Abstract
We report a cell-free system for the high-throughput synthesis and screening of gene products. The system, based on the eukaryotic translation apparatus of wheat seeds, has significant advantages over other commonly used cell-free expression systems. To maximize the yield and throughput of the system, we optimized the mRNA UTRs, designed an expression vector for large-scale protein production, and developed a new strategy to construct PCR-generated DNAs for high-throughput production of many proteins in parallel. The resulting system achieves high-yield expression and can maintain productive translation for 14 days. Additionally, in the integration of a PCR-directed system for template creation, at least 50 genes can be translated in parallel, yielding between 0.1 and 2.3 mg of protein by one person within 2 days. Assessment of correct protein folding by the products of this high-throughput protein-expression system were performed by enzymatic assays of kinases and by NMR spectroscopic analysis. The cell-free system, reported here, bypasses many of the time-consuming cloning steps of conventional expression systems and lends itself to a robotic automation for the high-throughput expression of proteins.
With the sequencing of the genomes of various species, attention has turned to the structure, properties, and functional activities of proteins. However, rapid progress in the study of proteins encoded by the newly discovered genes is based on the availability of sufficient amounts of a large number of proteins. Currently, three strategies are being used for protein production: chemical synthesis, in vivo expression, and cell-free protein synthesis. The first two methods have severe limitations: chemical synthesis is not practical for the synthesis of long peptides (1), and in vivo expression can produce only those proteins that do not affect the physiology of the host cell (2). Cell-free translation systems, in contrast, can synthesize large proteins with a speed and accuracy that approach those of in vivo translation (3, 4), and they can express proteins that would otherwise interfere with host cell physiology. However, these systems are reputed to be unstable and thus inefficient (5). One of the most convenient and promising eukaryotic cell-free translation systems is based on wheat embryos that store all of the components of translation in a concentrated dried state, ready for protein synthesis as soon as germination starts. Recently, we found that conventional wheat germ extracts contain the RNA N-glycosidase tritin and other inhibitors of translation such as thionin, ribonucleases, deoxyribonucleases, and proteases. These inhibitors originate from the endosperm (6, 7). Extensive washing of wheat embryos to eliminate endosperm contaminants has resulted in extracts with a high degree of stability and activity. By using mRNA having 5′-m7GpppG (cap) and a poly(A)-tail (pA) with this extract, the translation reaction proceeds for >60 h, when it is performed in a dialysis bag with a continuous supply of substrates and the removal of small byproducts (8), yielding mg quantities of enzymatically active proteins per ml of reaction volume (7). This high efficiency could be a result of the formation of polysomes during the translation reaction that was revealed by an ultracentrifuge analysis (7). However, to turn the cell-free system into one that addresses the high-throughput needs of modern proteomics, several critical improvements were needed. The system described here is suitable for high-throughput protein expression and incorporates several important design features: (i) optimized 5′- and 3′-RNA untranslated regions, which play a crucial role in the regulation of gene expression, with a concomitant elimination of the 5′-7mGpppG and poly(A)-tail; (ii) a new expression vector, which is specialized for generating high yields of protein; (iii) a specialized set of primers that generate transcription templates by PCR directly from Escherichia coli cells carrying cDNAs, thus bypassing time-consuming cloning steps.
The resulting system exhibits the following attractive features. (i) It is stable over long periods of time and, with the new expression vector, it can produce proteins in amounts of several milligrams per ml of reaction volume. (ii) It is amenable to high-throughput parallel protein synthesis. At least 50 genes can be translated in quantities of hundreds of micrograms by one person within 2 days. (iii) Both functional and structural tests on some of the protein products suggest that the system can produce proteins that fold into their natural states. These features may prove to be essential for the systematic study of protein structure–function relations and a series of applications delineating the scope of modern proteomics, including the following: high-throughput enzymatic testing of a large number of genomic expression products, high-throughput crystallization of proteins and identification of their three-dimensional structure through NMR or x-ray diffraction, rapid evolutionary design of proteins, construction of protein–protein interaction systems, and industrial-scale protein production.
Materials and Methods
Cell-Free Protein Synthesis.
Purification of wheat embryos and preparation of the cell-free extract were performed as described (7) with slight modifications. Batch-wise reaction mixture (25 μl) contained 6 μl of extract. Final concentrations of the various ingredients in translation solution are 24 mM Hepes/KOH, pH 7.8/1.2 mM ATP/0.25 mM GTP/16 mM creatine phosphate/10 μg creatine kinase/20 units of RNasin as ribonuclease inhibitor/2 mM DTT/0.4 mM spermidine/0.3 mM each of the 20-aa, 2.7 mM magnesium acetate/100 mM potassium acetate/5 μg deacylated tRNA prepared from wheat embryos/0.05% Nonidet P-40/and 0.005% NaN3 and mRNA. Incubations were done at 26°C.
