Abstract
The functional groups found among the RNA bases and in the phosphoribose backbone represent a limited repertoire from which to construct a ribozyme active site. This work investigates the possibility that simple RNA phosphodiester and hydroxyl functional groups could catalyze amide bond synthesis. Reaction of amine groups with activated esters would be catalyzed by a group that stabilizes the partial positive charge on the amine nucleophile in the transition state. 2′-Amine substitutions adjacent to 3′-phosphodiester or 3′-hydroxyl groups react efficiently with activated esters to form 2′-amide and peptide products. In contrast, analogs in which the 3′-phosphodiester is replaced by an uncharged phosphotriester or is constrained in a distal conformation react at least 100-fold more slowly. Similarly, a nucleoside in which the 3′-hydroxyl group is constrained trans to the 2′-amine is also unreactive. Catalysis of synthetic reactions by RNA phosphodiester and ribose hydroxyl groups is likely to be even greater in the context of a preorganized and solvent-excluding catalytic center. One such group is the 2′-hydroxyl of the ribosome-bound P-site adenosine substrate, which is close to the amine nucleophile in the peptidyl synthesis reaction. Given ubiquitous 2′-OH groups in RNA, there exists a decisive advantage for RNA over DNA in catalyzing reactions of biological significance.
Natural RNA molecules catalyze reactions at phosphodiester centers (1) and, in the case of the ribosome large subunit, form the catalytic active site for amide bond synthesis (2, 3). In vitro selection methods yield RNAs that catalyze diverse reactions, including amide bond formation (4–6). Catalysis at any RNA active site must reflect chemical limitations imposed by having four relatively similar building blocks and lacking functional groups with pKas in the physiological range.
Within this set of chemical constraints, catalytic RNAs employ strategies including juxtaposition and orientation of substrates (7, 8), incorporation of metal ion prosthetic groups (9, 10), perturbing the pKas of titratable base positions (11, 12), and positioning 2′-hydroxyl groups to stabilize an oxyanion (13, 14). The RNA phosphoribose backbone is also likely to play indirect roles in catalysis by functioning, for example, to position catalytic metal ions or to perturb the local environment at RNA bases.
2′-Amine Acylation and Nucleotide Flexibility
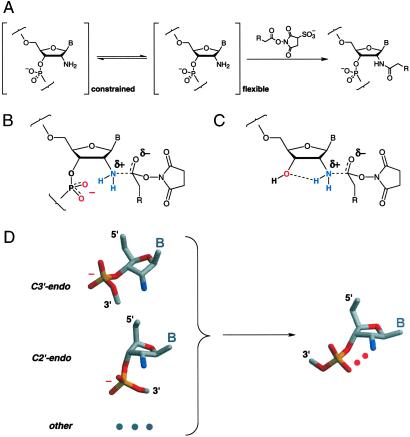
A robust approach for monitoring local flexibility in RNA and DNA is based on substituting the unreactive 2′-hydroxyl with a more nucleophilic amine (15–17). The 2′-amine group serves as a chemical handle that can be selectively modified by using an activated ester. This amide-forming reaction is gated by the underlying nucleic acid structure such that 2′-amine substitutions at flexible nucleotides are more reactive than amine groups at constrained positions (Fig. 1A). For example, 2′-amine substitutions in the anticodon loop of tRNAAsp react efficiently to form the 2′-amide, whereas 2′-amine positions that form stable base pairs, base triples, or tertiary interactions are relatively unreactive in the folded RNA (15). Analogously, 2′-amine substitutions that form base pair mismatches are more reactive than 2′-amino nucleotides constrained by canonical base pairing in DNA–DNA and DNA–RNA helices (16, 17).
Fig 1.
Intramolecular catalysis and 2′-amine acylation. (A) Locally flexible 2′-amine substituted nucleotides react with activated esters to form the 2′-amide. In this work, R is either a biotinamido butyl or fluorescein-5-(or 6)-carboxamido butyl group. B indicates a nucleobase. Intramolecular catalysis of 2′-amine acylation by a 3′-phosphodiester (B) or 3′-hydroxyl (C). (D) In C3′-endo (RNA-like) and C2′-endo (DNA-like) and, presumably, other conformations, the 3′-phosphodiester must undergo a conformational change to attain a catalytic geometry.
Nucleophilic attack by amines at esters is characterized by a Brønsted β value of ≈+0.8 (18, 19). This large, positive β value suggests that there is extensive bond formation and a significant positive charge on the amine nucleophile in the transition state (19). 2′-Amine acylation would be catalyzed by a group that stabilizes the partial positive charge on the nitrogen nucleophile, thereby lowering the activation barrier for this reaction. In the context of RNA, the neighboring 3′-phosphodiester anion or 3′-OH might catalyze amide bond formation, if properly positioned (Fig. 1 B and C).
