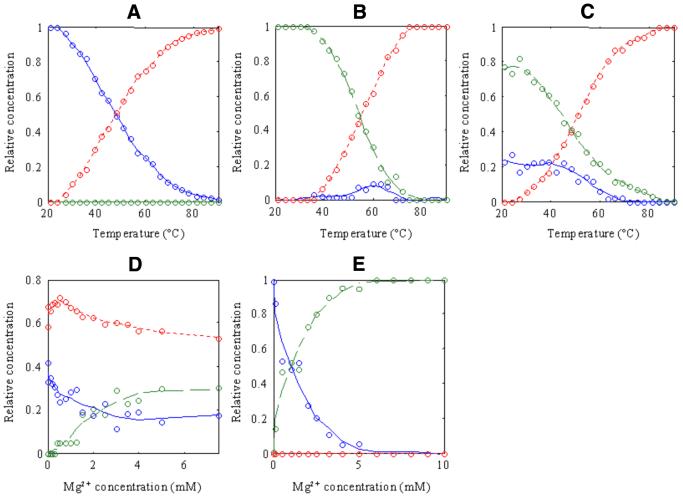
Figure 3.
MCR-ALS-resolved concentration profiles for the simultaneous data analysis of melting and salt titration experiments. (A) Melting at pure water [Cm,0], (B) melting at 10 mM PIPES pH 7, 10 mM MgCl2 and 100 mM NaCl [Cm,10], (C) melting at 10 mM PIPES pH 7, 2 mM MgCl2 and 100 mM NaCl [STUV], (D) salt titration at 54°C [Ct,54] and (E) salt titration at 21°C [Ct,21]. Blue continuous line, monomeric dumbbell conformation; green dashed line, dimeric bi-loop conformation; red dotted line, random coil conformation. Circles represent experimental points.
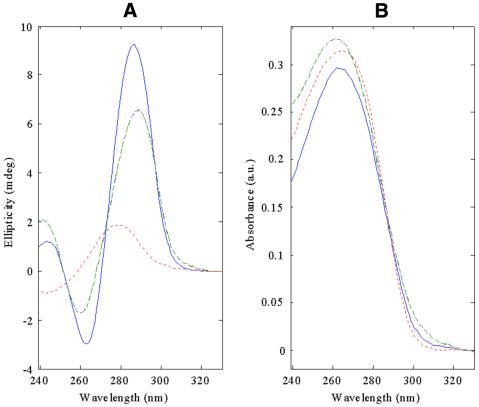
Figure 4.
MCR-ALS-resolved pure spectra for the three detected conformations in equilibria in melting and salt titration experiments. (A) CD pure spectra, (B) UV molecular absorption spectra. Lines and symbols as in Figure 3.
In experiment 1 (Fig. 3A), in pure water, only two conformations were present and assigned respectively to the monomeric dumbbell conformation (Tm = 48.1°C) and to the random coil conformations. This result agrees with the one previously obtained in the individual analysis of CD and UV absorption data from experiment 1 (Fig. 1). There, the system did not show any factor analysis ambiguity and gave equal results regardless of the model used. In experiment 2, however, three conformations were now detected and resolved (Fig. 3B). At the lowest temperatures only the conformation previously related to the dimeric bi-loop structure was detected (Tm = 54.8°C). But when the temperature was increased, and in addition to the formation of the random coil, a new minor conformation was now resolved with a spectrum matching that previously obtained in pure water at low temperatures (assigned to monomer dumbbell conformation). The presence of this minor conformation would now clearly explain the anomalous behavior observed in absorption melting curves at a single wavelength. Hence, the decrease of absorbance at temperatures <60°C would be related to the higher UV molecular absorptivity of the bi-loop in relation to the dumbbell and random coil conformations (Fig. 4B). At higher temperatures the dumbbell conformation will also disappear to yield the random coil conformation and the total absorbance increases slightly accordingly until the end of the melting process. This result was now achieved because the proposed simultaneous analysis allowed a better resolution of the minor dumbbell conformation (in experiment 2), not detected previously in the individual analysis of this experiment. Notoriously (and also differently to what was obtained when analyzed individually), three conformations were now perfectly resolved for experiment 3 (Fig. 3C) at 2 mM MgCl2. Dimeric bi-loop and monomeric dumbbell conformations were simultaneously present in this experiment from the beginning of the titration at the lowest temperatures, both decreasing their concentration when temperature was raised because of thermal denaturation. The corresponding concentration profiles of these two conformations could not be distinguished in the individual analysis of experiment 3 due to the so-called rank deficiency problems (35).