For the dialysis system, 500 μl of reaction mixture contained 300 μl of the extract and the same ingredients as described above. The dialysis bag was immersed in 5 ml of a solution containing all described ingredients except for creatine kinase (substrate solution). The reaction was started at 26°C in the presence of mRNAs (92 μg for GFP and 76 μg for FT-protein), and every 48 h the same amount of GFP mRNA was supplemented. The substrate solution also was replaced every 48 h. The reaction using a dialysis cup (molecular weight cutoff 12,000; Biotech International, Perth, Australia) were carried out the same way as the dialysis system, except that the volume of the reaction mixture was reduced to 50 μl, as explained below. Methods for determination of protein synthesis and for the analyses of the translation products have been described (7).
Preparation of mRNAs.
The mRNAs for optimization of 5′- and 3′-UTRs were synthesized by in vitro transcription of linearized pSP65-derived vector which carries a luciferase gene downstream of the SP6 RNA polymerase promoter (7). The mRNAs consist of the sequence cap-AAUACACGGAAUUCGAGCUCGCCCGGGAAAUCUCAAUG (the underlined sequence is the initiation codon) at its 5′-end, a coding sequence, and a 3′-noncoding region of a 549-nt sequence from a type-l dihydrofolate reductase (dhfr) gene of E. coli “549” sequence followed by pA of 100 nts. The templates for the Ω-containing mRNAs were created by introduction of the sequence into the above luciferase plasmids. They contain combinations of the following elements: the cap structure (cap), the original Ω sequence with the GUA trinucleotide at the 5′-end (GUAΩ), Ω with the GAA trinucleotide at the 5′-end (GAAΩ) as for 5′-UTRs; the downstream 18-nt sequence originating from E. coli dhfr, the 549-nt sequence from E. coli dhfr (549), 100-nt pA (pA), 549 plus a 629-nt sequence from pSP65 (1178), 549 plus 1077 nts from pSP65 (1626), and 1626 nts from a pSP65 construct containing the gene in opposite orientation (1626*), as for 3′-UTRs. Thus for example, cap/549 represents the mRNA with the 5′-cap structure and the 549-nt sequence but without pA. The mRNAs for the large-scale protein synthesis were prepared by transcription of the pEU derivatives without linearization. The main product had an ≈1,600-nt long 3′-UTR that could caused by a strong transcription stop. For the purification of the mRNA, transcription solution was passed through Sephadex G50, ethanol precipitated, washed with 70% ethanol, and dissolved in the substrate solution.
The “Split-Primer” PCR.
Nucleotide sequences of the primers used for the split-primer strategy were: primer-1, 5′-GCGTAGCATTTAGGTGACACT (the underlined sequence is the 5′-half of the promoter); primer-2: 5′-GGTGACACTATAGAAGTATTTTTACAACAATTACCAACAACAACAACAAACAACAACAACATTACATTTTACATTCTACAACTACCACCCACCACCACCAATG (the underlined sequence is the 3′-half of the promoter and the sequence in italic denotes the complementary region of primer-1); primer-3: 5′-CCACCCACCACCACCAatgnnnnnnnnnnnnnnnn (the 5′-coding region of a given gene is in lowercase letters); primer-4: 5′-AGCGTCAGACCCCGTAGAAA. For the PCR based on reported procedures (9–11), a mixture containing template (50 pg/μl plasmid or 3 μl of E. coli culture per 60-μl reaction), 200 μM of each dNTP, 25 units/ml ExTaq DNA polymerase (Takara), 10 nM primer-3 and primer-4, and the buffer supplied by the manufacturer was set on a Takara PCR Thermal Cycler MP (4-min denaturation at 94°C followed by 30 cycles of amplification: 98°C for 10 s, 60°C for 30 s, and 72°C for 5 min). After this, the first PCR amplification was saturated and a 3-μl aliquot was used for a second PCR (30 μl) in the presence of 100 nM primer-1 and primer-4 and 1 nM primer-2 with the same program.
Parallel Cell-Free Protein Synthesis.
