Abstract
The macrolide antibiotic tylosin has been used extensively in veterinary medicine and exerts potent antimicrobial activity against Gram-positive bacteria. Tylosin-synthesizing strains of the Gram-positive bacterium Streptomyces fradiae protect themselves from their own product by differential expression of four resistance determinants, tlrA, tlrB, tlrC, and tlrD. The tlrB and tlrD genes encode methyltransferases that add single methyl groups at 23S rRNA nucleotides G748 and A2058, respectively. Here we show that methylation by neither TlrB nor TlrD is sufficient on its own to give tylosin resistance, and resistance is conferred by the G748 and A2058 methylations acting together in synergy. This synergistic mechanism of resistance is specific for the macrolides tylosin and mycinamycin that possess sugars extending from the 5- and 14-positions of the macrolactone ring and is not observed for macrolides, such as carbomycin, spiramycin, and erythromycin, that have different constellations of sugars. The manner in which the G748 and A2058 methylations coincide with the glycosylation patterns of tylosin and mycinamycin reflects unambiguously how these macrolides fit into their binding site within the bacterial 50S ribosomal subunit.
Species within the actinomycetes group of Gram-positive bacteria produce most of the antibiotics that are known to target the bacterial ribosome. To protect their own ribosomes from inhibition during antibiotic production, actinomycetes possess an arsenal of defense mechanisms (1). These defenses include methylation of key rRNA nucleotides at the drug target site, drug modification, and active efflux of the drug from the cell. Many of these resistance mechanisms have been recruited by pathogenic bacteria and severely compromise the efficacy of antibiotics in the clinical treatment of infection.
The actinomycete Streptomyces fradiae is the producer of the macrolide antibiotic tylosin. Tylosin has been widely used as a feed additive for promoting animal growth and remains in common veterinary use against bacterial dysentery and respiratory diseases in poultry, swine, and cattle (2). Tylosin exerts its antimicrobial action by binding in the peptide exit tunnel of the bacterial 50S ribosomal subunit, where it inhibits protein synthesis by interfering with peptide bond formation as well as by blocking the passage of the nascent peptide chain through the tunnel. S. fradiae has four resistance gene products (TlrA, TlrB, TlrC, and TrlD) to protect itself against these inhibitory effects (3, 4). TlrA and TlrD are members of the Erm family of methyltransferases (and for the sake of consistency in the nomenclature, they have recently been renamed, Table 1). TlrD attaches a single methyl group to the N6 position of nucleotide A2058 in 23S rRNA (5), whereas TlrA dimethylates the same position (6). Monomethylation at A2058 confers high resistance to lincosamides, but low resistance to macrolides and streptogramin B antibiotics, whereas dimethylation gives high resistance to each of the three drug types and confers what is commonly termed the MLSB (macrolide, lincosamide, and streptogramin B antibiotics) phenotype (7, 8). Erm dimethyltransferases have become one of the most prevalent forms of MLSB resistance in pathogenic bacteria (9–12). TlrC is an efflux pump that confers modest resistance to tylosin (13–15), whereas TlrB is a methyltransferase that confers moderately high resistance to tylosin in several Streptomyces species (16–19).
Table 1.
Nomenclature of the rRNA methyltransferases referred to in the text
| Original gene name (original host) | Target nucleotide in 23S rRNA | Reference | New gene name | Reference |
|---|---|---|---|---|
| rrmA (E. coli) | G745 | 56 | rlmAI | 21 |
| tlrB (S. fradiae) | G748 | 18 | rlmAII | 21 |
| myrA (M. griseorubida) | G748 | 18, 53 | rlmAII | 21 |
| tlrD (S. fradiae) | A2058 monomethylation | 5 | ermN | 57 |
| myrB (M. griseorubida) | A2058 monomethylation | 53; M.L., unpublished work | ermW | 57 |
| lrm (S. lividans) | A2058 monomethylation | 25 | ermO | 57 |
| tlrA (S. fradiae) | A2058 dimethylation | 6 | ermS | 57 |
| ermE (Saccharopolyspora erythaea) | A2058 dimethylation | 58 | ermE (unchanged) |
To aid readability the original Streptomyces tlr gene names are retained in the text, whereas the new nomenclature is used for any recently discovered homologues in other organisms.
The target for methylation by S. fradiae TlrB has recently been identified as the base of nucleotide G748 in 23S rRNA (18). After the function of TlrB was defined, numerous other bacteria have been shown to possess homologues of this protein (20, 21). The rRNA target of the homologues from Gram-positive bacteria is identical to that of TlrB, and these enzymes have been collectively renamed RlmAII (Table 1). Intriguingly, however, the tylosin-resistance phenotype that is conferred by TlrB (and also by other RlmAII homologues) when expressed in Streptomyces could be duplicated neither in other Gram-positive bacteria nor in a susceptible strain of the Gram-negative bacterium Escherichia coli. These latter strains remained sensitive to tylosin despite complete methylation of G748 by TlrB. Clearly some additional factor was playing a role in the drug-resistance TlrB phenotype observed in Streptomyces.