Intramolecular catalysis readily accounts for the exquisite sensitivity (15–17) of 2′-amine acylation toward local nucleotide flexibility in RNA and DNA structures. For both C3′-endo (RNA-like) and C2′-endo (DNA-like) ribose conformations, the 3′-nonbridging oxygen groups are directed away from the 2′-ribose position. Thus, a conformational change is required to attain the correct catalytic geometry for 2′-amine acylation (Fig. 1D). 2′-Amine acylation is linked to local nucleotide flexibility (15), because flexible residues can more readily attain the catalytic conformation.
Using a series of nucleotide analogs, we show that reaction of a 2′-amine group to form the 2′-amide is strongly modulated by both the chemical nature and positioning of a catalytic group at the 3′-ribose position. These studies show that unperturbed RNA phosphodiester and ribose hydroxyl groups can directly facilitate catalysis.
Materials and Methods
General.
Most nucleotide analogs were resolved by electrophoresis on 30% polyacrylamide gels (0.75 mm × 28.5 cm × 23 cm; 29:1 acrylamide:bisacrylamide in 90 mM Tris-borate, 2 mM EDTA; 30–60 min at 30 W). Before electrophoresis, samples were heated (90°C, 3 min) in an equal volume of 80% formamide, 1 mM EDTA. For purification, nucleotides were excised from the gel, eluted in H2O (overnight at 4°C), and separated from solid acrylamide by microfiltration (EZ spin columns, Millipore).
Analog Synthesis, Purification, and Verification.
Analog 3 (Table 1) and the 5′-hydroxyl precursors of analogs 1, 2, and 6–8 (lacking 5′-32P-phosphorylation) were synthesized by Dharmacon Research (Boulder, CO) and verified by matrix-assisted laser desorption ionization (MALDI) or electrospray mass spectrometry. Analogs 1, 2, and 6–8 were generated by 5′-phosphorylating the appropriate precursor by using gel-purified [γ-32P]ATP and T4 polynucleotide kinase. pU2′-NH2 (10) was generated by treating the reference dinucleotide (1) with nuclease P1 (20); pU2′-NH2 (10) has an electrophoretic mobility identical to nuclease P1-cleaved pU2′-OHpU (2). cAMP2′-NH2 (9) was generated from 2.1 mmol pppA2′-NH2 (5) by using adenylyl cyclase (21, 22), was purified by TLC (2:5, 1 M ammonium acetate:EtOH on PEI-cellulose F, EM Science), was eluted from the silica matrix in H2O, and was verified by MALDI mass spectroscopy (M+ at m/z 329). cAMP2′-NH2 (9) has a mobility similar to cAMP2′-OH by both TLC and gel electrophoresis. The structure of the phosphotriester (8) was verified by three approaches. Hydrolysis of the phosphotriester (1M NaOH; 70°C; 60 min) yields products with electrophoretic mobilities identical to analogs 1 and 7 plus a third, a product with a mobility similar to 6, as expected (23). Treatment of the triester (8) with nuclease P1 (5 units in 10 μl) yields <15% cleaved product after 4 h, whereas analogs 1 and 2, containing authentic phosphodiester linkages, are completely cleaved in 5 min. Finally, the phosphotriester (8) migrates more slowly than the reference dinucleotide (1) by electrophoresis (Fig. 2A), as expected given the charge difference of 1. 3′-Carboxyphenyl-xyl-T2′-NH2 (ChemStar, Moscow) was deprotected by aminolysis (50% methanol; 60°C) to yield 12, was purified by HPLC, and was confirmed by electrospray mass spectroscopy.
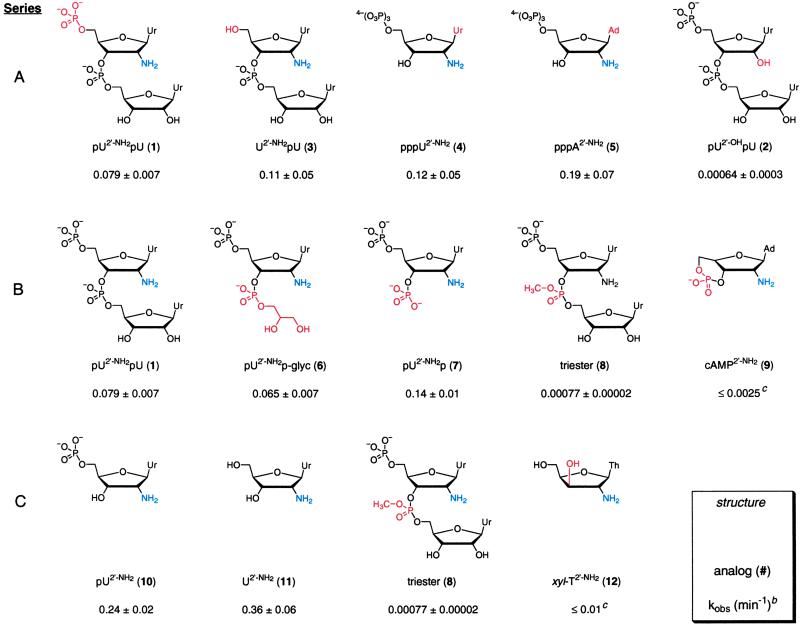
Table 1.
