Abstract
A43, an essential subunit of yeast RNA polymerase I (pol I), interacts with Rrn3, a class I general transcription factor required for rDNA transcription. The pol I–Rrn3 complex is the only form of enzyme competent for promoter-dependent transcription initiation. In this paper, using biochemical and genetic approaches, we demonstrate that the A43 polypeptide forms a stable heterodimer with the A14 pol I subunit and interacts with the common ABC23 subunit, the yeast counterpart of the ω subunit of bacterial RNA polymerase. We show by immunoelectronic microscopy that A43, ABC23, and A14 colocalize in the three-dimensional structure of the pol I, and we demonstrate that the presence of A43 is required for the stabilization of both A14 and ABC23 within the pol I. Because the N-terminal half of A43 is clearly related to the pol II Rpb7 subunit, we propose that the A43–A14 pair is likely the pol I counterpart of the Rpb7–Rpb4 heterodimer, although A14 distinguishes from Rpb4 by specific sequence and structure features. This hypothesis, combined with our structural data, suggests a new localization of Rpb7–Rpb4 subunits in the three-dimensional structure of yeast pol II.
The yeast Saccharomyces cerevisiae possesses three forms of nuclear RNA polymerase (pol I, II, and III), which are distinct by their subcellular localization, chromatographic behavior, subunit composition, sensitivity to α-amanitine, and promoter/template specificity.
The unique essential function of pol I is to transcribe multiple ribosomal DNA units to generate the 35S ribosomal precursor (1), which is subsequently matured into the functional 18S, 5.8S, and 25S RNA species (2). Yeast pol I contains 14 subunits that include a core of five subunits (A190, A135, AC40, AC19, and ABC23) related to the β′βα2ω eubacterial core enzyme (3, 4). In addition to ABC23, four subunits (ABC27, ABC14.5, ABC10α, and ABC10β) are shared by the three forms of enzyme (5). Finally, five other polypeptides (A49, A43, A34.5, A14, and A12.2) are subunits of pol I (6–10).
Although requirement of such a complex structure is still an open question, substantial amounts of data have highlighted functional properties of subunits. The active site is carried out by the two large subunits in bacteria, archaebacteria and eukaryotes (11–14). Additional studies have provided insights into the function of smaller subunits of pol I. The nonessential A34.5 subunit may help the enzyme to overcome the topological constraints imposed on rDNA by transcription (8). The dispensable A14 subunit might cooperate with A34.5 subunit in this process (9) and is important for the stabilization of subunits A43 and ABC23 within the pol I (9). A49 subunit displays ribonuclease H activity (15), whose involvement in pol I transcription is still elusive. A12.2 subunit is the pol I counterpart of pol II and pol III subunits involved in RNA cleavage activity of both enzymes (16, 17), suggesting that A12.2 may be implicated in the retraction and/or termination of pol I. Finally, the essential A43 subunit (7), which is not required for catalytic activity of pol I (18), is the interaction target of Rrn3, a general transcription factor necessary for rDNA transcription (19). This interaction is critical for the formation of the pol I–Rrn3 complex, which is the only form of enzyme competent for promoter-dependent transcription initiation (20). The key role in the transcriptional process of the A43 polypeptide led us to further investigate its structure–function relationships.
In this paper, using a variety of biochemical and genetic approaches, we demonstrate that A43 interacts with subunits A14 and ABC23. Biochemical data indicate that subunit A43 is important for stabilization of subunits A14 and ABC23 within the pol I. Based on sequence analysis, we propose that the A43–A14 pair is the pol I counterpart of the pol II Rpb7–Rpb4 heterodimer. This hypothesis, combined with immunoelectronic microscopy data localizing the subunits A43 and A14 within the pol I, suggests a new localization of Rpb7–Rpb4 subunits in the three-dimensional structure of yeast pol II.
Materials and Methods
Plasmids, Strains, and Media.
Standard yeast genetic techniques and media were used (21). Strains harboring mutant alleles of the RPA43 gene (rpa43-4, rpa43-6, and rpa43-18) have been described (19). For in vitro transcription–translation, RPA14, RPA43, or genes encoding A43 subunit truncated at the N or C terminus were cloned in pRSET5d plasmid.
Two-Hybrid Assay.
The two-hybrid screening was performed as described (22). Strain Y190 transformed with pAS-RPA43 was transformed with a DNA genomic library (23). Transformants were selected on 50 and 75 mM 3AT-containing medium and tested for activation of LacZ reporter gene. RPA43 or genes corresponding to A43 subunit truncated at the N or C terminus were cloned in pGBT9 plasmid in frame with the Gal4 DNA binding domain, and activation of the reporter genes was detected as described (22).
Synthesis of 35S-Labeled Subunits.
Labeled subunits were synthesized in a wheat germ extract (Promega) supplemented with T7 RNA polymerase (Promega), [35S]methionine (0.8 mCi/ml; 1 Ci = 37 GBq), and 1 μg of the different plasmids.
Coimmunoprecipitation Experiments.
Labeled subunits (104 cpm) were preincubated in 100 μl of IP buffer (50 mM Hepes, pH 7.5/50 mM NaCl/20% glycerol) for 3 h at 10°C. Reactions were incubated for 3 h at 10°C with 20 μl of Dynal Panrabbit or Panmouse IgG beads (Dynal, Great Neck, NY), preloaded with primary antibodies, and were washed with IP buffer containing 1% milk and 0.1% Nonidet P-40. Proteins bound to the beads were separated by SDS/PAGE and visualized by autoradiography.