Here we show that interpretation of the resistance mechanism involving TlrB has been clouded by the presence of a tlrD homologue on the chromosome of many Streptomyces species, including common laboratory strains of Streptomyces lividans (Table 1). After inactivation of the tlrD homologue, tlrB no longer confers the tylosin-resistant phenotype in S. lividans. Tylosin resistance is reestablished in the S. lividans tlrD− strain after transformation with plasmids that express both the TlrB and TlrD methyltransferases. Neither of these two methyltransferases acting on its own confers any appreciable resistance to tylosin. These results have been consistently duplicated in a tylosin-susceptible strain of E. coli. In addition, the Gram-positive bacterium Corynebacterium glutamicum, which possesses its own tlrB (rlmAII) but has no intrinsic resistance to tylosin, becomes resistant on transformation with tlrD. The requirement for methylation at two distinct sites in the rRNA represents a novel type of resistance mechanism. This synergistic mechanism by which the single methyl groups at G748 and A2058 confer resistance could provide S. fradiae with an effective means of fine-tuning its response to varying intracellular tylosin concentrations during its life cycle.
Materials and Methods
Bacterial Strains and Plasmids.
The bacterial strains and plasmids used in this study are listed in Table 4, which is published as supporting information on the PNAS web site, www.pnas.org. Briefly, the E. coli strain DH10B was used for cloning procedures; the drug-permeable strains E. coli AS19(rlmAI−), S. lividans 1326 (WT), and S. lividans(lrm−mgt−) were used for the determination of the minimal inhibitory concentrations (MICs) of macrolide and lincosamide antibiotics and are described below. For plasmid maintenance, ampicillin was used at 100 μg/ml, kanamycin at 25 μg/ml, tetracycline at 12.5 μg/ml, apramycin at 100 μg/ml, and thiostrepton at 10–50 μg/ml. For MIC determinations, the macrolide and lincosamide antibiotics listed in Tables 2 and 3 were serially diluted in 2-fold steps.
Table 2.
MICs in mg/l for the S. lividans lrm–mgt knockout strain
| Antibiotic
|
S. lividans (lrm−mgt−) | ||||||
|---|---|---|---|---|---|---|---|
| Control | rlmAI (G745) | rlmAII (G748) | tlrD (A2058) | rlmAI + tlrD | rlmAII + tlrD | ermE (2A2058) | |
| Tylosin | 2 | 2 | 8 | 8 | 8 | 512 | 8,000 |
| Erythromycin A | 2 | 2 | 2 | 64 | 32 | 64 | 8,000 |
| Lincomycin | 32 | 32 | 32 | 16,000 | 16,000 | 16,000 | 16,000 |
Cells contained the plasmid-encoded methyltransferases rlmAI (Acinetobacter), rlmAII (S. fradiae), tlrD (S. fradiae), and ermE (Saccharopolyspora erythraea), and combinations of these genes. Other homologues of the rlmA genes gave the same MIC values. Coexpression of any of the rlmA genes with ermE did not alter the ermE-resistance levels. Controls contain the empty pHJL 401 plasmid. MICs were additionally measured in S. lividans WT cells (see Table 5, which is published as supporting information on the PNAS web site).
Table 3.
MICs in mg/l for E. coli AS19(rlmAI−)
| Antibiotic
|
E. coli AS19(rlmAI−) | ||||||
|---|---|---|---|---|---|---|---|
| Control | rlmAI (G745) | rlmAII (G748) | tlrD (A2058) | rlmAI + tlrD | rlmAII + tlrD | ermE (2A2058) | |
| Tylosin | 1 | 2 | 2 | 4 | 16 | 256 | 2,048 |
| Mycinamycin II | 1 | 1 | 2 | 2 | 8 | 256 | 1,024 |
| Erythromycin A | 0.5 | 2 | 2 | 128 | 256 | 256 | 2,048 |
| Clarithromycin | 0.25 | 1 | 1 | 32 | 128 | 128 | 512 |
| Spiramycin | 8 | 16 | 16 | 256 | 512 | 512 | 2,048 |
| Carbomycin A | 0.5 | 1 | 1 | 16 | 16 | 16 | >128 |
| Lincomycin | 16 | 64 | 64 | 16,000 | 16,000 | 16,000 | 16,000 |
| Clindamycin | 2 | 4 | 4 | 512 | 512 | 512 | 512 |
Cells contained the plasmid-encoded methyltransferases rlmAI (E. coli), rlmAII (S. fradiae), tlrD (S. fradiae), and ermE (S. erythraea). All cells contained two compatible plasmids, one based on pUHE24-2 (28) for the rlmA genes and the other on pSD184 (27) for the tlrD and ermE genes (Table 4). Controls contain the two empty plasmids. Other homologues of the rlmA genes gave the same MIC values. Coexpression of any of the rlmA genes with ermE did not alter the ermE-resistance levels. Tylosin, erythromycin, spiramycin, and lincomycin were obtained from Sigma, clarithromycin was from Abbott, and carbomycin was from Pfizer Diagnostics.
Inactivation of lrm and mgt Genes in S. lividans.