RNA analog structures and acylation ratesa
The amine nucleophile is blue, and structural variation in a series is highlighted in red. Ur, uracil; Ad, adenine; Th, thymine; U, uridine; A, adenosine; T, thymidine; P, phosphate; glyc, glyceryl. For clarity, some analogs are included in more than one series.
kobs are reported at 20 mM succinimidyl ester.
Rate is an upper limit; no product formation was detected. Two unreactive analogs, cAMP2′-OH and U2′-OH, were also tested but are not shown (kobs ≤ 0.002).
Fig 2.
Reactivity of RNA and RNA analogs. (A) Relative electrophoretic mobilities of 2′-amino and 2′-hydroxyl nucleotides visualized by autoradiography of 5′-32P-end-labeled molecules (Left) or UV shadowing (Right). (B) 2′-Amine acylation reactions for the reference dinucleotide pU2′-NH2pU (1), the all-ribose control pU2′-OHpU (2), and the phosphotriester (8), resolved by gel electrophoresis. Filled arrows indicate 2′-acylated product, and open arrows designate background modification for 2 and a degradative by-product for 8. (C) Reaction of a succinimidyl ester with RNA is selective for a 2′-amine substitution and is enhanced in flexible structures. A 20-mer single-stranded oligonucleotide (15) containing a 2′-amine substitution at position 10 (open circles, kobs = 0.077 min−1) reacts ≈5-fold and 20-fold faster than if constrained in a mismatch-containing or duplex helix (open diamonds and squares, respectively). (D) Reaction of cAMP2′-NH2 (9) with a succinimidyl ester is undetectable, as judged by UV shadowing. In contrast, U2′-NH2pU (3) reacts to form the 2′-amide product (arrow) under the same conditions. (E) Reaction of xyl-T2′-NH2 (12) and U2′-NH2 (11) monitored by formation of an anionic fluorescein adduct by using gel electrophoresis. U and − indicate reactions performed in the presence of the all-ribose uridine nucleoside (U2′-OH) and in the absence of nucleoside, respectively. Arrows indicate two product bands for U2′-NH2, corresponding to regioisomers of the fluorescein-conjugated succinimidyl ester; asterisks indicate background bands observed in the no nucleoside (−) control. The fluorescently labeled xyl-T2′-NH2 product is undetectable.
Reaction Kinetics.
Reactions were performed in 100 mM Hepes/Mes/sodium acetate buffer (pH 8.0) at 37°C, were initiated by adding one-tenth vol of 200 mM succinimidyl-6-(biotinamido)hexanoate (Pierce) or succinimidyl-6-fluorescein-5-(or 6)-carboxamido hexanoate (Molecular Probes) in DMSO, and were quenched by adding ≥10-fold excess DTT or glycine. No reaction controls were generated by adding DTT or glycine before the succinimidyl ester. Rates were determined by using a pseudo-first-order equation that accounts for reagent hydrolysis (ref. 15; khydrolysis = 0.03 min−1). Several methods were used to detect reaction products, and an overlapping set of analogs was evaluated by multiple methods; rates are reported as the mean ± SD from multiple measurements or as ± curve fitting error. Reactions of 5′-32P-end-labeled analogs (1, 2, 6–8, and 10) were separated by electrophoresis and quantified using a PhosphorImager (Molecular Dynamics). Reactions of analogs 3 and 11 (500 μM analog, 100 μl) were monitored by reverse phase HPLC (detection at 260 nm; C18 column in ammonium acetate:acetonitrile). Reactions of cAMP2′-NH2 (9) and, for comparison, analogs 3–5 (5 mM analog; 5 μl; quenched with glycine) were resolved by electrophoresis, were detected by UV shadowing (254 nm; PEI-cellulose F silica plate; detection limit ≥5% product), and were quantified by Gaussian band fitting (Kodak EDAS 120 camera and software). Acylation products for xyl-T2′-NH2 (12) and, for comparison, U2′-NH2 (11) were monitored by reaction with a fluorescein-conjugated succinimidyl ester and quantified by fluorescence (250 μM nucleoside, excitation at 450 nm, PhosphorImager).
Phe-AMP reactions (20 μl) contained analog 1, 8, or 10, 100 mM Hepes/Mes/sodium acetate buffer (pH 7.25), and 20 mM Phe-AMP; were quenched by adding 4 μl of 500 mM DTT, 43% formamide; and resolved by gel electrophoresis. Reactions were initiated immediately on solubilizing Phe-AMP. The sequence, synthesis, purification, and succinimidyl ester reactions of the 20-mer oligoribonucleotide substrates were performed as described (15); melting temperatures were determined from differential denaturation curves (16).
Results
Probing Mechanistic Requirements for 2′-Amine Acylation in RNA by a Small Analog Approach.
To investigate the molecular linkage between 2′-amine acylation and local nucleotide flexibility in RNA, we studied acylation reactions for a series of nucleotide analogs. The analogs are identified by a number and are given simplified symbols that describe the structure in the 5′ to 3′ direction (Table 1). For example, pU2′-NH2pU (1) contains a 5′-uridine monophosphate (pU), a phosphodiester linkage between the nucleotides (UpU), and a 2′-amine substitution on the 5′-nucleotide (U2′-NH2).
