Abstract
MrgA1 and MrgC11 belong to a recently identified family of orphan G-protein coupled receptors, called mrgs (mas-related genes). They are only expressed in a specific subset of sensory neurons that are known to detect painful stimuli. However, the precise physiological function of Mrg receptors and their underlying mechanisms of signal transduction are not known. We therefore have screened a series of neuropeptides against human embryonic kidney (HEK) 293 cells that stably express either MrgA1 or MrgC11 to identify ligands and/or agonists. MrgA1- or MrgC11-specific agonists stimulated dose-dependent increases in intracellular free Ca2+ in a pertussis toxin-insensitive manner, but failed to alter basal or forskolin-stimulated levels of intracellular cAMP. Furthermore, studies using embryonic fibroblasts derived from various Gα protein knockout mice demonstrated that both the MrgA1 and MrgC11 receptors are coupled to the Gαq/11 signaling pathway. Screening of neuropeptides identified surrogate agonists, most of these peptides included a common C-terminal -RF(Y)G or -RF(Y) amide motif. Structure-function studies suggest that endogenous ligands of Mrg receptors are likely to be RF(Y)G and/or RF(Y) amide-related peptides and that postprocessing of these peptides may serve to determine Mrg receptor-ligand specificity. The differences in ligand specificity also suggest functional diversity amongst the Mrg receptors.
G protein-coupled receptors (GPCRs) constitute the largest family of cell surface receptors (1, 2). Agonist binding to GPCRs initiates an intracellular signaling pathway. Subsequent effectors are activated through heterotrimeric G proteins comprised of α, β, and γ subunits, leading to an appropriate physiological response.
Over the last decade, cloning and genome sequencing efforts have revealed the existence of hundreds of orphan GPCRs. Many of them are presumably receptors for as yet unidentified biologically active substances such as hormones and neuropeptides. Thus, the identification of an endogenous ligand for an orphan GPCR may reveal new signaling pathways and signaling substances. We have recently reported the identification of a novel family of orphan GPCRs, called mrgs (mas-related genes) in mice and humans (3). This family can be divided into three major homology groups (MrgA, MrgB, and MrgC) and is comprised of 32 murine and 4 human genes (hMrgX1–hMrgX4) with intact coding sequences, and additional related pseudogenes (4). Some Mrg receptors (17 MrgAs and MrgD) are particularly intriguing because their expression is restricted to specific subsets of sensory neurons that detect painful stimuli (3). Moreover, we found that some of these receptors, such as MrgA1, MrgA4, and MAS1, were distinctively activated by RF-amide (RFa) neuropeptides, of which the prototypic member is the molluscan peptide FMRF-amide (FMRFa) (3).
In general, the FMRFa-related peptides constitute a large family of neuropeptides that are widely and abundantly distributed in invertebrates functioning as neurotransmitters and neuromodulators (5, 6). In the genome of Caenorhabditis elegans, more than 20 RFa peptide genes, encoding over 50 distinct RFa peptides, have been found (6, 7). However, in vertebrates, only a few RFa peptides have been identified, such as NPFF and NPAF (8, 9), the prolactin-releasing peptide (10), the two RFRPs (11), the kisspeptin (12), and γ1-MSH. Their corresponding receptors have been identified or deduced by pharmacological techniques or by a mining genome databases and their functional significance has been well documented (9–11, 13–17).
A recent study by Lembo et al. (18) showed that human MrgX1 (hMrgX1; SNSR3) is expressed solely in dorsal root ganglia (DRG) neurons and potently activated by the preproenkephalin products, in particular, adrenal medulla peptide 22 (BAM-22P). Based on sequence homology analysis, hMrgX1 is classified as being between the mouse MrgA and MrgC subfamilies (3). We initially identified MrgC11 as a member of a subfamily of Mrg pseudogenes (3). In this work, we found that MrgC11 has an intact coding sequence and is expressed in a specific subpopulation of nociceptor neurons in the DRG. To address the questions of the ligand selectivity of these receptors and the mechanism of coupling to cellular signaling, we established human embryonic kidney (HEK) 293 cells stably expressing either MrgA1 or MrgC11 and have screened neuropeptides to identify ligands and/or agonists. We have investigated signal transduction events in terms of calcium signaling and Gα coupling specificity. In addition, we investigated the structural requirements for agonists by examining structure–activity relationships. These studies will facilitate the eventual identification of the natural ligands and physiological roles for a subset of these Mrg receptors and help clarify the function of these receptors.