The gene pair lrm and mgt (approximately 2.2 kb) was amplified by PCR using the primers 5′-ATAGAATTCTCCTGTCACGGAAGACAAGGACCG and 5′-ATCAAGCTTGTCTGACCGGCACGCCGTCG. The PCR product was cloned into pUC19 at the EcoRI and HindIII sites, after restricting at the sequences underlined. The lrm and mgt genes were then cut with SexA1 to remove a fragment composed of 180 bp from the 3′ end of lrm and 360 bp from the 5′ end of mgt, and an apramycin-resistance gene cassette (22) was inserted into the gap to create plasmid pSD113. A 3.4-kb fragment from the E. coli–Streptomyces shuttle vector pGM160, containing the pSG5 temperature-sensitive replicon and the thiostrepton-resistance gene (23), was then inserted into NcoI/EcoRI sites of pSD113, generating pSD113–160. Plasmid pSD113–160 was used to inactivate the lrmand mgt genes by a double-crossover event using standard methods (24, 25). Thiostrepton-sensitive/apramycin-resistant colonies were selected, and the integrity of the chromosomal replacement was checked by PCR.
Determination of MICs.
The MICs of macrolide and lincosamide antibiotics were determined for both S. lividans and E. coli (Tables 2 and 3). S. lividans strains were transformed with derivatives of plasmid pHJL401 (26) that contained either single or dual methyltransferase genes under control of the ermE promoter. S. lividans spores were spread onto R5 plates containing 10 μg/ml thiostrepton and MLSB antibiotics (Table 2), and growth was evaluated after 3–5 days of incubation at 30°C.
For studies on E. coli, a hyperpermeable strain AS19(rlmAI−), in which the rRNA methyltransferase gene rlmAI (formerly termed rrmA, see Table 1) had been inactivated (27), was transformed with two compatible plasmids. One was chosen from a set of derivatives based on the ColE1 plasmid pUHE24–2 (28) and contained either a Gram-negative rlmAI G745 methyltransferase gene or a Gram-positive rlmAII G748 methyltransferase gene (see Table 4). The second plasmid was under control of the p15A replicon and contained either the S. fradiae tlrD A2058 monomethyltransferase gene or the S. erythraea ermE A2058 dimethyltransferase gene. All of the methyltransferase genes in the E. coli vectors were under the control of the lac promoter. MICs were measured by using an agar dilution method (29). Approximately 104 E. coli cells were applied to the surface of LB agar plates (30), which contained ampicillin, tetracycline, 1 mM isopropyl β-d-thiogalactoside, and serial dilutions of the macrolide and lincosamide drugs (Table 3). Growth was evaluated after ≈20 h of incubation at 37°C. All MIC values were measured a minimum of three times and were highly reproducible.
Quantification of Base Methylation.
The levels of methylation at G745 and G748 by the RlmA methyltransferases and dimethylation at A2058 by ErmE were measured by reverse transcriptase primer extension procedures (18, 31–33). Monomethylation at the N6 of adenine cannot be estimated by this method, and the degree of TlrD methylation was inferred indirectly by comparison of the lincosamide-resistance profiles of TlrD+ and ErmE+ cells (27).
Computer Simulation of the Ribosome–Tylosin Interaction.
The docking of tylosin within the exit channel of the 50S subunit was modeled from the resistance patterns conferred by the TlrB and TlrD methylations presented here, together with chemical footprinting data of tylosin and related macrolides on the 23S rRNA (34). The positions of the neutral and amino-sugars on the macrolide rings influence whether the base methylations confer resistance, as well as determining which 23S rRNA nucleotides the drugs protect from chemical modification. These data enabled us to orient unambiguously tylosin within the ribosomal MLSB site.
Results
TlrB (RlmAII) Methyltransferases and Tylosin Resistance.
We reported recently that the S. fradiae and Micromonospora griseorubida rlmAII (tlrB) gene products are methyltransferases that specifically target guanine 748 within the loop of 23S rRNA hairpin 35. A causal link between this methylation and tylosin resistance was established in S. lividans (18). We became aware that the mechanism by which TlrB confers tylosin resistance was more complex than first anticipated while studying C. glutamicum. The strain we used has an rlmAII homologue on a plasmid (pAG1) (35); this gene is constitutively expressed and leads to complete methylation of nucleotide G748. Remarkably, however, C. glutamicum is sensitive to tylosin. Expression of the C. glutamicum plasmid copy of rlmAII in S. lividans confers the same level of resistance as S. fradiae tlrB. Clearly, Streptomyces has some component that is augmenting the effect of tlrB, and this component is missing from C. glutamicum. The obvious place to look was at any additional genes in S. lividans that had been implicated in macrolide resistance.
Inactivation of the lrm and mgt Genes in S. lividans.
Standard laboratory strains of S. lividans, such as TK21 and ATCC 1326, exhibit low-level macrolide resistance. Drug resistance is presumably conferred by the chromosomal gene pair lrm and mgt (25), where lrm (recently reclassified as ermO, Table 1) is functionally identical to the S. fradiae tlrD (ermN) gene, the product of which monomethylates the N6 position of adenine 2058 of 23S rRNA (5). This methyltransferase confers high-level resistance to lincomycin and lower levels of resistance to some macrolides (36). The mgt gene is immediately downstream of lrm and encodes a macrolide glycosyltransferase that inactivates a variety of macrolides, including erythromycin and tylosin (37).