The 2′-amine analogs have electrophoretic mobilities consistent with their size and net charge (Fig. 2A). For example, the phosphotriester (8) migrates more slowly than the reference dinucleotide (1), because of incorporation of a methyl group and a −1 loss in charge. 3′-5′-Cyclization yields a product (cAMP2′-NH2, 9) that migrates more slowly than pA2′-OH, reflecting the (−1) charge difference. 2′-Amine substituted analogs (1 and 9) migrate similarly to the corresponding 2′-hydroxyl analogs (Fig. 2A).
Acylation rates were determined by treating each nucleotide analog with an activated (succinimidyl) ester (Fig. 1A) at a pH (8.0) above the 2′-amine pKa for all analogs to ensure that the amine group was uncharged and nucleophilic (data not shown; see also refs. 24–26). Reaction products were resolved by polyacrylamide gel electrophoresis or HPLC, and 2′-amine acylation rates were obtained by fitting the fraction-acylated product to a pseudo-first-order equation that accounts for parallel hydrolysis of the succinimidyl ester (15).
pU2′-NH2pU (1, reference dinucleotide) was designed to model flexible nucleotides in large RNAs. This analog reacts to form the 2′-amide at a rate (kobs ≈ 0.079 min−1) identical to that characteristic of flexible single-stranded RNAs (Fig. 2B Top; compare filled and open circles in Fig. 2C). Acylation is selective for the 2′-amine substitution because the all-ribose analog, pU2′-OHpU (2), lacking a 2′-amine group, reacts slowly (kobs ≈ 0.00064 min−1; Fig. 2B Middle). The difference between acylation of the reactive pU2′-NH2pU (1) and nonnucleophilic pU2′-OHpU (2) analogs represents the dynamic range of our experiments, equal to 120-fold. Acylation rates for the reference dinucleotide (1) are identical, within error, for Mg2+ concentrations from 0–100 mM and for EDTA concentrations spanning 0–50 mM (data not shown). Thus, 2′-amine acylation is independent of divalent metal ions.
As a benchmark for 2′-amine acylation in larger RNAs, we reanalyzed (15) amide bond formation for 20-mer oligoribonucleotide substrates containing a single 2′-amino-cytidyl substitution at position 10 (see Fig. 2C). 2′-Amine acylation was monitored for a flexible single-stranded oligomer, for a completely base-paired duplex, and for a duplex containing a single, flexible C2′-NH2⋅A mismatch. Reactions were performed at 37°C, where both perfect and mismatched duplexes are intact (melting temperatures ≥45°C). The flexible, single-stranded substrate reacts 20- and 5-fold faster than the perfectly paired and mismatched duplex substrates, respectively (Fig. 2C, open symbols).
The reference dinucleotide (1) reacts at a rate within 2-fold of its 5′-OH counterpart, U2′-NH2pU (3). Analogs containing a 3′-OH adjacent to the 2′-amine group also form the 2′-amide efficiently, and pU2′-NH2 (10), U2′-NH2 (11), pppU2′-NH2 (4), and pppA2′-NH2 (5) are acylated at rates within 3-fold of each other (Table 1). Thus, 2′-amine acylation chemistry is independent of both the 5′-ribose substituent and the identity of the base.
Acylation as a Function of 3′-Phosphodiester-Like Substituents.
The rate of acylation for the less sterically hindered 3′-phospho-glyceryl-diester, pU2′-NH2p-glyc (6), is identical, within error, to the reference dinucleotide (1), suggesting that local steric differences do not significantly modulate reactivity at solvent-accessible (15) 2′-amine groups. The 3′-phospho-monoester dianion, pU2′-NH2p (7), reacts 2-fold more rapidly than the reference dinucleotide (Table 1, series B).
We then evaluated the effect of neutralizing the −1 charge at the 3′-position using the uncharged 3′-phosphotriester (8) (Table 1, series B). At long time points, reaction with the succinimidyl ester yields two distinguishable products. The more rapidly migrating product (Fig. 2B Bottom, open arrow) has a mobility identical to that of the reference dinucleotide (1) and likely reflects reaction of the readily formed (23) phosphodiester hydrolysis product. The second band (Fig. 2B Bottom, filled arrow) represents acylation of the triester either at the 2′-amino group or elsewhere in the nucleotide. Thus, this reaction (kobs = 0.00077 min−1) represents an upper limit on reactivity of the triester and is 100-fold slower than reaction of the reference dinucleotide (1).
An independent prediction of the proposal that 2′-amine acylation is catalyzed by the neighboring 3′-phosphodiester is that nucleotides containing a phosphodiester constrained in a non-catalytic conformation should be unreactive. cAMP2′-NH2 (9) contains an intact 3′-phosphodiester held in a conformation distant from the 2′-amine group (Table 1, series B). Recall that the identity of the base has no effect on 2′-amine acylation (see analogs 4 and 5; Table 1, series A).