Figure 3D shows the evolution of the three conformations in equilibria, dumbbell, bi-loop and random coil at 54°C when salt concentration changes. The resolution of the concentration profiles of these three conformations was not possible when experiment 4 was analyzed individually because the experimental spectra were always a complex mixture of conformations. The resolved concentration profiles in Figure 3D showed that at 0 M salt and 54°C, a mixture of random coil (major conformation) and dumbbell (minor conformation) was present. This agreed with the values of their resolved concentration profiles at the same temperature of 54°C in melting experiment 1 (Fig. 3A). However, salt titration at 54°C could not be carried out up to 10 mM MgCl2 because of experimental difficulties and precipitation problems. The trend observed up to 8 mM showed a decrease of the random coil and dumbbell conformations and an increase of bi-loop conformation, which would also be in agreement with the values of concentration profiles in melting experiment 2 at the same temperature of 54°C (Fig. 3B). Additionally, the values of the concentration profiles at 2 mM MgCl2 in Figure 3D would agree with the values of concentration profiles in experiment 3 at 54°C (Fig. 3C) where random coil is the major conformation. At 21°C (Fig. 3E), no random coil conformation was detected at appreciable concentrations, and only the dumbbell to bi-loop transition was observed. The resolved concentration profile confirmed that at 2 mM MgCl2 and 21°C, a mixture of bi-loop (major conformation) and dumbbell (minor conformation) was present at the beginning of experiment 3 (Fig. 3C). Figure 3E also confirmed that at 10 mM MgCl2 and 21°C the oligonucleotide was only present in its bi-loop form, which was also observed at the beginning of experiment 2 in Figure 3B.
Finally, in Figure 4, pure CD and UV absorption spectra for the MCR-ALS resolved conformations are shown. Spectral features agreed with those observed experimentally. Hence, CD maxima for bi-loop conformations are located at 286 and 263 nm, for dumbbell at 289 and 260 nm and for random coil at 279 nm. The different intensities of the dumbbell and bi-loop conformation spectra explain the experimental spectra in all the experiments, including melting experiments 2 and 3 at 10 and 2 mM MgCl2 and salt titration experiment 4 at 54°C, which could not be satisfactorily explained by single wavelength analysis or by MCR-ALS multiwavelength analysis of their respective individual experiments.
Thermodynamic study of melting and of dumbbell to bi-loop dimerization equilibria
As stated in the Materials and Methods section, in order to complete the study of the equilibria between conformations, melting experiments at different oligonucleotide concentrations were performed in pure water and salt media. MCR-ALS results of melting experiments in water showed that Tm values were independent of oligomer concentration (Table 1). This behavior was consistent with a monomolecular process, such as the intra-molecular unfolding of the dumbbell conformation. The calculated mean value for this equilibrium ΔG at 25°C, –5 ± 1 kJ/mol matches well with the values for previously reported univariate NMR and CD measurements (23). On the other hand, MCR-ALS results of melting experiments in salt media at several oligomer concentrations showed that Tm values depended on oligomer concentration. This behavior is in agreement with an equilibrium between conformations of different nuclearity, as in dimerization equilibria. The mean value for ΔG at 25°C, –65 ± 10 kJ/mol obtained from that value (Table 1) are in good agreement with results previously found (23).
CONCLUSIONS
MCR-ALS methodology was used to study the equilibrium between the different conformations of d<pTGCTCGCT>. The MCR-ALS procedure is seen to be a powerful tool for the analysis of multiwavelength spectroscopic data in cases where multiple species in equilibrium are present. The application of this method allowed the identification and resolution of three different conformations of d<pTGCTCGCT>: dimeric ‘bi-loop’, dumbbell and random coil structures. The pure CD and UV spectra of each of these species were unambiguously resolved, and thermodynamic data describing the different equilibria were obtained. This methodology can be easily applied to other cases where an equilibrium between multiple nucleic acid species prevents a direct analysis of the spectroscopic data.
Acknowledgments
ACKNOWLEDGEMENTS
J.J. acknowledges a PhD Grant from the Spanish Ministerio de Ciencia y Tecnología. This research was supported by the Spanish MCYT (BQU2000-0788 and BQU2001-3693) and the Generalitat de Catalunya (SGR-0048, SGR-0049, SGR-00056 and Centre de Referència de Biotecnologia).