The unique primer for the split-primer method (primer-3) for each of the 14 DNAs from GenBank or Munich Information Center for Protein Sequences Arabidopsis thaliana database (MAtDB in Table 1) was designed according to the sequence in the database. For the other 13, the sequences were randomly cloned from commercially available phage cDNA libraries and were sequenced partially to identify the gene. Each specific primer was then designed according to the databases. For the introduction of the GST sequence, a double-stranded DNA with the 3′-portion of SP6 promoter followed by GAAΩ, the GST ORF, and 5′-CCACCCACCACCACCA was generated by PCR, purified, and used (0.2 μM) instead of primer-2. Three microliters of E. coli suspension grown overnight in a 96-well titer plate was used for the 60-μl PCR (11), where the amplification reaction reaches the saturation level, and the resulting DNA was transcribed as above. The transcript in each well of the titer plate was precipitated with ethanol and was spun down by centrifugation of the plate using a Hitachi R10H rotor to collect precipitates. Each washed mRNA (usually 30–35 μg) was transferred into the dialysis cup that contained 50 μl of the translation mixture. The cups then were dipped into the substrate solution (1 ml) in a 24-hole plate (Whatman) and incubated for 36 h.
Table 1.
An example of proteins synthesized in the PCR-directed wheat germ cell-free system
| Proteins encoded by cDNAs
|
Annotation no.
|
M.W.
|
Authentic | Fusion | Clone name
|
||
|---|---|---|---|---|---|---|---|
| Total, mg | Sup, % | Total, mg | Sup, % | ||||
| Arabidopsis (from GenBank and MIPS) | |||||||
| Chlorophyll a/b-binding protein | X56062 | 25,995 | 0.2 | 30 | 0.5 | 90 | At01 |
| Agamous-like gene 9 (AGL9) | AF015552 | 29,065 | 0.7 | 30 | 0.8 | 90 | At02 |
| Flowering locus T (FT) | AF152096 | 19,808 | 0.3 | 100 | 0.8 | 100 | At03 |
| HY5 | AB005456 | 18,462 | 0.4 | 90 | 1.5 | 90 | At04 |
| Flowering locus F (FLF) | AF116527 | 21,864 | 0.2 | 100 | 0.4 | 100 | At05 |
| Hypothetical protein (from a commercial cDNA library) | At1g69630† | 11,311 | 0.1 | 40 | 0.1 | 100 | At06 |
| Putative heat-shock protein 40 | AL021749 | 38,189 | 1.8 | 30 | 1.0 | 80 | At07 |
| Heat-shock protein 70-3 | Y17053 | 71,144 | 0.9 | 100 | — | — | At08 |
| Putative s-adenosylmethionine synthetase | AY037214 | 42,793 | 1.5 | 100 | 0.6 | 100 | At09 |
| NADPH thioredoxin reductase | Z23108 | 40,635 | 0.1 | 10 | 0.5 | 20 | At10 |
| Putative ACC oxidase | AF370155 | 36,677 | 1.0 | 10 | 1.2 | 100 | At11 |
| Putative fructokinase | AF387001 | 35,276 | 1.0 | 10 | 0.6 | 100 | At12 |
| Rubisco activase | X14212 | 51,981 | 0.4 | 20 | 0.6 | 80 | At13 |
| Glutaredoxin | At4g15660† | 11,311 | — | — | 0.4 | 80 | At14 |
| Chlorophyllase 2 | AF134302 | 34,902 | 0.2 | 10 | 0.4 | 70 | At15 |
| Human (from GenBank) | |||||||
| Neuron-specific γ-2 enolase | M22349 | 47,266 | 1.0 | 100 | 0.5 | 100 | Hs01 |
| ζ-crystallin/quinone reductase | L13278 | 35,205 | 2.3 | 80 | 1.3 | 100 | Hs02 |
| X11-like protein | AB014719 | 82,480 | 0.5 | 100 | 0.2 | 80 | Hs03 |
| Importin α 1 | NM_002266 | 57,859 | — | — | 0.2 | 30 | Hs04 |
| Glyceraldehyde-3-phosphate dehydrogenase | M17851 | 36,051 | 0.4 | 70 | 0.9 | 100 | Hs05 |
| Enolase 3 | NM_001976 | 46,956 | 1.7 | 80 | 0.9 | 100 | Hs06 |
| APBA3 | NM_004886 | 61,451 | 0.9 | 100 | 0.2 | 100 | Hs07 |
| JAK-binding protein (from a commercial cDNA library) | NM_003745 | 23,550 | 0.4 | 30 | 0.2 | 100 | Hs08 |
| Phosphoglycerate kinase 1 | XM_010102 | 43,965 | 1.0 | 100 | 0.7 | 100 | Hs09 |
| β-actin | X00351 | 41,735 | 1.3 | 100 | 0.3 | 100 | Hs10 |
| Hypothetical protein FLJ10652 | XM_006938 | 41,539 | — | — | 0.2 | 10 | Hs11 |
| Hypothetical protein FLJ10559 | XM_001479 | 35,237 | 0.7 | 50 | 1.0 | 70 | Hs12 |
λ ZAP II Library, product of Stratagene #937010.