Membrane-Immobilized Protein Interaction Assay.
Far Western experiments were performed as described (24).
Expression and Copurification of ABC23 and A43.
Escherichia coli BL21 (DE3) strain was transformed with pET15b-ABC23 encoding a His-6 N-terminally tagged form of ABC23 and with pACYC184-11b encoding a wild-type version of A43. A control strain was only transformed with pACYC184-11b. Cells were grown, induced, collected, and lysed as described (19). Crude extracts corresponding to cells expressing either A43 and tagged ABC23, or only A43 were supplemented with 5 mM imidazole, and loaded onto a 5-ml Hi-Trap nickel column equilibrated with Ni buffer (20 mM Tris⋅HCl, pH 8/500 mM KOAc/5 mM Mg(Oac)2/20% glycerol/0.1% Tween 20). Bound proteins were eluted with 200 mM imidazole in Ni buffer, separated by SDS/PAGE, and revealed by Western blotting with anti-A43 antibodies and by Ponceau red staining.
Sequence and Structure Analysis.
Searches with protein databases were performed using PSI-BLAST (25). The secondary structure organization was visualized by using hydrophobic cluster analysis (26, 27), which also allowed the refinement of sequence alignments. Three-dimensional structures were manipulated by using the swiss-pdb viewer (28).
Results
The A43 Subunit Interacts with the A14 Subunit.
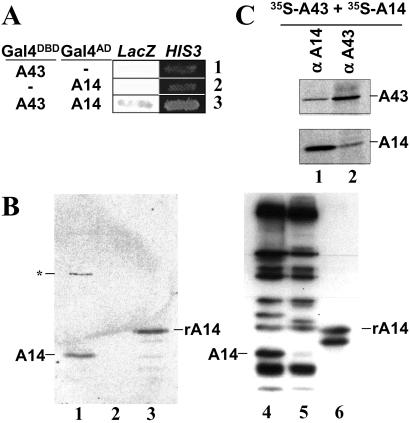
To identify the yeast pol I subunits interacting with the A43 subunit, we performed a two-hybrid screen of a yeast genomic library (22). The only pol I subunit identified in this screen was A14, which corresponded to five independent clones. Consistently, we observed the activation of transcription of the two reporter genes (LacZ and HIS3) in a yeast strain transformed by two plasmids driving the expression of the A43–Gal4DBD and the A14–Gal4AD fusion proteins, respectively (Fig. 1A, lane 3). No activation was obtained in control experiments where one of the two fusion proteins was omitted (Fig. 1A, lanes 1 and 2). This observation strongly suggests that A43 subunit interacts with A14 subunit.
Fig 1.
Subunit A43 interacts with subunit A14. (A) Two-hybrid. Y190 yeast strain was transformed by two plasmids, one driving the expression of a Gal4DBD fusion protein, the other driving the expression of a Gal4AD fusion protein (as indicated). Activation of the LacZ and HIS3 reporter genes was monitored by blue color in the presence of X-gal and by growth in the presence of 50 mM 3-aminotriazol, respectively. (B) Far Western blot. Ten micrograms each of pol I (lanes 1 and 4) and pol IΔ (lanes 2 and 5) and 1 μg of purified recombinant, tagged A14 subunit (lanes 3 and 6) were subjected to SDS/PAGE and transferred onto poly(vinylidene difluoride) membranes. The membrane was incubated with 35S-labeled A43 (lanes 1–3), and radiolabeled proteins were visualized by autoradiography. The asterisk indicates a nonspecific signal that does not correspond to a pol I subunit. Subunits of pol I, pol IΔ, and recombinant A14 subunit were identified by Western blot using anti-pol I antibodies (lanes 4–6, respectively). (C) Coimmunoprecipitation. 35S-A43 and 35S-A14 polypeptides were preincubated and subjected to immunoprecipitation by antibodies raised against subunit A14 or A43 (αA14 and αA43). Immunoprecipitated proteins were separated by SDS/PAGE and identified by autoradiography.
To confirm this result, we used a membrane-immobilized protein interaction assay (Far Western blotting). Purified pol I subunits were resolved by SDS/PAGE, and the separated subunits were transferred onto two poly(vinylidene difluoride) membranes. Subunits transferred onto the membrane were identified by Western blot analysis using antibodies raised against native pol I (Fig. 1B, lanes 4–6). The other membrane was incubated with 35S-labeled A43, and the interacting subunits were revealed by autoradiography (Fig. 1B, lanes 1–3). As shown in Fig. 1B, among the different pol I subunits, only the 14-kDa polypeptide was labeled (lane 1). When the same experiment was performed with an incomplete form of enzyme lacking subunits A14, ABC23, and A43 (pol IΔ) (lane 5), none of the subunits was labeled (lane 2). Finally, 35S-labeled A43 strongly bound to full-length recombinant A14 (lane 3), but only weakly to a higher electrophoretic mobility form of recombinant A14, probably proteolyzed at the C terminus (compare lanes 3 and 6). These observations suggest that A43 subunit directly interacts with A14 subunit.
We next investigated if A14 and A43 polypeptides could form a stable binary complex. 35S-labeled-A43 and -A14 polypeptides were independently synthesized in vitro in wheat germ extracts, preincubated, and immunoprecipitated using anti-A43 or anti-A14 antibodies. We previously checked that antibodies exclusively precipitated the subunit against which they were raised (data not shown). As shown in Fig. 1C, A43 was coprecipitated by anti-A14 antibodies (lane 1). Consistently, A14 was coprecipitated by anti-A43 antibodies (lane 2). Altogether, these results demonstrate that subunits A43 and A14 form a stable binary complex.