We disrupted the lrm and mgt genes in S. lividans 1326 to facilitate unambiguous interpretation of the tlrB resistance data. Putative lrm–mgt disruption mutants were randomly chosen and were shown to have significantly reduced resistance to erythromycin and tylosin, with MICs falling from 32–64 μg/ml in the WT to 2 μg/ml in the mutants (Table 2). These levels of resistance are consistent with a report for disruption of the same gene pair with a hygromycin-resistance cassette in S. lividans strain TK21 (25). Two putative S. lividans(lrm−mgt−) clones were selected, and the correctness of the knockout event was confirmed by PCR analysis of the chromosomes of both clones.
Methylation in 23S rRNA Hairpin 35 Is Not Sufficient to Confer Tylosin Resistance.
Expression of TlrB (or any of the other RlmAII methyltransferases) in S. lividans(lrm−mgt−) in the absence of a TlrD homologue conferred only a slight increase in tolerance to tylosin (Table 2), confirming the observations made with C. glutamicum. Expression of TlrD on its own has the same minor effect on tylosin tolerance (Table 2). Of the other methyltransferases investigated, the TlrB orthologue RlmAI had no effect on macrolide resistance, whereas cells with the A2058 dimethyltransferase, ErmE, were highly resistant to all of the MLSB drugs used in this study. Reverse transcriptase analyses of rRNAs from these cells (not shown) demonstrated that G745 and G748 were completely methylated by RlmAI and RlmAII, respectively, and ErmE dimethylation at A2058 was consistently >80%. TlrD monomethylation at A2058 cannot be measured directly by reverse transcription, although the high levels of resistance to the lincosamide drugs lincomycin and clindamycin indicated that methylation at A2058 approached stoichiometric levels (ref. 27; Table 2).
The E. coli mutant strain AS19 is susceptible to most MLSB drugs including tylosin (27). This strain, like many other Gram-negative bacteria (21), has a functional rlmAI gene. This finding establishes, in agreement with the results for S. lividans described above, that G745 methylation does not interfere with binding of tylosin to the ribosome. Nevertheless, we inactivated rlmAI in AS19 to have a clean genetic background in which the phenotypes of novel combinations of methyltransferases could be examined.
E. coli AS19(rlmAI−) was transformed with plamid-encoded copy of the rlmAII homologue from S. fradiae, Bacillus subtilis, C. glutamicum, Streptococcus pneumoniae, and M. griseorubida. In parallel sets of transformations, a copy of rlmAI from E. coli, Acinetobacter, or Shewanella was introduced on similar plasmids (see Table 4). The tlrD and ermE genes were introduced on plasmids that are compatible with the rlmA plasmids. Expression of each of the methyltransferase genes, alone or in combination, resulted in essentially the same high levels of rRNA methylation in E. coli as observed in S. lividans, and the MIC levels obtained in both bacteria are comparable (Tables 2 and 3). The E. coli transformants were then screened with an extended range of drugs, including the additional 16-membered ring macrolides mycinamycin, carbomycin, and spiramycin. The results demonstrate that the G745-specific rlmAI, the G748-specific rlmAII, or the A2058-specific tlrD monomethyltransferase genes are insufficient on their own to confer tylosin resistance.
TlrB Acts Synergistically with TlrD to Confer Tylosin Resistance.
Coexpression of tlrB and tlrD in S. lividans(lrm−mgt−) produces the same tylosin-resistant phenotype that was originally reported for WT S. lividans strains with plasmid-encoded tlrB (16–19). Simultaneous expression of both genes results in an increased resistance to tylosin that corresponds to eight dilution steps (256-fold) compared with an increase of only two dilution steps (4-fold) by either gene on its own. These effects were duplicated in the E. coli AS19(rlmAI−) system, with the combination of tlrB and tlrD conferring 256-fold resistance to both tylosin and the structurally similar macrolide mycinamycin (Table 3). The effects of concomitant expression of tlrB and tlrD on the other macrolide and lincosamide drugs were at best additive. Dimethylation at A2058 by ErmE conferred high resistance to all of the drugs tested here, and no increase was seen after combining ErmE with any of the other methyltransferases.
Discussion
Here we describe a resistance mechanism that is specific for tylosin and the structurally related macrolide mycinamycin, but does not confer resistance to other 16-membered macrolides such as carbomycin and spiramycin, 14-membered ring macrolides, or lincosamide antibiotics. This form of resistance differs from previously described mechanisms in that it requires methylation at two distinct positions, G748 and A2058, within the drug binding site. Neither methylation on its own confers more than a marginal increase in tolerance to tylosin or mycinamycin. The methylated bases are widely separated in the primary structure of the rRNA, but are folded to lie within 15 Å of each other in the peptide exit tunnel of the 50S ribosomal subunit (38–41).