† MIPS Arabidopsis thaliana database MAtDB (www.mips.biochem.mpg.de/proj/thal/).
Below the detectable level.
These genes were cloned from tissue (heart, brain, kidney, liver, placenta) cDNAs (BioChain Institute, #0516001).
Amyloid β (A4) precursor protein-binding, family A, member 3.
λ ZAP II Library, product of Stratagene #936204.
Analysis of Enzyme Activity of the Products.
Five cDNA clones containing sequences for protein kinase domain annotated were selected from MAtDB. cDNAs were cloned into pCRII plasmid (Invitrogen) from the Arabidopsis cDNA library (Stratagene #937010). PCR-aided introduction of streptavidin sequence into each gene at 5′-site was carried out in a manner similar to the construction of GST-fused genes described above. After the transcription, each streptavidin-fused protein kinase was synthesized in the dialysis cup for 12 h. For the purification of the product, 2 μl of the reaction mixture was mixed with 8 μl of biotin magnetic beads (Spherotech, Libertyville, IL) and was washed twice with protein kinase buffer (50 mM Tris⋅HCl, pH 7.6/100 mM potassium acetate/10 mM MgCl2/1 mM DTT; refs. 12 and 13). Each protein retained on beads was treated with 20 units of λ protein phosphatase in 20 μl of commercial buffer (New England Biolabs; ref. 14) at 30°C for 30 min and then was suspended in the kinase buffer and washed twice. Beads were suspended in the kinase buffer and the reaction mixture (10 μl) containing [γ-32P]ATP as the phosphate donor was incubated at 30°C for 30 min. Samples were boiled in the SDS-denaturing buffer, and proteins were separated on SDS/polyacrylamide gel. Autophosphorylation activity of the products was determined by autoradiography.
Heteronuclear Sequential Quantum Correlation (HSQC) Spectrum.
The FT protein was synthesized in the same way as in Fig. 2b except that Ala, Leu, Gly, and Gln in both translation and substrate mixture were replaced with their 15N-labeled forms (Isotec). After incubation for 48 h, the reaction mixture (1 ml) was dialyzed against 10 mM phosphate buffer (pH 6.5) overnight, then centrifuged at 30,000 × g for 10 min. The supernatant containing 30 μM of the protein was directly subjected to NMR spectroscopy. The spectrum was recorded on a Bruker DMX-500 spectrometer at 25°C, and 2,048 scans were averaged for the final 1H-15N HSQC spectrum (15).
Fig 2.
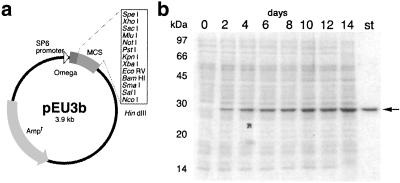
A cell-free expression vector and its performance. (a) Schematic illustration of pEU. (b) SDS/PAGE analysis of GFP produced during 14 days of reaction. mRNA produced by transcription of circular pEU was used for the translation reaction in the dialysis membrane system and was added every 48 h. A 0.1-μl aliquot of the mixture was run on the gel, and protein bands were stained with CBB. The arrow shows GFP; “st” designates an authentic GFP band (0.5 μg).
Results
Optimization of the 5′- and 3′-UTRs.
The 5′-cap plays a crucial role in eukaryotic translation initiation but is problematic in cell-free translation systems. When RNA template is prepared in vitro, the cap structure is introduced by using an RNA polymerase that incorporates a modified free dinucleotide (7-mG-5′-ppp-5′G) at the end of the RNA polynucleotide chain. However, the efficiency of the incorporation is low, and there is inevitably an excess of free modified nucleotide remaining in the product mixture. The free modified nucleotide binds competitively to the cap-binding protein, eIF4E, and thereby inhibits translation. Complete removal of the free nucleotide is tedious and not easily accomplished in the context of a high-throughput system. In addition, cap-containing RNA fragments are unavoidably generated by ribonucleases in the reaction mixture, and they inhibit translation initiation. Furthermore, there is a very narrow range of concentration of capped RNA that gives efficient translation; this range is best determined empirically because the exact concentration of effectively capped RNA is difficult to determine. To maintain the RNA in that range, continuous supplementation of capped RNA is required (7). This optimization is problematic when proteins are to be synthesized from a large number of genes (RNAs) in parallel. The poly(A) tail at the 3′-end of RNA is also a problem in preparing templates for in vitro translation, because long plasmid poly dT/dA sequences are unstable during replication of plasmids in host cells.