Genetic Interactions Between RPA43 and RPA14.
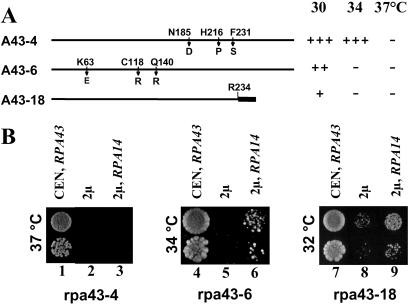
We previously isolated thermosensitive yeast strains harboring three different mutant rpa43 alleles (rpa43-4, rpa43-6, and rpa43-18) (19). The rpa43-18 allele encoded a truncated polypeptide, in which the 92 C-terminal residues were replaced by a short divergent sequence, whereas the rpa43-4 and rpa43-6 alleles harbored each a triple point mutation (Fig. 2A).
Fig 2.
Genetic interactions between RPA43 and RPA14 genes. (A) Positions and nature of the mutated residues in the rpa43 thermosensitive mutant are indicated. In A43-18, the C terminus of the protein is replaced by a short divergent amino acid sequence (thick line). Growth phenotype at different temperatures conferred by these different mutant alleles is summarized on the right. (B) Multicopy suppression. The rpa43-6, rpa43-18, and rpa43-4 mutant strains were transformed with a centromeric plasmid carrying the wild-type RPA43 gene (lanes 1, 4, and 7), with the empty pFL44 multicopy plasmid as a control (lanes 2, 5, and 8), or with the pFL44 vector harboring the RPA14 gene (lanes 3, 6, and 9). Growth of two dilutions of transformants was monitored at restrictive temperature.
To determine whether overexpression of A14 could suppress the thermosensitive phenotype of these mutant strains, each of them was transformed with a high copy number plasmid harboring the RPA14 gene or with the empty plasmid. As shown in Fig. 2B, the rpa43-6 and rpa43-18 strains transformed with the multicopy plasmid harboring RPA14 grew at restrictive temperature (34°C or 37°C, lanes 6 and 9), albeit more slowly than a wild-type strain (lanes 4 and 7). No suppression by overexpression of A14 subunit was observed for the rpa43-4 mutant strain (Fig. 2B, lane 3), indicating that the suppression effect observed for the rpa43-6 and rpa43-18 strains was allele specific. These data suggested that the conditional phenotypes of these two strains resulted, at least partially, from a defect in the association within the pol I of the mutant A43 subunit that can be overcome by increasing the cellular concentration of A14 subunit. In agreement with this hypothesis, A43 polypeptide was present in substoichiometric amounts in pol I purified from the rpa43-6 strain (19). These results demonstrated genetic interactions between the RPA43 and RPA14 genes and confirmed the biochemical data described above.
Interaction Domains of the A43 Subunit.
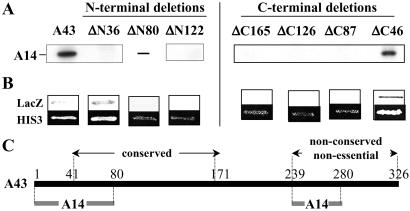
To delineate domains of A43 involved in these interactions, we generated a series of plasmids encoding A43 polypeptides progressively truncated at the N terminus (A43-ΔN36, A43-ΔN80, and A43-ΔN122 lacking the 36, 80, and 122 N-terminal residues, respectively) and at the C terminus (A43-ΔC46, A43-ΔC87, A43-ΔC126, and A43-ΔC165 lacking the 46, 87, 126, and 165 C-terminal residues, respectively). Each ORF encoding a truncated form of A43 in fusion with the hemagglutinin epitope (located at the C terminus for polypeptides truncated at the N terminus, and vice versa) was cloned into a vector under the control of the T7 RNA polymerase.
The interaction of the truncated forms of A43 with A14 subunit was first investigated by coimmunoprecipitation experiments. For this purpose, 35S-labeled A43 polypeptide variants and A14 subunit were independently synthesized in vitro in wheat germ extracts. SDS/PAGE analysis indicated that all A43 variants but the A43-ΔN80 polypeptide were correctly synthesized by in vitro transcription–translation. Each truncated form of A43 was next incubated with untagged A14 subunit, and coimmunoprecipitation experiments were performed with anti-hemagglutinin antibodies. As expected, full-length A43 subunit interacted with A14 subunit, whereas we could not detect a stable association of A14 with a truncated form of A43 lacking the N-terminal 36 residues, or a more extended deletion of 122 residues (Fig. 3A). Deleting the 46 C-terminal residues of A43 did not affect its interaction with A14, on the contrary to any further C-terminal deletion of A43 (Fig. 3A).
Fig 3.