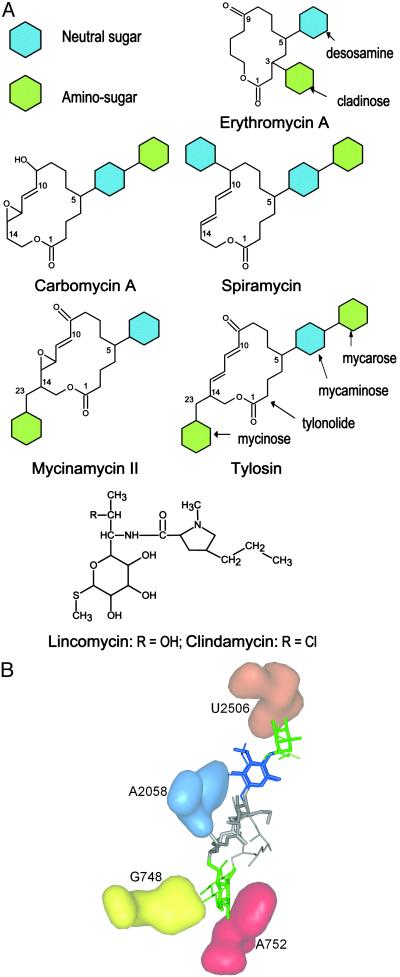
Tylosin and mycinamycin have unique glycosylation patterns among the macrolides tested here, with a mycaminose sugar at C5 of the lactone ring and a mycinose sugar extending from C14 (Fig. 1). The constellation of sugars around the macrolactone ring determines how a macrolide interacts with the ribosomal MLSB site and whether base methylations within this site interfere with drug binding. We used these data to model tylosin into the ribosomal MLSB site. First, the tylosin 5-mycaminose was superimposed onto the erythromycin/clarithromycin 5-desosamine in the crystal structure (40), placing the 2′-OH of mycaminose within hydrogen bonding distance of the N6 of A2058; mycinamycin fits into the MLSB site in the same manner although it lacks the mycarose moiety of the tylosin 5-disaccharide (Fig. 1A), which reaches toward the peptidyl transferase center (42) to make an additional interaction at U2506. This latter interaction is also facilitated by the 5-disaccharides of spiramycin and carbomycin (34) and is probably linked to ability of these drugs to inhibit the peptidyl transferase reaction. Consistent with this finding, the macrolides in Fig. 1A with a monosaccharide at the 5-position do not directly inhibit peptide bond formation to any appreciable extent (34, 43, 44).
Fig. 1.
(A) Structures of the antibiotics used in this study. Clarithromycin is the 6-methoxy derivative of erythromycin A. (B) Comparison of the structures of ribosome-bound tylosin obtained by crystallography (thick lines; ref. 52) and modeling (thin lines; this study). The positions of the tylosin 5-mycaminose (blue) amino sugar and the 5-mycarose (green) and 23-mycinose (green) neutral sugars are color-coded as in A and can be seen to interact with A2058 (blue), U2506 (orange), and G748 (yellow), respectively. The 23-mycinose is positioned where it additionally interacts with A752 (red), protecting the base from chemical modification (34).
The placement of the 5-sugar moieties orients tylosin and mycinamycin in the MLSB site so that the mycinose sugar, attached to C23 via C14 on the other side of the tylonolide ring (Fig. 1A), can be positioned to interact with the base of G748. This puts the mycinose sugar within 4 Å of the base of nucleotide A752, and, consistent with this, both tylosin and desmycosin (which is structurally equivalent to mycinamycin) protect A752 from chemical modification (34). In contrast, nucleotide A752 is not protected by macrolides such as erythromycin, carbomycin, and spiramycin, which lack a 14-[23-]sugar moiety (34, 45, 46), nor by the smaller lincosamide structures (47). The importance of the mycinosyl moiety of tylosin for antimicrobial activity has been demonstrated (48–51).
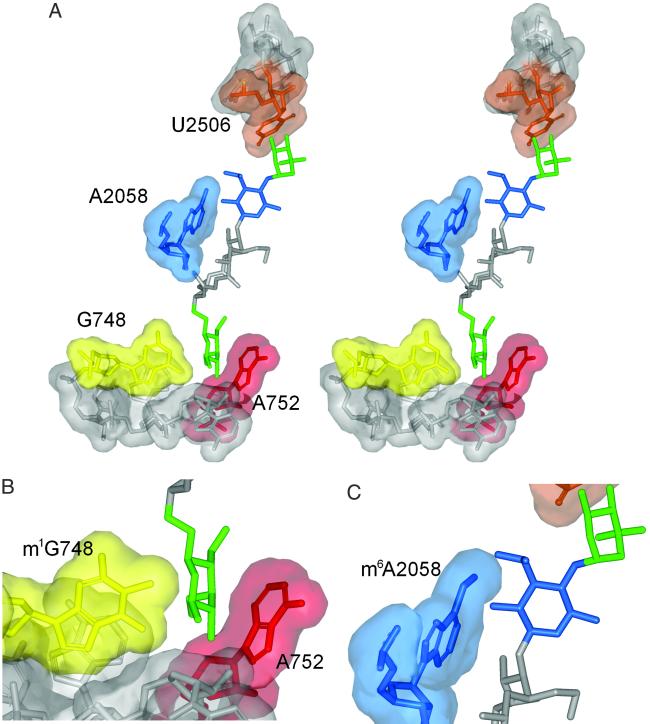
Since we submitted this article, a high-resolution crystal structure of tylosin bound to the 50S subunit of Haloarcula marismortui has been published (52). The positions of the three sugars in the tylosin crystal structure fit remarkably well with the model described above. However, apart from the 5- and 14-positions, we lacked any reliable criteria to model the rest of the tylonolide ring and, taking a cue from the erythromycin-50S structure (40), this ring was put into a tensed conformation, rather than in the lower energy form found in the tylosin-50S crystal structure (52). We have left an outline of our original model in Fig. 1B as a tribute to the strengths and pitfalls of molecular modeling. The stereo picture of tylosin in its binding site (Fig. 2A) is a representation of the crystal structure (52) in which key bases (Fig. 2 B and C) have been substituted for those found in Streptomyces and Escherichia ribosomes.