Acylation of cAMP2′-NH2 (9) does not produce a detectable product band, as monitored by UV shadowing (Fig. 2D), whereas reaction of the dinucleotide U2′-NH2pU (3) is readily detected under identical conditions (t1/2 ≈ 7 min; Fig. 2D). The upper limit for reaction of the cyclic analog is 0.0025 min−1. Comparison of this upper limit with reaction of the pppU2′-NH2 (4), pppA2′-NH2 (5), and U2′-NH2 (11) analogs (Table 1) indicates that constraining the 3′-phosphodiester in a 3′-5′-cyclic linkage slows acylation by at least 100-fold.
Ribose Hydroxyl Contributions to 2′-Amine Acylation.
The transition state for amide bond synthesis at a 2′-amino group could also be stabilized via a hydrogen bonding interaction with an adjacent 3′-hydroxyl (Fig. 1C). The nucleotide pU2′-NH2 (10) reacts 3-fold more rapidly than the reference dinucleotide (compare 1 and 10, Table 1). Reaction of the pU2′-NH2 nucleotide can, first, be compared with the lack of reactivity observed for the triester (8), in which the 3′-OH is blocked by substitution of the neutral triester ligand. Because steric effects at the 3′-position do not play a major role in 2′-amine acylation (see 1, 6, and 7; Table 1, series B), these data emphasize an important role for the hydrogen bond acceptor activity of the 3′-OH in catalysis.
Second, we tested reactivity of a xylose (xyl) stereoisomer in which the 3′-OH is trans to the 2′-amine across the ring C2′ and C3′ carbons (see xyl-T2′-NH2 (12); Table 1, series C). Reaction of the xyl analog (12) was compared directly with U2′-NH2 (11), monitored by reaction with a succinimidyl ester tethered to a fluorescent R group (see Fig. 1A). As expected, U2′-NH2 reacts efficiently to yield low-mobility fluorescently labeled products (arrows in Fig. 2E). In strong contrast, reaction of xyl-T2′-NH2 does not yield a fluorescent product, beyond those observed in the presence of buffer alone [compare xyl-T2′-NH2 lanes with the no nucleoside (−) and U lanes in Fig. 2E]. The rate of 2′-amine acylation is reduced by at least 30-fold for the xyl isomer (Table 1, series C).
2′-Amine Acylation Using an Amino Acid Adenylate.
Succinimidyl esters are simple molecules in which reactivity at a carbonyl center is enhanced by coupling to a good leaving group. This strategy is also common in biological systems, although the leaving groups are different. We therefore tested the ability of RNA phosphodiester and hydroxyl groups to catalyze synthesis of an authentic peptide bond by reacting 2′-amine analogs with phenylalanyl adenylate (Phe-AMP; Fig. 3A).
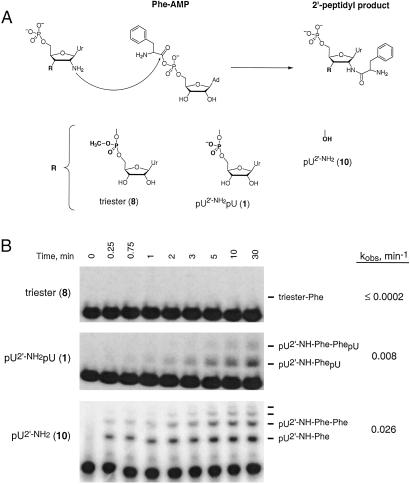
Fig 3.
Peptide bond formation at 2′-amine groups. (A) Reaction of 2′-amine substituted nucleotides with the amino acid adenylate, Phe-AMP, to form a 2′-peptide bond. (B) Reaction of an amino acid adenylate with a 2′-amine requires an intact 3′-phosphodiester or 3′-hydroxyl. The reference dinucleotide (1) and pU2′-NH2 (10) form both single and multiple phenylalanyl adducts (6). Rates of 2′-peptide product formation were determined (15) by taking into account hydrolysis of Phe-AMP [khydrolysis = 0.12 min−1 (6)].
The reference dinucleotide (1), the uncharged phosphotriester (8), and pU2′-NH2 (10) were treated with Phe-AMP. The reference dinucleotide (1) is aminoacylated at least 40-fold faster than the phosphotriester, and pU2′-NH2 (10) reacts ≥130-fold faster than the triester (Fig. 3B). Analogs containing 3′-OH groups adjacent to the 2′-amine reproducibly react 3-fold faster than analogs containing 3′-phosphodiester groups with both Phe-AMP (Fig. 3) and succinimidyl esters (Table 1). Juxtaposition of an RNA ribose hydroxyl or phosphodiester in an otherwise unremarkable environment is an effective strategy for catalyzing peptide bond synthesis.
Both the reference dinucleotide (1) and pU2′-NH2 (10) form higher order products reflecting reaction of additional equivalents of Phe-AMP (Fig. 3B). This observation is consistent with prior work showing that polymerization of aminoacyl adenylates to form peptides is efficient in the absence of added catalyst (6, 27). As an example of an apparent exception that proves the rule, efficient polymerization of aminoacyl adenylates may reflect intramolecular activation of the amine nucleophile by the adenylate phosphodiester.