Interaction domains of subunit A43 with subunit A14. (A) Interaction of A14 subunit with hemagglutinin-tagged A43 polypeptides progressively truncated at the N terminus (A43-ΔN) or at the C terminus (A43-ΔC) (the number of deleted residues is indicated). Each truncated form was synthesized in vitro in the presence of [35S]methionine (no protein was obtained for the ΔN80 construction), and their interaction with 35S-A14 was monitored by immunoprecipitation with anti-hemagglutinin antibodies. Immunoprecipitated proteins were separated by SDS/PAGE and visualized by autoradiography. (B) The ORFs encoding the truncated versions of subunit A43 were cloned in fusion with Gal4DBD in pGBT9 vector. Their ability to interact with A14 fused to Gal4AD was monitored in Y190 strain. Activation of the LacZ and HIS3 reporter genes was monitored by staining in the presence of X-gal and by analysis of growth in the presence of 50 mM 3-aminotriazol, respectively. (C) Schematic representation of the interaction domains of A43 with A14. The conserved domain of A43 (residues 42–168), present in putative orthologues of this subunit in Schizosaccharomyces pombe, Candida albicans, and higher eukaryotes (see ref. 22), is indicated. The 87 C-terminal residues of A43 are not conserved through evolution (data not shown). These residues are not essential for cell viability (see Fig. 2) but contain an interaction domain with A14.
Next, we confirmed these data by the two-hybrid method. Each ORF encoding a truncated version of A43 subunit was cloned in pGBT9 vector in fusion with the Gal4DBD. Using this approach, the 36 N-terminal residues of A43 were found to be dispensable for the interaction with A14, whereas deletions of the 80 or the 122 N-terminal residues abolished the interaction (Fig. 3B). These results are in agreement with the coimmunoprecipitation data except for the first 36 residues of A43, which are required in vitro but not in vivo for the interaction with A14 subunit. Although not strictly required, these residues may be important for the stability of the A43–A14 complex. Interaction data observed with C-terminal truncated forms of A43 fully confirmed the coimmunoprecipitation data (Fig. 3 A and B); deletion of the 46 C-terminal residues of A43 did not affect the interaction with A14, whereas no interaction was observed with larger C-terminal deletions.
Altogether, the coimmunoprecipitation and two-hybrid data suggested that residues situated at the N terminus (residues 1–80) and at the C terminus (residues 239–280) of A43 subunit are important for its interaction with A14 (Fig. 3C).
The A43 Subunit Stabilizes the A14 Subunit Within the pol I.
Upon purification from yeast cells disrupted for the RPA14 gene (9), we previously characterized an incomplete form of pol I, named pol IΔ, lacking subunits A43, ABC23, and A14. To determine what was the effect of the absence of A43 subunit on the stability of the pol I complex, we analyzed the subunit composition of the enzyme purified from a mutant yeast strain lacking the RPA43 gene (strain D128, ref. 7). The lethal phenotype of this deletion was rescued by the multicopy plasmid pNOY102 harboring the 35S rDNA gene under the control of a pol II galactose-inducible promoter (1). Crude extracts were prepared from the mutant strain and from an isogenic wild-type strain grown in parallel in a galactose-containing medium, and pol I was partially purified according to the procedure set up by Nomura and colleagues (1). The presence of pol I in the partially purified fractions was analyzed by Western blot with polyclonal antibodies raised against the enzyme (29). Whereas complete enzyme was present in the WT fraction, the Δ43 fraction contained a form of pol I lacking A43 and A14 polypeptides and containing a substoichiometric amount of ABC23 subunit (Fig. 4). The remaining pol I subunits were stably associated into a complex because they coeluted when subjected to permeation chromatography or to sedimentation on glycerol gradient (data not shown). This result showed that A43 subunit was critical for the stable association of A14 subunit within the pol I complex. These data, and the symmetrical result described above (i.e., A14 subunit stabilizes A43 subunit within the pol I), indicate that subunits A14 and A43 form a binary complex, which is important for the stable association of both subunits within the pol I complex.
Fig 4.
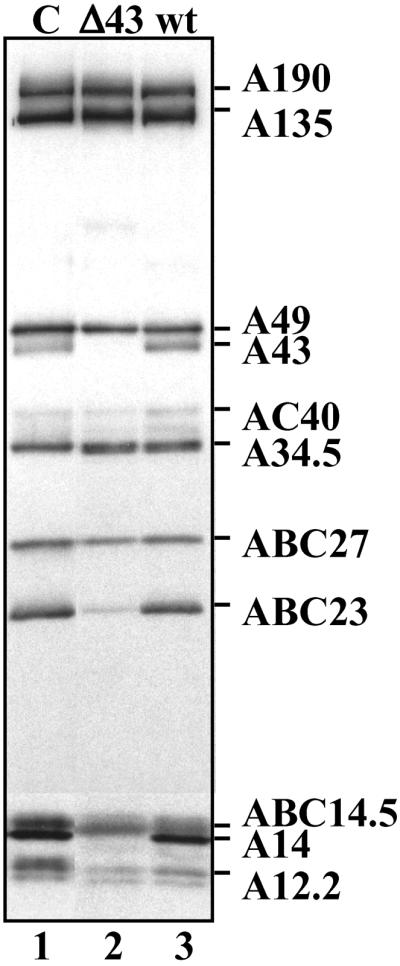
Subunit composition of pol I partially purified from the rpa43-Δ strain. The subunit composition of pol I partially purified from WT (lane 3) and rpa43-Δ strains (lane 2) was analyzed by Western blot using antibodies raised against yeast pol I (lane 1, purified pol I).
The A43 Subunit Interacts with the ABC23 Subunit.