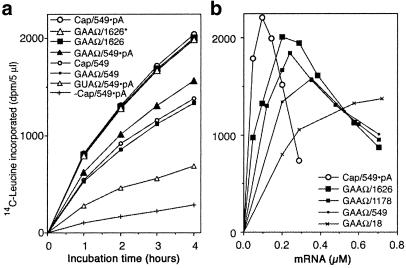
To solve these problems, we developed new 5′- and 3′-UTRs that enhance mRNA translation in the absence of cap and pA. As a replacement of the 5′-cap, we first tested luciferase mRNA with a 71-base naturally occurring translational enhancer (16, 17), the omega (Ω) sequence from tobacco mosaic virus. This RNA had a much higher activity than uncapped mRNA lacking the Ω sequence (Fig. 1a). Next, we examined the effect of Ω with three additional bases at the 5′-terminus and found that the mRNA with a 5′-terminal GAA sequence (GAAΩ549⋅pA) had the highest activity of all 64 possible three-base sequences in that position (data not shown). The activity of the GAAΩ549⋅pA mRNA was ≈75% of that of the capped RNA (Fig. 1a).
Fig 1.
Template activities of mRNAs having different UTRs. For each construct, the 5′ component (cap or Ω) is indicated first, followed by the 3′ component [length of UTR and presence of poly(A)]. Protein synthesis activity was measured as hot-trichloroacetic acid-insoluble radioactivity by using luciferase mRNA. (a) mRNA concentration was 0.1 μM for cap/549⋅pA and cap/549 and 0.2 μM for the other mRNAs. (b) Incubations were done for 4 h.
The pA sequence at the 3′-end of natural mRNAs is reported to be dispensable in yeast translation (18). By varying the 3′-UTR of mRNAs with GAAΩ as the 5′-UTR, we found that translation does not depend on a specific sequence or pA but depends only on the length of the 3′-UTR (Fig. 1b). For example, the mRNA with a 3′-UTR of 1,626 nts (GAAΩ/1,626) has a comparable activity as the capped, polyadenylated mRNA and the GAAΩ/1,626* mRNA that has a 3′-UTR from another region and the other strand of pSP65 (Fig. 1a). These results can be explained if the mRNAs in our system are primarily degraded from their 3′-end by exonucleases such as the exosome complex (19).
We also examined the dependence of protein yield on the concentration of the mRNAs. The capped mRNA showed the expected narrow optimum (Fig. 1b) that is probably caused by the competition described above. In contrast, the GAAΩ-containing mRNAs had a wider range of concentration. There was a direct correlation between the yield and the length of the 3′-UTR. The results indicate that the GAAΩ mRNAs with a longer 3′-UTR can be used in relatively large amounts over a broad range of concentration. Based on these findings, we constructed an efficient expression vector, pEU (plasmid of Ehime University, Matsuyama, Japan; Fig. 2a). By adding mRNA transcribed from circular pEU into the dialysis cell-free system, a large amount of protein (9.7 mg of GFP in 1 ml) was produced. The translation reaction continued to produce protein for surprisingly long periods, up to 14 days (Fig. 2b, arrow). Although supplementation of mRNA was required every 2 days, the amount of protein produced was more than that of the endogenous proteins. The result supports our previous notion that the translation machinery itself is inherently robust and stable (7).
PCR-Directed Cell-Free Protein Synthesis.
Although pEU is useful for large-scale production of a specific protein, laborious molecular cloning steps are required to generate the template for translation. This labor limits the throughput of the cell-free protein production system in the parallel expression of many different proteins or sequence variants of the same protein. Therefore, we searched for a PCR primer set with which the above UTR elements can be introduced into any given cDNA sequence, and the PCR product can be directly used for the transcription reaction without purification. Ideally, the upstream primer should have the SP6 promoter and GAAΩ. We started with a conventional PCR method using four primers, one of which had the complete SP6 promoter sequence (11, 12). But, the resulting products were not good templates for transcription when used without a gel-purification. In fact, this primer system gave mostly short transcripts and resulted in the production of small peptides (data not shown). This could be caused by the generation of “primer dimer” (9) artifacts.