Fig. 2.
(A) Stereoview of tylosin in its binding site. The figure is based on the crystal structure (52), in which key bases are substituted for those in the Escherichia and Streptomyces rRNA sequences. (B and C) Enlargement of the targets for TlrB and TlrD methyltransferases at nucleotides G748 and A2058, respectively, showing how methylation here impinges on tylosin's binding space. The clouds indicate the van der Waals contact radii around the nucleotides atoms (not including protons).
From Fig. 2C, one can envisage the effect of monomethylation at N6 of A2058 on tylosin binding. The single methyl group at A2058 encroaches on the tylosin binding space but can also freely rotate around the adenine C6-N6 bond bringing it away from the macrolide. Adjustment of an A2058 methyl group might be facilitated while other rRNA contacts, such as at G748 in domain II and between the tylonolide ring and the hydrophobic surface at C2 of A2058 and A2059 (52), hold tylosin within the MLSB site. Similarly, methylation of G748 alone (Fig. 2B) does not appreciably disrupt drug binding. Conceivably, the 5-mycaminose and tylonolide contacts hold the drug in its binding site while placement of the 23-mycinose moiety is accommodated around the G748 methyl group. However, concomitant methylation at both G748 and A2058 would hinder the fit of either tylosin or mycinamycin within the MLSB site, consistent with the MIC data for strains methylated at both of these nucleotides.
The recent crystallography study also describes the structures of carbomycin, spiramycin, and an erythromycin derivative within the MLSB site (52). Although the crystal structures indicate that neither the G745 nor the G748 methylation would contact these drugs, we note that each of these methylations slightly increases the MICs of these drugs (Table 3) for reasons that we do not presently understand. With the exception of tylosin and mycinamycin, monomethylation of A2058 confers a distinct phenotype to all of the drugs in Table 3. For example, the lincosamides make more than one H-bond to the N6 of A2058 (40), and addition of a single methyl group is sufficient to confer maximal resistance (Tables 2 and 3), as reported in ref. 36. A clear effect of A2058 monomethylation was also observed for erythromycin and clarithromycin and to a lesser extent for spiramycin and carbomycin (Table 3). This finding fits with the lack of any substantial interaction between these drugs and domain II of the rRNA, which could assist binding when A2058 is monomethylated. Dimethylation at A2058 is uncompromisingly consistent in conferring high-level resistance to all MLSB drugs and does so by removing the hydrogen-bonding potential of the N-6 position while sterically hindering access to the drug site.
S. fradiae possesses an array of resistance determinants, which could enable the organism to respond to varying levels of tylosin during mycelial differentiation. The tlrC-encoded efflux and tlrD-encoded A2058 monomethylation are constitutively expressed, providing protection from low concentrations of tylosin diffusing in from older regions of the mycelium. When local regions of the mycelium begin to produce tylosin, increasing intracellular drug concentrations will induce tlrB expression, leading to G748 methylation and thereby enhancing the effect of tlrD. Higher drug concentrations will ultimately induce tlrA expression and A2058 dimethylation, maximizing resistance and ensuring the continuance of protein synthesis during endogenous tylosin production. A similar stepwise response to drug concentrations possibly also occurs in the mycinamycin producer M. griseorubida, which is presently the only other organism that has been shown to possess both a tlrB homologue and a (putative) tlrD homologue (ref. 53 and unpublished data).
Questions remain concerning how these resistance mechanisms evolved, and why many Gram-positives possess a tlrB (rlmAII) homologue (21), despite the fact that only a few of these bacteria synthesize drugs, and most of them lack an accompanying tlrD homologue. A potential clue comes from recent studies showing that the constricted G748/A2058 region of the ribosome tunnel can discern specific sequences in the nascent peptide chain (54, 55). Possibly, the selection for methylation in this region might originally have been connected with how the ribosome modulated the extrusion of peptides through the tunnel. As macrolide drugs bind at this same site, resistance could have been an incidental spin-off, which led to the subsequent recruitment of methyltransferases into roles as drug-resistance determinants.
Supplementary Material
Acknowledgments
We are grateful to Masaharu Inouye and Kazumi Hasegawa of Asahi Kasei Corporation for providing mycinamycin, Donnie Owens of Pfizer Inc. for carbomycin, Wolfgang Wohlleben for plasmid pGM160, and Jacob Poehlgaard for modeling tylosin on 50S and preparing the figures. We thank Eric Cundliffe for discussions and comments on the manuscript. The research was supported by the Danish Biotechnology Instrument Centre (DABIC), the European Commission's Fifth Framework Program (Grant QLK2-CT2000-00935), and the Nucleic Acid Centre of the Danish National Research Foundation.