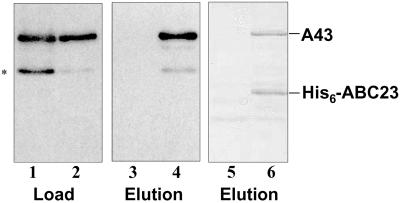
The biochemical data obtained with yeast strains disrupted for RPA14 or RPA43 genes confirmed the relationship between subunits A14 and A43 and, in addition, indicated that the association of the ABC23 subunit within the pol I was somehow affected by the absence of A43 and/or A14 subunits, suggesting that the interaction of ABC23 with one or these two polypeptides stabilizes its association within the complex. Yet, in vitro, ABC23 is able to reassociate very efficiently to the pol IΔ, in the absence of subunits A14 and A43 (18). To clarify the link between these three subunits, we explored the physical interactions between ABC23 subunit and A43 or A14 subunit. Whatever the method used, no interaction was detected between A14 and ABC23 subunits (data not shown). On the contrary, we isolated a stable binary complex after coexpression of A43 and ABC23 subunits in E. coli. Crude extracts were prepared from a bacterial strain coexpressing A43 and His-6-tagged ABC23. The extract was loaded onto a Nickel column, bound proteins were eluted by competition with imidazole, and the presence of A43 subunit in the elution fraction was checked by Western blot analysis using polyclonal anti-A43 antibodies. As shown in Fig. 5, A43 subunit was detected in the elution fraction (lane 4). Ponceau red staining on the membrane showed that approximately stoichiometric amounts of A43 polypeptide and tagged ABC23 were eluted from the column (lane 6). A43 subunit from an extract prepared from a control strain did not bind by itself to the Nickel column (Fig. 5, lanes 3 and 5).
Fig 5.
The A43 subunit interacts with the ABC23 subunit. Presence of A43 subunit was checked by Western blot using anti-A43 antibodies in crude cell extracts prepared from an E. coli strain coexpressing A43 and His-6-tagged ABC23 (lanes 2, 4, and 6), and from a control strain expressing only A43 subunit (lanes 1, 3, and 5). The asterisk indicates a proteolyzed form of A43. Lanes 3 and 4, extracts were loaded onto a Nickel column, bound proteins were eluted by competition with 200 mM imidazole, and the presence of A43 subunit in the elution fractions was checked by Western blot analysis using anti-A43. Lanes 5 and 6, Ponceau red staining on the membrane after transfer of the proteins of the elution fractions.
Subunits A43, A14, and ABC23 Colocalize Within the pol I.
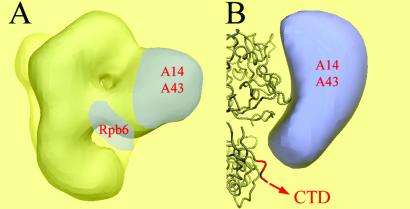
The above data demonstrate that subunits A43 and A14 directly interact and suggest a physical interaction between subunits ABC23 and A43. To determine if this interaction network may occur in the pol I, immunoelectron microscopy experiments were performed on the enzyme. Image analysis of the enzyme labeled with IgG resulted in an average image in which the binding site of the antibodies could be identified by comparison with the native enzyme. The binding sites were then situated on a previously determined three-dimensional enzyme model (30). At 25-Å resolution, the structure of pol I is very similar to the atomic structure of pol IIΔ 4/7 but differs by the presence of a stalk of protein density protruding from the main body of the enzyme. Immunoelectron microscopy experiments mapped the position of subunits A14 and A43 within this stalk (Fig. 6A) and confirmed earlier observations showing that the stalk disappears in the pol IΔ (18). Importantly, docking of the atomic model of yeast pol IIΔ 4/7 into the three-dimensional model of the native pol I, assuming that all common subunits are similarly positioned in both enzymes (31), shows that ABC23 subunit is located in close proximity to the A43–A14 subcomplex (Fig. 6A). These observations strongly reinforce the interactions observed between subunits ABC23, A43, and A14.
Fig 6.
Colocalization of subunits A43, ABC23, and A14 within the pol I. (A) Structure of the yeast pol I as determined by cryoelectron microscopy at 25 Å resolution (gray shading) (31). Blue tags represent the positions of subunits A14 and A43, mapped by immunolabeling (19, 31), and of Rpb6, inferred from the docking of the atomic structure of pol IIΔ 4/7 (43) into the pol I envelope. These results show that subunits A14 and A43 form a stalk near Rpb6. (B) Close-up view of the interaction interface of A14 and A43 with the core enzyme. The blue volume represents the additional density due to A14 and A43 present in pol I. The aligned atomic structure of pol II shows that the C-terminal repeats of Rpb1 (CTD) start next to the interaction site of the two pol I–specific subunits.
Discussion
The results described in this study demonstrate a direct interaction between pol I subunits A43 and A14: (i) both subunits interact in a Far Western experiment, (ii) a stable heterodimer can be assembled from in vitro translated subunits, (iii) the two full-length proteins interact by the two-hybrid method, and (iv) overexpression of A14 partially suppresses the conditional phenotypes associated with the rpa43-6 and rpa43-18 alleles. This biochemical and genetic evidence indicates that subunits A43 and A14 form a stable heterodimer. In addition, we show that A43 subunit is critical for the stable association of A14 subunit within the pol I complex.