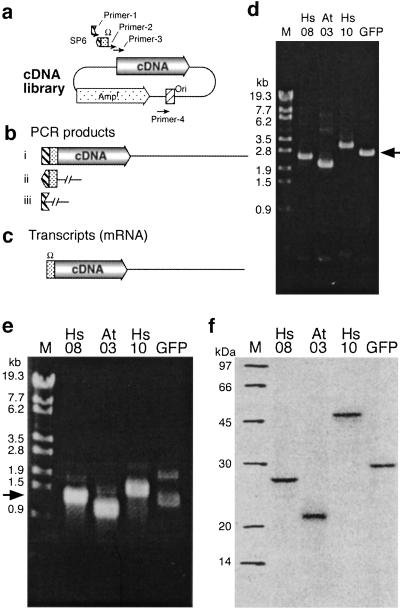
Therefore, we designed a set of primers in which the complete promoter sequence is not contained in any one of the primers but is generated only when the sequences of two primers are joined correctly (Fig. 3 a and b), leading to the production of only complete mRNA (Fig. 3c). Experimental results showed each PCR product of this split-primer method produced DNA (Fig. 3d) and mRNA (Fig. 3e) of expected size and gave a single radioactive protein band of the expected size on autoradiograms (Fig. 3f).
Fig 3.
Parallel expression of cDNA into proteins by using the split-primers PCR technique. (a) Design of the split-type primers for the introduction of the required UTRs into cDNA sequences. (b and c) Expected PCR-generated DNAs and mRNA, respectively. (d) Split-primer PCR-generated products. (e) Transcription was carried out with 10-μl aliquots from the PCR samples in a 100-μl reaction. Transcripts were visualized by electrophoresis. (f) All of the transcript was used for batch-mode translation (50 μl, 4 h), and the protein products were analyzed by SDS/PAGE autoradiography.
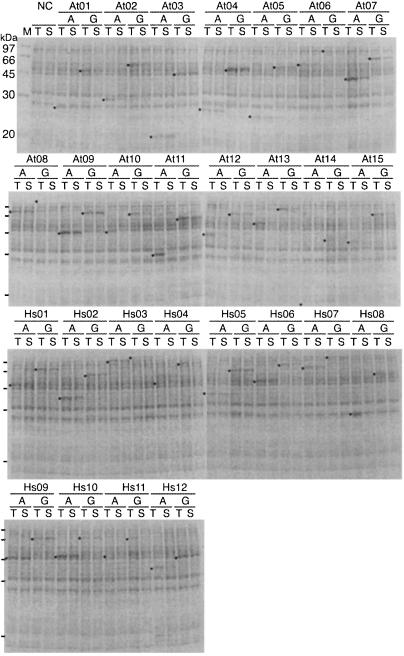
To evaluate the performance of the method in high-throughput screening, we carried out parallel cell-free protein synthesis, starting with E. coli cells carrying cDNAs (Table 1 and Fig. 4). Each cDNA sequence was amplified by the split-primer method. In addition, we prepared the template of a GST-fusion protein for each cDNA in the same way. In 50 of these 54 cases, a clearly visible stained protein band was seen (Fig. 4a, asterisks). Both forms of the products, authentic (A) and GST fusion (G), were correct in size as estimated from their mobility, and the yield was from 0.1 to 2.3 mg per ml of reaction volume in 36 h, as estimated by densitometric scanning of the bands (Table 1). Some gene products were recovered in the supernatant (S), and others were in the precipitate phase after centrifugation. We could not detect any dependence of the yield and solubility of the proteins on the gene sources or the vector sequences.
Fig 4.
A high-throughput production for screening proteins from cDNA libraries. Authentic (A) and GST-fused (G) proteins in the reaction mixtures after a semiautomated PCR/transcription and translation from 54 different cDNAs separated by SDS/PAGE and stained with CBB. T and S mark total translation product and the supernatant fraction, respectively, after centrifugation at 30,000 × g for 15 min.
It is important to note here that all of the 54 genes were amplified under the same condition, so that the PCR products were transcribed under the same conditions without purification, and that transcription products were translated under the same conditions again without purification. Furthermore, the determination of mRNA content for each gene was not necessary before translation, because the yields in the PCRs were nearly constant among genes and because the optimum concentration of mRNAs are fairly broad (see above). All of these features are important for the parallel production of many proteins in moderate amounts sufficient for rapid screening. One can increase the number of samples up to 96 or more and finish the experiment in less than 2 days: 9 h for PCR, 3 h for transcription, and up to 36 h for protein synthesis. Because this protocol bypasses the time-consuming steps of recombinant technologies, the method lends itself to a fully automated high-throughput synthesis of gene products from large cDNA libraries.
Quality of the Proteins Synthesized in the Wheat Cell-Free System.