Abbreviations
MLSB, macrolide, lincosamide, and streptogramin B antibiotics
MIC, minimal inhibitory concentration
References
- 1.Cundliffe E. (1989) Annu. Rev. Microbiol. 43, 207-233. [DOI] [PubMed] [Google Scholar]
- 2.Bryskier A. J., Butzler, J. P., Neu, H. C. & Tulkens, P. M., (1993) Macrolides: Chemistry, Pharmacology, and Clinical Uses (Arnette Blackwell, Paris).
- 3.Baltz R. H. & Seno, E. T. (1988) Annu. Rev. Microbiol. 42, 547-574. [DOI] [PubMed] [Google Scholar]
- 4.Bate N., Butler, A. R., Gandecha, A. R. & Cundliffe, E. (1999) Chem. Biol. 6, 617-624. [DOI] [PubMed] [Google Scholar]
- 5.Zalacain M. & Cundliffe, E. (1991) Gene 97, 137-142. [DOI] [PubMed] [Google Scholar]
- 6.Zalacain M. & Cundliffe, E. (1989) J. Bacteriol. 171, 4254-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leclercq R. & Courvalin, P. (1991) Antimicrob. Agents Chemother. 35, 1267-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weisblum B. (1995) Antimicrob. Agents Chemother. 39, 577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farrell D. J., Morrissey, I., Bakker, S. & Felmingham, D. (2001) J. Antimicrob. Chemother. 48, 541-544. [DOI] [PubMed] [Google Scholar]
- 10.Johnston N. J., De Azavedo, J. C., Kellner, J. D. & Low, D. E. (1998) Antimicrob. Agents Chemother. 42, 2425-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leclercq R. & Courvalin, P. (2002) Antimicrob. Agents Chemother. 46, 2727-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sutcliffe J. A. & Leclercq, R. (2002) in Milestones in Drug Therapy: Macrolide Antibiotics, eds. Schönfeld, W. & Kirst, H. A. (Birkhäuser, Basel), pp. 281–317.
- 13.Ouellette M., Legare, D. & Papadopoulou, B. (1994) Trends Microbiol. 2, 407-411. [DOI] [PubMed] [Google Scholar]
- 14.Rosteck P. R., Reynolds, P. A. & Hershberger, C. L. (1991) Gene 102, 27-32. [DOI] [PubMed] [Google Scholar]
- 15.Schöner B., Geistlich, M., Rosteck, P., Jr., Rao, R. N., Seno, E., Reynolds, P., Cox, K., Burgett, S. & Hershberger, C. (1992) Gene 115, 93-96. [DOI] [PubMed] [Google Scholar]
- 16.Birmingham V. A., Cox, K. L., Larson, J. L., Fishman, S. E., Hershberger, C. L. & Seno, E. T. (1986) Mol. Gen. Genet. 204, 532-539. [DOI] [PubMed] [Google Scholar]
- 17.Fouces R., Mellado, E., Diez, B. & Barredo, J. L. (1999) Microbiology 145, 855-868. [DOI] [PubMed] [Google Scholar]
- 18.Liu M., Kirpekar, F., van Wezel, G. P. & Douthwaite, S. (2000) Mol. Microbiol. 37, 811-820. [DOI] [PubMed] [Google Scholar]
- 19.Wilson V. & Cundliffe, E. (1999) J. Antibiot. 52, 288-296. [DOI] [PubMed] [Google Scholar]
- 20.Bujnicki J. M., Blumenthal, R. M. & Rychlewski, L. (2002) J. Mol. Microsc. Biotechnol. 4, 93-99. [PubMed] [Google Scholar]
- 21.Liu M. & Douthwaite, S. (2002) Mol. Microbiol. 44, 195-204. [DOI] [PubMed] [Google Scholar]
- 22.Blondelet-Rouault M. H., Weiser, J., Lebrihi, A., Branny, P. & Pernodet, J. L. (1997) Gene 190, 315-317. [DOI] [PubMed] [Google Scholar]
- 23.Muth G., Nuβbaumer, B., Wohllebenund, W. & Pühler, A. (1989) Mol. Gen. Genet. 219, 341-348. [Google Scholar]
- 24.Kieser T., Bibb, M. J., Buttner, M. J., Chatter, K. F. & Hopwood, D. A., (2000) Practical Streptomyces Genetics (John Innes Foundation, Norwich, CT).
- 25.Pernodet J. L., Fish, S., Blondelet-Rouault, M. H. & Cundliffe, E. (1996) Antimicrob. Agents Chemother. 40, 581-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larson J. L. & Hershberger, C. L. (1986) Plasmid 15, 199-209. [DOI] [PubMed] [Google Scholar]
- 27.Liu M. & Douthwaite, S. (2002) Antimicrob. Agents Chemother. 46, 1629-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bujard H., Gentz, R., Lanzer, M., Stuber, D., Muller, M., Ibrahimi, I., Hauptle, M. T. & Dobberstein, B. (1987) Methods Enzymol. 155, 416-433. [DOI] [PubMed] [Google Scholar]
- 29.National Committee for Clinical Laboratory Standards, (1993) Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, M7–A3 (National Committee for Clinical Laboratory Standards, Villanova, PA).