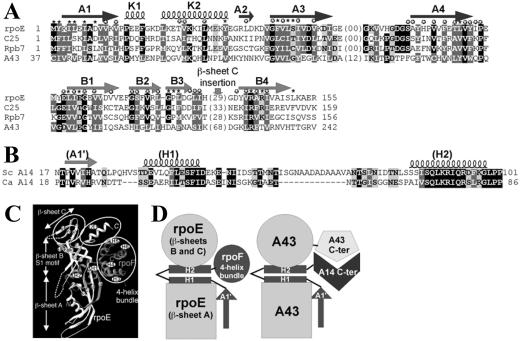
The A43–A14 pair share common properties with Rpb7 and Rpb4, two subunits of yeast pol II: (i) the two subunits of each couple can form a stable heterodimer in vitro (see refs. 32–34), (ii) purification of pol II or pol I from a yeast strain disrupted for the nonessential gene RBP4 or RPA14 generates incomplete forms of enzyme-lacking subunits Rpb7 and A43, respectively (9, 35), (iii) the RNA polymerases lacking either Rpb7–Rpb4 or A43–A14 subunits are active in a nonspecific transcription assay (18, 32) but do not support in vitro specific transcription (19, 32), and (iv) on the basis of weak sequence similarities, it has been suggested that subunit A43 was homologous to Rpb7 and to the pol III subunit C25 (36, 37). The observation that rpoE and rpoF, two subunits of the archaeal RNA polymerase, are homologous to subunits Rpb7 and Rpb4 (38, 39) supports the idea that an Rpb4–Rpb7-like heterodimer is present in each form of eukaryotic RNA polymerase. In agreement with this hypothesis, it was recently shown, based on experimental interaction data and sequence analysis, that yeast subunits C17 and C25 are the pol III counterpart of the Rpb4–Rpb7 heterodimer (ref. 40 and Fig. 7A for the sequence alignment between C25 and Rpb7).
Fig 7.
The A43/A14 subunits belong to the rpoE/rpoF family. (A) Sequence alignment of S. cerevisiae A43, C25, and Rpb7 subunits, and Methanococcus jannaschii rpoE subunit, whose secondary structures are reported (see text). Sequence identities are indicated on a black background; similarities are gray shaded (white letters, hydrophobic amino acids or amino acids that can substitute them in some circumstances). RpoE residues with <10% of their surface accessible to solvent in the rpoE/rpoF structure are indicated with stars (open stars for residues participating in the rpoE hydrophobic core and filled stars for residues involved in contacts with rpoF). (B) Sequence alignment of the conserved motifs of the S. cerevisiae A14 subunit (133 residues) and of its putative homologue from C. albicans (177 residues). Predicted secondary structures are shown up to the sequences and labeled according to their putative correspondence to the rpoF structure. (C) Ribbon representation of the rpoE–rpoF dimer (41). Circled regions represent regions predicted to be different in the A43–A14 subunit. (D) A possible model for the domain structure of the A43–A14 subunit (see text).
The A14–A43 Heterodimer Is the Possible pol I Counterpart of the Rpb4–Rpb7 Pair.
PSI-BLAST searches using the A43 sequence as query revealed marginal similarities with those of Rpb7, C25, and RpoE. The proposed sequence relationship was further confirmed and refined using hydrophobic cluster analysis, a bidimensional method that focuses on the fold invariant features (26, 27), and allows to add to the lexical analysis a description of the two-dimensional structure. Hydrophobic clusters forming the core of the rpoE structure (38) were observed with a good correspondence in Rpb7, C25, and the N-terminal half of A43 (data not shown), most of the residues participating in this core formation being conserved in these three sequences (open stars in Fig. 7A). Moreover, residues present at the heterodimer interface (buried only in the heterodimer structure, filled stars in Fig. 7A) are also well conserved, suggesting a similar way of A43 for interacting with its partner than what was observed for the rpoE/rpoF couple (see below). As shown in Fig. 7C, the observed similarities are higher in the first half of the rpoE-like domain, corresponding to the β-sheet A as inferred from the experimental structure of rpoE (41). The second half of the rpoE-like domain, encompassing β-sheets B and C, is more divergent among the Rpb7 family. The sequences of a three-stranded β-sheet C, representing an insertion relatively to typical S1 motifs (circled in Fig. 7C), are poorly conserved, precluding an accurate alignment for this region between A43 and the other sequences. Similarly, strand B4 was tentatively aligned with the cluster centered on residue 230, and no accurate alignment could be found for the C-terminal part of the S1 motif (encompassing strands B5 and B6, as well as helix K4) (circled in Fig. 7C). Moreover, an additional extension, which is located after, or even substitute, helix K4, and which is absent in the other members of the Rpb7 family, forms the C-terminal part of the A43 sequence (residues 260–326). This C-terminal sequence contains regular secondary structures (β strands) but does not share any obvious similarity with other proteins.
On the contrary to what is observed for the A43 polypeptide, sequence of the A14 subunit (ScA14) does not display significant similarities with any yeast RNA polymerase subunit. We looked for the existence of potential homologues in other genomes and found only in the C. albicans databank a hypothetical protein of 177 residues (CaA14), which displays highly conserved motifs with A14 in the 100 N-terminal residues (Fig. 7B). This observation suggests that A14 sequences have strongly diverged during evolution. hydrophobic cluster analysis fully confirmed the similarity between the C. albicans and the S. cerevisiae sequences (data not shown). These two A14 sequences do not match the sequence profile defined for the rpoF family, indicating that the three-dimensional structure of the A43 partner in pol I might differ from rpoF (Fig. 7C). An interesting alternative possibility is that only a part of the rpoF-fold is maintained in A14, corresponding to the three first regular secondary structures that make direct contact with the rpoE subunit (β-strand A′, contributing to the rpoF β-sheet A, followed by the two helices H1 and H2, which forms a semicircular belt around rpoE, at the interface of β-sheets A and B). The two A14 sequences are indeed compatible with this particular succession of secondary structures (Fig. 7B). Regarding this hypothesis, the C-terminal four-helix bundle (circled in Fig. 7C) present in rpoF, Rpb4, and C17, should not be present in A14. According to the two hypotheses mentioned above, we suggest that the difference observed in the C-terminal region of A43 (additional domain) relatively to the archaeal rpoE structure could be correlated to a difference in the structure of its partner A14 relatively to the rpoF structure, encompassing the whole protein or limited to its C-terminal region (Fig. 7D). In agreement with these hypotheses, our biochemical data indicate that the C terminus of A43 (residues 239–280) is one of the two interacting regions of this polypeptide with A14. Genetic data reinforce this conclusion: the phenotype conferred by the A43-18 subunit, which is mutated in this C-terminal segment, is suppressed by A14 overexpression, whereas the phenotype conferred by the A43-4 mutant, which contains mutations only in the central part of the subunit, is not suppressed by A14 overexpression.