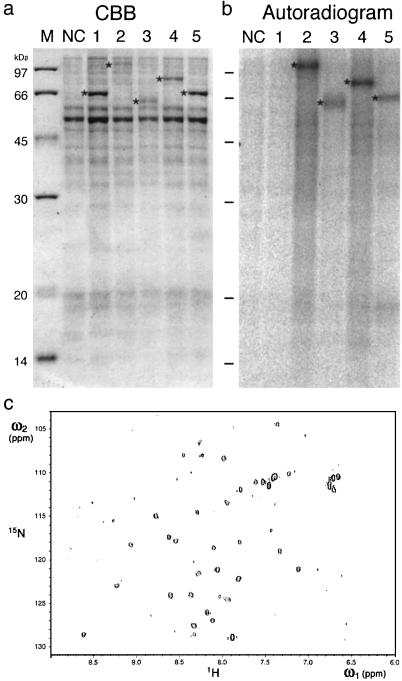
The utility of any protein expression system is related to the degree to which the protein products resemble the naturally folded states. We have assessed this by testing a subset of proteins for their functional integrity as enzymes. As shown in Fig. 5a, each of five Arabidopsis protein kinases were synthesized effectively at the expected size (asterisk). Four of five of these proteins displayed autophosphorylation activity after incubation with [γ-32P]ATP and magnesium (Fig. 5b). These results suggest that at least the kinase domains of these products are folded into active forms in the cell-free system.
Fig 5.
Activity and folding of the cell-free-produced polypeptides. (a and b) Autophosphorylation activity of five Arabidopsis protein kinases. SDS/PAGE and CBB-stained gel of the partially purified products (a) and the autoradiogram (b). Lanes 1–6 in a and b represent At1g07150, At5g49760, At2g02800, At5g62710, and At4g35500, respectively. NC denotes samples from the reaction mixture incubated in the absence of mRNA. M denotes protein size marker. (c) HSQC spectrum of the hypothetical protein of the flowering locus T protein produced in the cell-free.
Next, we examined the folding of proteins produced in the cell-free system. As an example, a cDNA coding for a hypothetical flowering locus T gene of Arabidopsis [At03 (20) in Table 1] was selected. Cell-free protein synthesis was carried out by using dialysis membrane for large-scale production of the protein in the presence of 15N-labeled amino acids (data not shown), yielding 0.6 mg of protein per ml in 48 h, a sufficient quantity for estimating the protein folding by NMR (21). Fig. 5b shows a reasonable number of signals well dispersed in the 1H–15N spectrum, suggesting that at least minimal criteria for proper folding have been met (21). Thus, an important feature of the system is that a dialyzed crude translation product is ready for the first screening of its “foldedness” by NMR.
Discussion
Cell-free systems lend themselves to high-throughput formats that are desirable for parallel production of many proteins. A number of reports have been published on cell-free systems from E. coli showing that it is possible to produce large amounts of proteins for NMR structural analyses (22). Several designs of E. coli systems have been used (23–25). E. coli systems, however, have two serious limitations: one is intrinsic and the other is technically unavoidable. Although the fundamental ability of prokaryotic cytosol to support effective cotranslational protein folding has been demonstrated, multidomain proteins, found more often in eukaryotes, tend to fold into their correct conformations much better in eukaryotic rather than prokaryotic translation systems (26, 27). In fact, we could show here that four of five genes encoding kinases gave proteins that have autophosphorylating activity. More recent systematic screening of protein kinase genes from Arabidopsis revealed that 233 of 530 (44%) had autophosphorylation activity (Y. Hasegawa, R.M., T.S., M. Seki, K. Shinozaki & Y.E., unpublished data). This percentage may be a minimum estimate because those proteins we have detected activity in the fixed condition, as we have not attempted to optimize the assay conditions with respect to ions and ATP. The second problem with E. coli systems for high-throughput cell-free expression is that PCR-generated DNA fragments are not transcribed and translated efficiently in E. coli systems in general, even though several PCR-based methods have been reported (28, 29). This is because contamination by the mRNA- and DNA-degradation enzymes originating from the cells decreases the stability of the templates and reduces yields. In contrast, the wheat system is of eukaryotic nature, and the added mRNAs are stable for long periods of time. Wheat seeds are available at low cost and may have minimal problems in bio-pollution and ethical issues because it is a foodstuff. Thus, the present study overcomes all of the limitations associated with the current E. coli cell-free systems.
In conclusion, we have described a system for high-throughput expression of proteins. The system has been designed for the facile transcription of cDNA to RNA and translation to protein without cumbersome cloning steps. The yields and qualities of eukaryotic protein products seem to be favorable when compared with other systems. Our system makes it possible to prepare large numbers of proteins in parallel and is particularly useful in the analysis of sequence variation on protein function or in the comparison of the properties of a set of proteins encoded by different genes.
Acknowledgments
We thank Dr. H. E. Morita for determination of the HSQC spectrum. This work was supported by Japan Society for the Promotion of Science Grant JSPS-RFTF 96100305 (to Y.E.).