Materials and Methods
Materials.
Wild type and Gα knockout (KO) mouse embryonic fibroblasts (MEFs) were prepared and cultured from embryonic day 8.5 to 9.5 embryos as described (19). HEK293 and Gα KO MEFs were cultured in DMEM/10% FBS. U73122, U73343, and thapsigargin (TG) were purchased from Calbiochem. Fura-2/AM was purchased from Molecular Probes. All other reagents were purchased from Sigma.
Cloning and Expression of the MrgC11 Receptor.
Nonisotopic in situ hybridization on frozen sections was performed as described by using cRNA probes (3). For double labeling with Griffonia simplicifolia IB4 lectin, sections were incubated with 12.5 μg/ml FITC-conjugated IB4 lectin (Sigma) after in situ hybridization. The full-length cDNA-encoding MrgC11 was cloned from a newborn (P0) mouse DRG cDNA library.
Cell Culture and Transfection.
HEK293 cells were transfected with cDNA encoding the MrgA1-GFP, mNPFF2-GFP, or MrgC11-GFP in pcDNA3.1/Zeo (+) plasmid (Invitrogen) using the FuGENE 6 transfection reagent (Roche Molecular Biochemicals). The transfected cells were selected with 400 μg/ml zeocin in DMEM supplemented with 10% FBS. Each cloned cell was further selected for membrane localization of receptor–GFP fusion proteins. The selected cells were maintained in the same medium supplemented with 200 μg/ml zeocin. We designated these established stable cell lines as HEK-MrgA1, HEK-NPFF2, and HEK-MrgC11. Expression of each receptor was confirmed by Western blotting using an anti-GFP monoclonal antibody (Santa Cruz Biotechnology).
Intracellular Calcium Release Assay.
HEK-MrgA1 or HEK-MrgC11 was plated in 96-black-well plates (Corning) and grown to confluence. After incubation with Fura-2/AM for >20 min, cells were washed and equilibrated for 20 min with HBSS (Hanks' balanced salt solution) assay buffer. The fluorescence emission caused by intracellular calcium mobilization elicited by agonists was determined by using a fluorometric imaging plate reader, Flexstation (Molecular Devices). All peptides were from Phoenix Pharmaceuticals (St. Joseph, MO), Bachem, American Peptide (Sunnyvale, CA), or Sigma.
Internalization.
MrgA1-GFP or MrgC11-GFP stably expressing HEK293 cells were grown in 35-mm glass-bottomed dishes (MatTek, Ashland, MA) in DMEM with 10% FBS. After 4- to 6-h serum starvation, cells were treated with agonists at 37°C for 30 min. Cells were washed with PBS and fixed with 3.7% paraformaldehyde in PBS. The subcellular localization of Mrg-GFP was visualized under a Leica confocal fluorescence microscope with a ×20 or ×40 lens.
Peptide Synthesis.
The peptides BAM-15, BAM15-amide (BAM15a), and Dynorphin-14 were synthesized and purified in the CalTech peptide facility, and peptide identities were confirmed by mass spectrometry.
cAMP Assay.
cAMP was measured by using a radioimmuno assay kit (Amersham Pharmacia) as described by the manufacturer. HEK-MrgA1 or HEK-MrgC11 were cultured in 6-well plates coated with matriGel for ≈16 h at 37°C in growth medium. After 4- to 6-h serum starvation, cells were stimulated with or without representative agonists in the presence or absence of 10 μM forskolin for 10 min. The cells were rapidly washed twice with PBS containing 200 μM Ro20-1724 and cAMP was extracted with 2 ml of cold 60% ethanol. Quantitation of cAMP was then performed by using a [3H] cAMP displacement assay (20).