Based on all observations indicated above, we propose that A14 subunit is the pol I equivalent of the pol II Rpb4 polypeptide and that the A43–A14 pair is the pol I counterpart of the pol II Rpb7–Rpb4 heterodimer.
A Model for the Localization of the Rpb4–Rpb7-Like Heterodimer in the Three-Dimensional Structure of RNA Polymerases.
Immunoelectronic microscopy experiments revealed that the A14–A43 heterodimer forms a stalk in the three-dimensional model of pol I. Docking of the atomic structure of pol IIΔ 4/7 into the three-dimensional model of pol I revealed the interface of the stalk with the conserved core enzyme. It interacts with the “linker” domain of the largest subunit, at the site where the C-terminal repeats are starting in pol II (Fig. 6B). The interface also contains part of the clamp domain in the C-terminal region of Rpb2, which forms the Zn7 binding site (β-sheets 43 and 44, residues 1165–1182). Finally, the additional density present in pol I contacts the C-terminal assembly region of Rpb6. The CTD repeats are absent in A190, and these results show that a dedicated structure in pol I containing the A14–A43 dimer is positioned at the same place as the CTD in pol II and might fulfill similar functions, at least by binding transcription factor (19).
Interestingly, a two-hybrid screen using C17 (the pol III counterpart of Rpb4) as a bait revealed an interaction of this subunit with the 67 C-terminal residues of C160, the largest pol III subunit (40). This observation supports the idea that the counterparts of the Rpb4–Rpb7 dimer in pol I and pol III (i.e., A14–A43 and C17–C25, respectively), are similarly localized in the three-dimensional structure of the enzymes and suggests that, in pol II, the Rpb4–Rpb7 complex is not part of the DNA-binding cleft, as proposed (42), and might be positioned close to the CTD.
Acknowledgments
We thank E. Favry and A. Chevalier for technical assistance, V. Goguel for discussions and corrections to the manuscript, P. Thuriaux for providing plasmids and strains, Claire Boschiero for help with two-hybrid screen and DNA sequencing, and Emilie Levivier and Jean-Paul Mornon for help in sequence analysis. This work was supported by the Human Frontier Science Program Organization.
Abbreviations
pol I, RNA polymerase I
References
- 1.Nogi Y., Vu, L. & Nomura, M. (1991) Proc. Natl. Acad. Sci. USA 88, 7026-7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Udem S. A. & Warner, J. R. (1972) J. Mol. Biol. 65, 227-242. [DOI] [PubMed] [Google Scholar]
- 3.Dequard-Chablat M., Riva, M., Carles, C. & Sentenac, A. (1991) J. Biol. Chem. 266, 15300-15307. [PubMed] [Google Scholar]
- 4.Minakhin L., Bhagat, S., Brunning, A., Campbell, E. A., Darst, S. A., Ebright, R. H. & Severinov, K. (2001) Proc. Natl. Acad. Sci. USA 98, 892-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carles C., Treich, I., Bouet, F., Riva, M. & Sentenac, A. (1991) J. Biol. Chem. 266, 24092-24096. [PubMed] [Google Scholar]
- 6.Liljelund P., Mariotte, S., Buhler, J.-M. & Sentenac, A. (1992) Proc. Natl. Acad. Sci. USA 89, 9302-9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thuriaux P., Mariotte, S., Buhler, J.-M., Sentenac, A., Vu, L., Lee, B. S. & Nomura, M. (1995) J. Biol. Chem. 270, 24252-24257. [DOI] [PubMed] [Google Scholar]
- 8.Gadal O., Mariotte-Labarre, S., Chedin, S., Quemeneur, E., Carles, C., Sentenac, A. & Thuriaux, P. (1997) Mol. Cell. Biol. 17, 1787-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smid A., Riva, M., Bouet, F., Sentenac, A. & Carles, C. (1995) J. Biol. Chem. 270, 13534-13540. [DOI] [PubMed] [Google Scholar]
- 10.Nogi Y., Yano, R., Dodd, J., Carles, C. & Nomura, M. (1993) Mol. Cell. Biol. 13, 114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grachev M. A., Lukhtanov, E. A., Mustaev, A. A., Rikhter, V. A. & Rabinov, I. V. (1987) Bioorg. Khim. 13, 552-555. [PubMed] [Google Scholar]
- 12.Grachev M. A., Mustaev, A. A., Zaychikov, E. F., Lindner, A. J. & Hartmann, G. R. (1989) FEBS Lett. 250, 317-322. [DOI] [PubMed] [Google Scholar]
- 13.Riva M., Carles, C., Sentenac, A., Grachev, M. A., Mustaev, A. A. & Zaychikov, E. F. (1990) J. Biol. Chem. 265, 16498-16503. [PubMed] [Google Scholar]