References
- 1.Blaschke U. K., Silberstein, J. & Muir, T. W. (2000) Methods Enzymol. 328, 478-496. [DOI] [PubMed] [Google Scholar]
- 2.Chrunyk B. A., Evans, J., Lillquist, J., Young, P. & Wetzel, R. (1993) J. Biol. Chem. 268, 18053-18061. [PubMed] [Google Scholar]
- 3.Kurland C. G. (1982) Cell 28, 201-202. [DOI] [PubMed] [Google Scholar]
- 4.Pavlov M. Y. & Ehrenberg, M. (1996) Arch. Biochem. Biophys. 328, 9-16. [DOI] [PubMed] [Google Scholar]
- 5.Roberts B. E. & Paterson, B. M. (1973) Proc. Natl. Acad. Sci. USA 70, 2330-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogasawara T., Sawasaki, T., Morishita, R., Ozawa, A., Madin, K. & Endo, Y. (1999) EMBO J. 18, 6522-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Madin K., Sawasaki, T., Ogasawara, T. & Endo, Y. (2000) Proc. Natl. Acad. Sci. USA 97, 559-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spirin A. S., Baranov, V. I., Ryabova, L. A., Ovodov, S. Y. & Alakhov, Y. B. (1988) Science 242, 1162-1164. [DOI] [PubMed] [Google Scholar]
- 9.Erlich H. A., Gelfand, D. & Sninsky, J. (1991) Science 252, 1643-1651. [DOI] [PubMed] [Google Scholar]
- 10.Gurevich V. V. (1996) Methods Enzymol. 275, 382-397. [DOI] [PubMed] [Google Scholar]
- 11.Sambrook J. & Russell, D. W., (2001) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 12.Shah K., Vervoort, J. & de Vries, S. C. (2001) J. Biol. Chem. 276, 41263-41269. [DOI] [PubMed] [Google Scholar]
- 13.Sakai T., Kagawa, T., Kasahara, M., Swartz, T. E., Christie, J. M., Briggs, W. R., Wada, M. & Okada, K. (2001) Proc. Natl. Acad. Sci. USA 98, 6969-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H. C. & Fecteau, K. A. (2000) J. Biol. Chem. 275, 25850-25857. [DOI] [PubMed] [Google Scholar]
- 15.Morita E. H., Kosada, T., Yamazaki, T., Kyougoku, Y. & Hayashi, H. (1998) J. Biomol. NMR 12, 453-454. [DOI] [PubMed] [Google Scholar]
- 16.Gallie D. R. & Walbot, V. (1992) Nucleic Acids Res. 20, 4631-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallie D. R. (1996) Plant Mol. Biol. 32, 145-158. [DOI] [PubMed] [Google Scholar]
- 18.Searfoss A. M. & Wickner, R. B. (2000) Proc. Natl. Acad. Sci. USA 97, 9133-9137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs J. S., Anderson, A. R. & Parker, R. P. (1998) EMBO J. 17, 1497-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi Y., Kaya, H., Goto, K., Iwabuchi, M. & Araki, T. (1999) Science 286, 1960-1962. [DOI] [PubMed] [Google Scholar]
- 21.Montelione G. T., Zheng, D., Huang, Y. J., Gunsalus, K. C. & Szyperski, T. (2000) Nat. Struct. Biol. 7, 982-985. [DOI] [PubMed] [Google Scholar]
- 22.Kigawa T., Yabuki, T., Yoshida, Y., Tsutsui, M., Ito, Y., Shibata, T. & Yokoyama, S. (1999) FEBS Lett. 442, 15-19. [DOI] [PubMed] [Google Scholar]
- 23.Noren C. J., Anthony-Cahill, S. J., Griffith, M. C. & Schultz, P. G. (1989) Science 244, 182-188. [DOI] [PubMed] [Google Scholar]
- 24.Hanes J. & Plukthun, A. (1997) Proc. Natl. Acad. Sci. USA 94, 4937-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson D. S., Keefe, A. D. & Szostak, J. W. (2001) Proc. Natl. Acad. Sci. USA 98, 3750-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolb V. A., Makeyev, E. V. & Spirin, A. S. (2000) J. Biol. Chem. 275, 16597-16601. [DOI] [PubMed] [Google Scholar]
- 27.Netzer W. J. & Hartl, F. U. (1997) Nature 388, 343-349. [DOI] [PubMed] [Google Scholar]
- 28.Lesley S. A., Brow, M. A. D. & Burgess, R. R. (1991) J. Biol. Chem. 266, 2632-2638. [PubMed] [Google Scholar]
- 29.Martemyanov K. A., Spirin, A. S. & Gudkov, A. T. (1997) FEBS Lett. 414, 268-270. [DOI] [PubMed] [Google Scholar]