REFERENCES
- 1.Lane A.N. and Jenkins,T.C. (2001) Structures and properties of multi-stranded nucleic acids. Curr. Org. Chem., 5, 845–869. [Google Scholar]
- 2.Gilbert D.E. and Feigon,J. (1999) Multistranded DNA structures. Curr. Opin. Struct. Biol., 9, 305–314. [DOI] [PubMed] [Google Scholar]
- 3.Lebrun A. and Lavery,R. (1997) Unusual DNA conformations. Curr. Opin. Struct. Biol., 7, 348–354. [DOI] [PubMed] [Google Scholar]
- 4.Frank-Kamenetskii M.D. and Mirkin,S.M. (1995) Triplex DNA structures. Annu. Rev. Biochem., 64, 65–95. [DOI] [PubMed] [Google Scholar]
- 5.Arthanari H. and Bolton,P.H. (2001) Functional and dysfunctional roles of quadruplex DNA in cells. Chem. Biol., 8, 221–230. [DOI] [PubMed] [Google Scholar]
- 6.Hélène C. (1991) The anti-gene strategy: control of gene expression by triplex-forming oligonucleotides. Anticancer Drug Des., 6, 569–584. [PubMed] [Google Scholar]
- 7.Jenkins T.C. (2000) Targeting multi-stranded DNA structures. Curr. Med. Chem., 7, 99–115. [DOI] [PubMed] [Google Scholar]
- 8.Mergny J.-L., Riou,J.-F., Mailliet,P., Teulade-Fichou,M.-P. and Gilson,E. (2002) Natural and pharmacological regulation of telomerase. Nucleic Acids Res., 30, 839–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Read M., Harrison,R.H., Romagnoli,B., Tanious,F.A., Gowan,S.H., Reszka,A.P., Wilson,W.D., Kelland,L.R. and Neidle,S. (2001) Structure-based design of selective and potent G-quadruplex-mediated telomerase inhibitors. Proc. Natl Acad. Sci. USA, 98, 4844–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riou J.F., Guittat,L., Mailliet,P., Laoui,A., Renou,E., Petitgenet,O., Mégnin-Chanet,F., Hélène,C. and Mergny,J.-L. (2002) Cell senescence and telomere shortening induced by a new series of specific G-quadruplex DNA ligands. Proc. Natl Acad. Sci. USA, 99, 2672–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marathias V.M. and Bolton,P.H. (1999) 6-Thioguanine alters the structure and stability of duplex DNA and inhibits quadruplex DNA formation. Nucleic Acids Res., 27, 2860–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K.Y., McCurdy,S., Shea,R.G., Swaminathan,S. and Bolton,P.H. (1993) A DNA aptamer which binds to and inhibits thrombin exhibits a new structural motif for DNA. Biochemistry, 32, 1899–1904. [DOI] [PubMed] [Google Scholar]
- 13.Jing N., Rando,R.F., Pommier,Y. and Hogan,M.E. (1997) Ion selective folding of loop domains in a potent anti-HIV oligonucleotide. Biochemistry, 36, 12498–12505. [DOI] [PubMed] [Google Scholar]
- 14.Wang E. and Feigon,J. (1998) Structure of nucleic acid triplexes. In Neidle,S. (ed.), Oxford Handbook of Nucleic Acid Structure. Oxford University Press, Oxford, pp. 355–381.
- 15.Patel D.J., Bouaziz,S. and Kettani,A. (1998) Structure of guanine rich and cytosine-rich quadruplexes formed in vitro by telomeric, centromeric, and triplet repeat disease DNA sequences. In Neidle,S. (ed.), Oxford Handbook of Nucleic Acid Structure. Oxford University Press, Oxford, pp. 389–449.