- 14.Mustaev A., Kashlev, M., Lee, J. Y., Polyakov, A., Lebedev, A., Zalenskaya, K., Grachev, M., Goldfarb, A. & Nikiforov, V. (1991) J. Biol. Chem. 266, 23927-23931. [PubMed] [Google Scholar]
- 15.Huet J., Buhler, J.-M., Sentenac, A. & Fromageot, P. (1977) J. Biol. Chem. 252, 8848-8855. [PubMed] [Google Scholar]
- 16.Awrey D. E., Weilbaecher, R. G., Hemming, S. A., Orlicky, S. M., Kane, C. M. & Edwards, A. M. (1997) J. Biol. Chem. 272, 14747-14754. [DOI] [PubMed] [Google Scholar]
- 17.Chedin S., Riva, M., Schultz, P., Sentenac, A. & Carles, C. (1998) Genes Dev. 12, 3857-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lanzendörfer M., Smid, A., Klinger, C., Schultz, P., Sentenac, A., Carles, C. & Riva, M. (1997) Genes Dev. 11, 1037-1047. [DOI] [PubMed] [Google Scholar]
- 19.Peyroche G., Milkereit, P., Bischler, N., Tschochner, H., Schultz, P., Sentenac, A., Carles, C. & Riva, M. (2000) EMBO J. 19, 5473-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milkereit P. & Tschochner, H. (1998) EMBO J. 17, 3692-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherman F., Fink, G. R. & Lawrence, C. W., (1979) Methods in Yeast Genetics: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 22.Flores A., Briand, J.-F., Gadal, O., Andrau, J.-C., Rubbi, L., Van Mullem, V., Boschiero, C., Goussot, M., Marck, C., Carles, C., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 7815-7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fromont-Racine M., Rain, J. C. & Legrain, P. (1997) Nat. Genet. 16, 277-282. [DOI] [PubMed] [Google Scholar]
- 24.Schnapp G., Santori, F., Carles, C., Riva, M. & Grummt, I. (1994) EMBO J. 13, 190-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaboriaud C., Bissery, V., Benchetrit, T. & Mornon, J. P. (1987) FEBS Lett. 224, 149-155. [DOI] [PubMed] [Google Scholar]
- 27.Callebaut I., Labesse, G., Durand, P., Poupon, A., Canard, L., Chomilier, J., Henrissat, B. & Mornon, J. P. (1997) Cell Mol. Life Sci. 53, 621-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guex N. & Peitsch, M. C. (1997) Electrophoresis 18, 2714-2723. [DOI] [PubMed] [Google Scholar]
- 29.Buhler J.-M., Huet, J., Davies, K. E., Sentenac, A. & Fromageot, P. (1980) J. Biol. Chem. 255, 9949-9954. [PubMed] [Google Scholar]
- 30.Schultz P., Celia, H., Riva, M., Sentenac, A. & Oudet, P. (1993) EMBO J. 12, 2601-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bischler N., Brino, L., Carles, C., Riva, M., Tschochner, H., Mallouh, V. & Schultz, P. (2002) EMBO J. 21, 4136-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edwards A. M., Kane, C. M., Young, R. A. & Kornberg, R. D. (1991) J. Biol. Chem. 266, 71-75. [PubMed] [Google Scholar]
- 33.Khazak V., Estojak, J., Cho, H., Majors, J., Sonoda, G., Testa, J. R. & Golemis, E. A. (1998) Mol. Cell. Biol. 18, 1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larkin R. M. & Guilfoyle, T. J. (1998) J. Biol. Chem. 273, 5631-5637. [DOI] [PubMed] [Google Scholar]
- 35.McKune K., Richards, K. L., Edwards, A. M., Young, R. A. & Woychik, N. A. (1993) Yeast 9, 295-299. [DOI] [PubMed] [Google Scholar]
- 36.Shpakovskii G. V. & Shematorova, E. K. (1999) Bioorg. Khim. 25, 791-796. [PubMed] [Google Scholar]
- 37.Sadhale P. P. & Woychik, N. A. (1994) Mol. Cell. Biol. 14, 6164-6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langer D., Hain, J., Thuriaux, P. & Zillig, W. (1995) Proc. Natl. Acad. Sci. USA 92, 5768-5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Werner F., Eloranta, J. J. & Weinzierl, R. O. (2000) Nucleic Acids Res. 28, 4299-4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siaut, M., Zaros, C., Levivier, E., Ferri, M. L., Court, M., Werner, M., Callebaut, I., Thuriaux, P., Sentenac, A. & Conesa, C. (2002) Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed]
- 41.Todone F., Brick, P., Werner, F., Weinzierl, R. O. & Onesti, S. (2001) Mol. Cell 8, 1137-1143. [DOI] [PubMed] [Google Scholar]
- 42.Jensen G. J., Meredith, G., Bushnell, D. A. & Kornberg, R. D. (1998) EMBO J. 17, 2353-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cramer P., Bushnell, D. A., Fu, J., Gnatt, A. L., Maier-Davis, B., Thompson, N. E., Burgess, R. R., Edwards, A. M., David, P. R. & Kornberg, R. D. (2000) Science 288, 640-649. [DOI] [PubMed] [Google Scholar]