Calcium Imaging.
Measurement of [Ca2+]i, at the individual cell level was performed as described (3). Briefly, MrgA1-GFP- or MrgC11-GFP-transfected cells were grown in specialized glass-bottom dishes (Bioptechs, Butler, PA) and loaded with fura-2/AM in Hepes-buffered saline. By using a dual wavelength spectrofluorometer coupled to an inverted fluorescence microscope, GFP-positive cells were identified by using an excitation wavelength of 488 nm, a dichroic 505 nm long-pass filter, and an emitter filter at band pass of 535 nm (Chroma Technology, Brattleboro, VT). Measurements of [Ca2+]i were performed on individual Mrg-GFP positive cells at excitation wavelength of 340 and 380 nm and an emission wavelength of 510 nm.
Results
Cloning and Expression Analysis of MrgC11.
We suggested that all of the mouse MrgC genes were nonfunctional pseudogenes based on the draft mouse genomic sequence data that was initially available (3). To determine whether any of the MrgC genes were indeed expressed in DRG neurons, we designed degenerate PCR primers specific for all members of the MrgC subfamily. After PCR amplification from a newborn (P0) DRG cDNA library, we identified sequences corresponding to MrgC11. No other MrgC gene products were identified, suggesting that MrgC11 is the only expressed MrgC gene in the mouse.
Next, a full-length MrgC11 cDNA was cloned from the newborn DRG cDNA library. Contrary to our original prediction that MrgC11 was a pseudogene, this experimentally verified transcript contains an intact ORF that is predicted by hydrophobicity analysis to contain seven transmembrane domains. The MrgC11 protein is 51% and 54% identical to the GPCRs MrgA1 and hMRGX1, respectively (Fig. 1A).
Fig 1.
Sequence comparisons of mouse MrgA1, MrgC11, and human MrgX1 and expression of MrgC11. (A) Alignment of the amino acid sequence of mouse MrgA1 (mMrgA1), mouse MrgC11 (mMrgC11), and human MrgX1 (hMrgX1). Residues shaded in black are identical in >50% of the proteins and residues shaded in gray indicate conservative substitutions. The 7 transmembrane domains (TM1–7) are over-lined. (B and C) In situ hybridization with cRNA riboprobes detecting mMrgC11 in newborn (B) and adult (C) DRG neurons. (D) Double label in situ with mMrgC11 probe (red) and staining with fluorescent lectin IB4 (green) in adult mouse DRG neuron.
We confirmed that MrgC11 is expressed in newborn and in adult DRG neurons by means of in situ hybridization (Fig. 1 B and C). MrgC11 is coexpressed in the small-diameter nociceptive neurons that contain IB-4 binding sites (Fig. 1D). Recently, the rat ortholog of MrgC11 was cloned and named Sensory Neuron-Specific GPCR Receptor (SNSR) because of its exclusive expression in small-diameter, IB-4-positive nociceptive neurons (18). Taken together, these data indicate that MrgC11 codes for a potentially functional GPCR in nociceptive neurons.