Discussion
Catalysis at any RNA or protein active site involves at least two fundamental features. First, the substrate molecules and catalytic groups must be held in a reactive or near-reactive conformation. Overwhelming evidence suggests that RNA is amply capable of binding and positioning substrates in an active site (1–6). For the nucleotide analogs studied in this work, this positioning function is fulfilled by constraining catalytic groups adjacent to the amine nucleophile in a ribose ring system. Second, active sites function to reduce the energetic costs for reaching a reaction transition state. For this second requirement, one strategy for stabilizing a transition state is to make use of ubiquitous functional groups in the RNA backbone.
Intramolecular Catalysis Gates Acylation Reactions at 2′-Amine Substitutions in RNA.
We were motivated to understand the catalytic potential of RNA backbone groups in an effort to explain the observation that acylation of 2′-amine-substituted nucleotides in RNA and DNA is strongly gated by the underlying nucleic acid structure (refs. 15–17; Fig. 1A). The strong correlation between 2′-amine acylation and local nucleotide flexibility is well explained by a transition state model in which amide bond formation is catalyzed by a neighboring 3′-phosphodiester or 3′-hydroxyl group (Fig. 1 B and C). In RNA nucleotides involved in pairing or tertiary interactions, non-bridging phosphate oxyanion groups are not appropriately positioned to catalyze reactions at a 2′-amine in either the common C3′- or C2′-endo ribose conformations. However, diverse studies (28–30) suggest that conformations enabling a phosphate diester–2′-ribose interaction are transiently accessible in RNA. Intramolecular catalysis of 2′-amine acylation is mechanistically linked to local dynamics because flexible nucleotides more readily attain the catalytic conformation (Fig. 1D).
Two local changes at the 3′-phosphodiester have dramatic effects on reactivity. When the 3′-phosphodiester charge is eliminated by methylation (triester, 8) or when the phosphodiester is constrained in a conformation distant from the 2′-amine (9), acylation rates are reduced by a factor of at least 100-fold (≈3 kcal/mol). 2′-Amine reactivity is recovered by removing the phosphotriester to leave a 3′-hydroxyl (pU2′-NH2, 10). Direct involvement of the 3′-OH in catalysis is specifically supported by the unreactive nature of the xyl (3′-OH trans to the 2′-amine) analog (12). Catalysis of a synthetic reaction by RNA backbone groups is general in that the 3′-phosphodiester and hydroxyl groups also catalyze authentic peptide bond formation with amino acid adenylates (Fig. 3).
There are two classes of mechanisms for phosphodiester catalysis of amide bond synthesis at a 2′-amine center. The nonbridging oxygen group may functions as a (weak, pKa ≈ 1.5, ref. 31) base to abstract a proton from the amine nucleophile. Alternatively or in concert, the anionic phosphodiester group may stabilize electrostatically the partial positive charge (δ+ in Fig. 1B) on nitrogen in the transition state. Stabilization of the 2′-amide transition state by a 3′-hydroxyl likely involves the 3′-OH accepting a hydrogen bond from the 2′-amine in the transition state (Fig. 1C). 3′-Hydroxyl groups may be well suited to function as a hydrogen bond acceptor in these analogs because the 3′-ribose position is sterically unhindered and 2′→3′ hydrogen bonding involves a favorable pseudo-5-membered ring (32).
Catalysis of Peptide Synthesis on the Ribosome by a 2′-Hydroxyl from the P-Site Substrate?
The amide synthesis reactions studied in this work are analogous to the peptidyl transferase reaction catalyzed by the ribosome. In the ribosome, a tRNA-linked amine nucleophile is bound in the A-site and attacks an activated aminoacyl ester in the P-site (Fig. 4A). pKa measurements (33, 34) emphasize that the chemical environment at 2′- and 3′-hydroxyl groups is similar. Thus, like a 3′-hydroxyl (Fig. 1C), a properly positioned 2′-hydroxyl could facilitate peptide bond formation by accepting a hydrogen bond from the amine nucleophile (Fig. 4A).
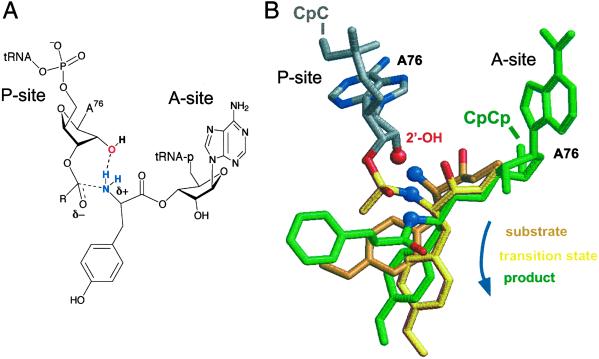
Fig 4.