- 30.Sambrook J., Fritsch, E. F. & Maniatis, T., (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 31.Hansen L. H., Kirpekar, F. & Douthwaite, S. (2001) J. Mol. Biol. 310, 1001-1010. [DOI] [PubMed] [Google Scholar]
- 32.Sigmund C. D., Ettayebi, M., Borden, A. & Morgen, E. A. (1988) Methods Enzymol. 164, 673-690. [DOI] [PubMed] [Google Scholar]
- 33.Vester B. & Douthwaite, S. (1994) J. Bacteriol. 176, 6999-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poulsen S. M., Kofoed, C. & Vester, B. (2000) J. Mol. Biol. 304, 471-481. [DOI] [PubMed] [Google Scholar]
- 35.Tauch A., Puhler, A., Kalinowski, J. & Thierbach, G. (2000) Plasmid 44, 285-291. [DOI] [PubMed] [Google Scholar]
- 36.Jenkins G., Zalacain, M. & Cundliffe, E. (1989) J. Gen. Microbiol. 135, 3281-3288. [DOI] [PubMed] [Google Scholar]
- 37.Jenkins G. & Cundliffe, E. (1991) Gene 108, 55-62. [DOI] [PubMed] [Google Scholar]
- 38.Ban N., Nissen, P., Hansen, J., Moore, P. B. & Steitz, T. A. (2000) Science 289, 905-920. [DOI] [PubMed] [Google Scholar]
- 39.Harms J., Schluenzen, F., Zarivach, R., Bashan, A., Gat, S., Agmon, I., Bartels, H., Franceschi, F. & Yonath, A. (2001) Cell 107, 679-688. [DOI] [PubMed] [Google Scholar]
- 40.Schlünzen F., Zarivach, R., Harms, J., Bashan, A., Tocilj, A., Albrecht, R., Yonath, A. & Franceschi, F. (2001) Nature 413, 814-821. [DOI] [PubMed] [Google Scholar]
- 41.Yusupov M. M., Yusupova, G. Z., Baucom, A., Lieberman, K., Earnest, T. N., Cate, J. H. & Noller, H. F. (2001) Science 292, 883-896. [DOI] [PubMed] [Google Scholar]
- 42.Nissen P., Hansen, J., Ban, N., Moore, P. B. & Steitz, T. A. (2000) Science 289, 920-930. [DOI] [PubMed] [Google Scholar]
- 43.Gale E. F., Cundliffe, E., Reynolds, P. E., Richmond, M. H. & Waring, M. J., (1981) The Molecular Basis of Antibiotic Action (Wiley, London).
- 44.Vázquez D., (1979) Inhibitors of Protein Biosynthesis (Springer, Berlin). [DOI] [PubMed]
- 45.Hansen L. H., Mauvais, P. & Douthwaite, S. (1999) Mol. Microbiol. 31, 623-632. [DOI] [PubMed] [Google Scholar]
- 46.Xiong L., Shah, S., Mauvais, P. & Mankin, A. S. (1999) Mol. Microbiol. 31, 633-639. [DOI] [PubMed] [Google Scholar]
- 47.Douthwaite S. (1992) Nucleic Acids Res. 20, 4717-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cacciapuoti A. F., Loebenberg, D., Moss, E. L. J., Menzel, F. W., Rudeen, J. A., Naples, L. R., Cramer, C. L., Hare, R. S., Mallams, A. K. & Miller, G. H. (1990) J. Antibiot. (Tokyo) 43, 1131-1136. [DOI] [PubMed] [Google Scholar]
- 49.Gotoh Y., Saitoh, H. & Miyake, T. (1998) Carbohydr. Res. 309, 45-55. [DOI] [PubMed] [Google Scholar]
- 50.Kiyoshima K., Sakamoto, M., Nomura, H., Yoshioka, T., Okamoto, R., Sawa, T., Naganawa, H. & Takeuchi, T. (1989) J. Antibiot. (Tokyo) 42, 1661-1672. [DOI] [PubMed] [Google Scholar]
- 51.Fish S. A. & Cundliffe, E. (1996) J. Antibiot. (Tokyo) 49, 1044-1048. [DOI] [PubMed] [Google Scholar]
- 52.Hansen J. L., Ippolito, J. A., Ban, N., Nissen, P., Moore, P. B. & Steitz, T. A. (2002) Mol. Cell 10, 117-128. [DOI] [PubMed] [Google Scholar]
- 53.Inouye M., Morohoshi, T., Horinouchi, S. & Beppu, T. (1994) Gene 141, 39-46. [DOI] [PubMed] [Google Scholar]
- 54.Nakatogawa H. & Ito, K. (2002) Cell 108, 629-636. [DOI] [PubMed] [Google Scholar]
- 55.Tenson T. & Ehrenberg, M. (2002) Cell 108, 591-594. [DOI] [PubMed] [Google Scholar]
- 56.Gustafsson C. & Persson, B. C. (1998) J. Bacteriol. 180, 359-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roberts M. C., Sutcliffe, J., Courvalin, P., Jensen, L. B., Rood, J. & Seppälä, H. (1999) Antimicrob. Agents Chemother. 43, 2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Skinner R., Cundliffe, E. & Schmidt, F. J. (1983) J. Biol. Chem. 258, 12702-12706. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.