- 16.Mills M., Arimondo,P.B., Lacroix,L., Garestier,T., Hélène,C., Klump,H. and Mergny,J.-L. (1999) Energetics of strand-displacement reactions in triple helices: a spectroscopic study. J. Mol. Biol., 291, 1035–1054. [DOI] [PubMed] [Google Scholar]
- 17.Kanehara H., Mizuguchi,M., Tajima,K., Kanaori,K. and Makino,K. (1997) Spectroscopic evidence for the formation of four-stranded solution structure of oligodoxycytidine phosphorothioate. Biochemistry, 36, 1790–1797. [DOI] [PubMed] [Google Scholar]
- 18.Lane A.N. and Jenkins,T.C. (2000) Thermodynamics of nucleic acids and their interactions with ligands. Q. Rev. Biophys., 33, 255–306. [DOI] [PubMed] [Google Scholar]
- 19.Salisbury S.A., Wilson,S.E., Powell,H.R., Kennard,O., Lubini,P., Sheldrick,G.M., Escaja,N., Alazzouzi,E., Grandas,A. and Pedroso,E. (1997) The bi-loop, a new general four-stranded DNA motif. Proc. Natl Acad. Sci. USA, 94, 5515–5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.González C., Escaja,N., Rico,M. and Pedroso,E. (1998) NMR structure of two cyclic oligonucleotides. A monomer–dimer equilibrium between dumbbell and quadruplex structures. J. Am. Chem. Soc., 120, 2176–2177. [Google Scholar]
- 21.Escaja N., Pedroso,E., Rico,M. and González,C. (2000) Dimeric solution structure of two cyclic octamers: four-stranded DNA structures stabilized by A:T:A:T and G:C:G:C tetrads. J. Am. Chem. Soc., 122, 12732–12742. [Google Scholar]
- 22.Zhang N., Gorin,A., Majumdar,A., Kettani,A., Chernichenko,N., Skripkin,E. and Patel,D.J. (2001) Dimeric DNA quadruplex containing major groove-aligned A:T:A:T and G:C:G:C tetrads stabilized by inter-subunit Watson–Crick A:T and G:C pairs. J. Mol. Biol., 312, 1073–1088. [DOI] [PubMed] [Google Scholar]
- 23.Escaja N. (1999) Síntesis de oligonucleótidos cíclicos y su utilización en el estudio de nuevos motivos estructurales del DNA. PhD Thesis, University of Barcelona, Spain.
- 24.Mergny J.L., Phan,A.T. and Lacroix,L. (1998) Following G-quartet formation by UV-spectroscopy. FEBS Lett., 435, 74–78. [DOI] [PubMed] [Google Scholar]
- 25.Tauler R., Smilde,A. and Kowalski,B.R. (1995) Selectivity, local rank, three way data analysis and ambiguity in multivariate curve resolution. J. Chemom., 9, 31–41. [Google Scholar]
- 26.Gargallo R., Tauler,R. and Izquierdo-Ridorsa,A. (1997) Application of multivariate curve resolution procedure to the analysis of second-order melting data of synthetic and natural polynucleotides. Anal. Chem., 69, 1785–1791. [DOI] [PubMed] [Google Scholar]
- 27.Malinowski E.R. and Howery,D.E. (1991) Factor Analysis in Chemistry, 2nd Edn. Wiley, New York, NY.
- 28.Alazzouzi E., Escaja,N., Grandas,A. and Pedroso,E. (1997) A straightforward solid-phase synthesis of cyclic oligonucleotides. Angew. Chem. Int. Ed., 36, 1506–1508. [Google Scholar]
- 29.Eckstein F. (ed.), (1991) Oligonucleotides and Analogues: A Practical Approach. IRL Press, Oxford.
- 30.Vives M., Gargallo,R. and Tauler,R. (2001) Analytical characterization of the conformational transitions of polynucleotides by means of different molecular spectroscopies and multivariate curve resolution. Anal. Biochem., 291, 1–10. [DOI] [PubMed] [Google Scholar]
- 31.Gargallo R., Vives,M., Tauler,R. and Eritja,R. (2001) Protonation studies and multivariate curve resolution on oligodeoxynucleotides carrying the mutagenic base 2-aminopurine. Biophys. J., 81, 2886–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Golub G.H. and van Loan,C.F. (1989) Matrix Computations, 2nd Edn. The John Hopkins Press Ltd, London, UK.
- 33.Maeder M. (1987) Evolving factor analysis for the resolution of overlapping chromatographic peaks. Anal. Chem., 59, 527–531. [Google Scholar]
- 34.Windig W. and Guilment,J. (1991) Interactive self-modeling mixture analysis. Anal. Chem., 63, 1425–1430. [Google Scholar]
- 35.Amhrein M., Srinivasan,B., Bonvin,D. and Shumacher,M.M. (1996) On the rank deficiency and rank augmentation of the spectral measurement matrix. Chemom. Intell. Lab. Syst., 33, 17–33. [Google Scholar]
- 36.Erie D., Sinha,N., Olson,W., Jones,R. and Breslauer,K. (1987) A dumbbell shaped, double-hairpin structure of DNA: a thermodynamic investigation. Biochemistry, 26, 7150–7159. [DOI] [PubMed] [Google Scholar]
- 37.Erie D.A., Jones,R.A., Olson,W.K., Sinha,N.K. and Breslauer,K.J. (1989) Melting behavior of a covalently closed, single-stranded, circular DNA. Biochemistry, 28, 268–273. [DOI] [PubMed] [Google Scholar]