Model for catalysis of peptidyl transfer at the ribosome. (A) Substrate-assisted catalysis of peptide bond formation by a P-site 2′-hydroxyl. The ribosome A-site and P-site bind the terminal CCA76 portions of the aminoacyl- and peptidyl-tRNAs, respectively. (B) Structure of the P-site A76 (gray) with A-site analogs of (i) the aminoacyl substrate (orange), (ii) a peptidyl transition state analog (yellow), and (iii) the dipeptide aminoacyl product (green). P-site and product structures are from 1KQS (3), and the A-site substrate is from 1FGO (2). The transition state is derived from the phosphate-puromycin portion of the Yarus (35) inhibitor [1FFZ (2)], after molecular mechanics minimization to regularize the bond between the phosphate tetrahedral center and the P-site adenosine containing an intact 2′-OH. This reorientation translates the tetrahedral center and amine nucleophile by ≈1 Å and shortens the distance between the P-site 2′-OH and A-site amine by 0.6 Å.
At the time of this writing, structures are available (2, 3) that illustrate ribosomal 50S subunit interactions with (i) the A-site substrate, (ii) an approximate transition state (35), and (iii) A- and P-site products (Fig. 4B). We obtained good superpositions of these structures in the context of the large ribosomal subunit by aligning on the A-site (dimethyl) adenosine. For both the A-site substrate (in orange in Fig. 4B) and transition state structures (in yellow), the amine nucleophile (blue sphere) is positioned very near (1.6 and 2.5 Å, respectively) the 2′-OH group from the P-site adenosine (red sphere in Fig. 4B).
Aligning the substrate, transition state-like, and product (in green in Fig. 4B) structures in this way suggests a possible trajectory for peptide synthesis on the ribosome (blue arrow in Fig. 4B). The amine nucleophile is close (≈2 Å) to the P-site 2′-OH early in this trajectory and moves away from this catalytic group (to ≈4 Å as visualized in the product structure) once peptide synthesis is complete. This model is consistent with the observation that P-site analogs lacking a 2′-OH group bind to the ribosome but do not support peptide synthesis (36, 37). For all structures shown in Fig. 4, the P-site 2′-OH is closer to the amine nucleophile than is the N3 of A2486, which has also been proposed to participate in catalysis (2, 3). The P-site hydroxyl may function in concert with this or other groups, especially given that the overall peptide synthesis reaction requires up to three proton transfer reactions. These transfers include deprotonation of the –NH2 in the transition state (Fig. 4), deprotonation of the –NH3+ in the ground state, and protonation of the 3′-OH leaving group.
Implications.
The significant catalytic effects of RNA phosphodiester and ribose hydroxyl groups observed for the nucleotide analogs (Table 1 and Fig. 3) must represent only a fraction of the potential for rate enhancement. Proper positioning of the phosphodiester groups in our analogs involves an entropic cost to form a pseudo-7-membered ring. Catalysis by RNA phosphodiester or hydroxyl groups, both in cis (Fig. 1) and in trans (Fig. 4), would be enhanced in the context of a rigid macromolecular structure. Moreover, transition state-specific electrostatic effects (see Fig. 1 B and C) would be amplified in a less polar active site that excludes solvent. This work demonstrates the substantial chemical utility of recruiting ubiquitous RNA phosphodiester and hydroxyl groups to stabilize directly transition states in RNA active sites.
Acknowledgments
We thank F. Eckstein, M. Yarus and M. Illangasekare, and A. Gilman and M. Hatley for gifts of U2′-NH2, Phe-AMP, and mammalian adenylyl cyclase, respectively; S. Scaringe, L. Weinstein, M. Hatley, and S. Danek for advice on analog synthesis; L. Perera for assistance with ribosome modeling; and G. Bassi, M. Forbes, M. Gagné, M. Waters, and P. Moore for helpful discussions. This work was supported by National Science Foundation Career Award MCB-9984289 (to K.M.W.).
References
- 1.Cech T. R. & Golden, B. L. (1999) in The RNA World, eds. Gesteland, R. F., Cech, T. R. & Atkins, J. F. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 321–349.
- 2.Nissen P., Hansen, J., Ban, N., Moore, P. B. & Steitz, T. A. (2000) Science 289, 920-930. [DOI] [PubMed] [Google Scholar]
- 3.Schmeing T. M., Seila, A. C., Jansen, J. L., Freeborn, B., Soukup, J. K., Scaringe, S. A., Strobel, S. A., Moore, P. B. & Steitz, T. A. (2002) Nat. Struct. Biol. 9, 225-230. [DOI] [PubMed] [Google Scholar]
- 4.Illangasekare M., Sanchez, G., Nickles, T. & Yarus, M. (1995) Science 267, 643-647. [DOI] [PubMed] [Google Scholar]
- 5.Zhang B. & Cech, T. R. (1997) Nature 390, 96-100. [DOI] [PubMed] [Google Scholar]
- 6.Illangasekare M. & Yarus, M. (1999) RNA 5, 1482-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weber A. L. & Orgel, L. E. (1980) J. Mol. Evol. 16, 1-10. [DOI] [PubMed] [Google Scholar]
- 8.Tamura K. & Schimmel, P. (2001) Proc. Natl. Acad. Sci. USA 98, 1393-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feig A. L. & Uhlenbeck, O. C. (1999) in The RNA World, eds. Gesteland, R. F., Cech, T. R. & Atkins, J. F. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 287–320.
- 10.Shan S., Kravchuk, A. V., Piccirilli, J. A. & Herschlag, D. (2001) Biochemistry 40, 5161-5171. [DOI] [PubMed] [Google Scholar]
- 11.Perrotta A. T., Shih, I. & Been, M. D. (1999) Science 286, 123-126. [DOI] [PubMed] [Google Scholar]
- 12.Nakano S., Chadalavada, D. M. & Bevilacqua, P. C. (2000) Science 287, 1493-1497. [DOI] [PubMed] [Google Scholar]
- 13.Strobel S. A. & Ortoleva-Donnelly, L. (1999) Chem. Biol. 6, 153-165. [DOI] [PubMed] [Google Scholar]
- 14.Yoshida A., Shan, S., Herschlag, D. & Piccirilli, J. A. (2000) Chem. Biol. 7, 85-96. [DOI] [PubMed] [Google Scholar]
- 15.Chamberlin S. I. & Weeks, K. M. (2000) J. Am. Chem. Soc. 122, 216-224. [Google Scholar]
- 16.John D. M. & Weeks, K. M. (2000) Chem. Biol. 7, 405-410. [DOI] [PubMed] [Google Scholar]
- 17.John D. M. & Weeks, K. M. (2002) Biochemistry 41, 6866-6874. [DOI] [PubMed] [Google Scholar]
- 18.Bruice T. C. & Lapinski, R. (1958) J. Am. Chem. Soc. 80, 2265-2267. [Google Scholar]
- 19.Jencks W. P. & Gilchrist, M. (1968) J. Am. Chem. Soc. 90, 2622-2637. [Google Scholar]
- 20.Romier C., Dominguez, R., Lahm, A., Dahl, O. & Suck, D. (1998) Proteins 32, 414-424. [PubMed] [Google Scholar]
- 21.Johnson R. A. & Salomon, Y. (1991) Methods Enzymol. 195, 3-21. [DOI] [PubMed] [Google Scholar]
- 22.Hatley M. E., Benton, B. K., Xu, J., Manfredi, J. P., Gilman, A. G. & Sunahara, R. K. (2000) J. Biol. Chem. 275, 38626-38632. [DOI] [PubMed] [Google Scholar]
- 23.Weinfeld M., Drake, A. F., Saunders, J. K. & Paterson, M. C. (1985) Nucleic Acids Res. 13, 7067-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verheyden J. P. H., Wagner, D. & Moffatt, J. G. (1971) J. Org. Chem. 36, 250-254. [DOI] [PubMed] [Google Scholar]
- 25.Miller P. S., Bhan, P. & Kan, L.-S. (1993) Nucleosides Nucleotides 12, 785-792. [Google Scholar]
- 26.Aurup H., Tuschl, T., Benseler, F., Ludwig, J. & Eckstein, F. (1994) Nucleic Acids Res. 22, 20-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewinsohn R., Paecht-Horowitz, M. & Katchalsky, A. (1967) Biochim. Biophys. Acta 140, 24-36. [Google Scholar]
- 28.Soukup G. A. & Breaker, R. R. (1999) RNA 5, 1308-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Egli M., Usman, N. & Rich, A. (1993) Biochemistry 32, 3221-3237. [PubMed] [Google Scholar]
- 30.Auffinger P. & Westhof, E. (1997) J. Mol. Biol. 274, 54-63. [DOI] [PubMed] [Google Scholar]
- 31.Kumler W. D. & Eiler, J. J. (1943) J. Am. Chem. Soc. 65, 2355-2361. [Google Scholar]
- 32.Yang J. & Gellman, S. H. (1998) J. Am. Chem. Soc. 120, 9090-9091. [Google Scholar]
- 33.Izatt R. M., Rytting, J. H., Hansen, L. D. & Christensen, J. J. (1966) J. Am. Chem. Soc. 88, 2641-2645. [DOI] [PubMed] [Google Scholar]
- 34.Velikyan I., Acharya, S., Trifonova, A., Földesi, A. & Chattopadhyaya, J. (2001) J. Am. Chem. Soc. 123, 2893-2894. [DOI] [PubMed] [Google Scholar]
- 35.Welch M., Chastang, J. & Yarus, M. (1995) Biochemistry 34, 385-390. [DOI] [PubMed] [Google Scholar]
- 36.Sprinzl M. & Cramer, F. (1979) Prog. Nucleic Acid Res. Mol. Biol. 22, 1-69. [DOI] [PubMed] [Google Scholar]
- 37.Quiggle K., Kumar, G., Ott, T. W., Ryu, E. K. & Chladek, S. (1981) Biochemistry 20, 3480-3585. [DOI] [PubMed] [Google Scholar]