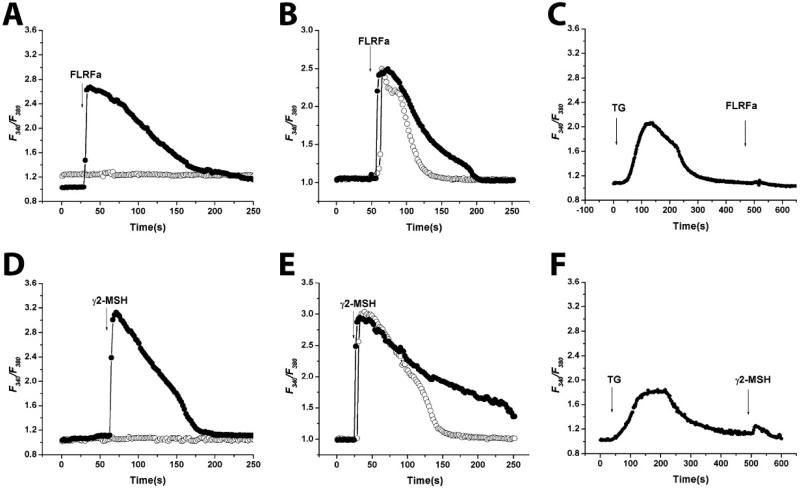
FLRFa and γ2-MSH Elicit Transient Intracellular Calcium Responses in a Receptor-Specific Manner.
We tested a variety of compounds in the ligand screen to determine whether they act as agonists and elicit receptor specific calcium responses in HEK-MrgA1 and HEK-MrgC11 cells. We used FLRFa or γ2-MSH to activate MrgA1 and MrgC11, respectively because these are the most potent agonists amongst the peptides tested for each receptor (see Table 1). Pretreatment of the cells for 10 min with a specific phospholipase C inhibitor, 10 μM U73122 completely inhibited the 3 μM FLRFa- or 1 μM γ2-MSH-induced calcium release (Fig. 2 A and D). In contrast, pretreatment of cells with 10 μM U73343 (an inactive analogue of U73122) did not significantly affect [Ca2+]i responses for both receptors (Fig. 2 A and D).
Table 1.
The EC50 values (in nM) of various peptides for HEK-MrgA1 and HEK-MrgC11 cells using FLEXstation assay
| Peptides | Sequences | MrgC11 | MrgA1 |
|---|---|---|---|
| AnthoRF- amide | pEGRFa | 16 ± 6 | Inactive |
| AF-2 | KHEYLRFa | 130 ± 24 | Inactive |
| ACEP-1 | SGQSWRPQGRFa | 46 ± 12 | Inactive |
| FLRF-amide | FLRFa | 157 ± 12 | 402 ± 21 |
| FMRF-amide | FMRFa | 114 ± 32 | 420 ± 71 |
| FMRF-OH | FMRF | 544 ± 117 | 8,204 ± 458 |
| Met-ENK-RF- amide | YGGFMRFa | 133 ± 20 | 5,252 ± 1,280 |
| Met-Enk-RF | YGGFMRF | 545 ± 19 | Inactive |
| γ1-MSH | YVMGHFRWDRFa | 17 ± 3 | Inactive |
| γ2-MSH | YVMGHFRWDRFG | 11 ± 5 | Inactive |
| BAM3200 | YGGFMRRVGRPEWWMDYQKRYGGFL | 300 ± 124 | >10,000 |
| BAM-22P | YGGFMRRVGRPEWWMDYQKRYG | 26 ± 10 | 2,542 ± 654 |
| BAM-15 | VGRPEWWMDYQKRYG | 53 ± 2 | 23,326 ± 1,866 |
| BAM-15- amide | VGRPEWWMDYQKRYa | 479 ± 14 | 8,773 ± 493 |
| Dynnorphin- 14 | IRPKLKWDNQKRYG | 22 ± 1 | Inactive |
| PrRP-20 | TPDINPAWYTGRGIRPVGRFa | 144 ± 18 | Inactive |
| Kiss(107–121) | KDLPNYNWNSFGLRFa | 102 ± 24 | Inactive |
| Kiss(112–121) | YNWNSFGLRFa | 50 ± 4 | Inactive |
| PQRF-amide | PQRFa | 126 ± 28 | >10,000 |
| NPFF | FLFQPQRFa | 54 ± 5 | 2,145 ± 245 |
| NPAF | AGEGLNSQFWSLAAPQRFa | 282 ± 30 | Inactive |
| RFRP-1 | MPHSFANLPLRFa | 1,245 ± 112 | Inactive |
| RFRP-3 | VPNLPQRFa | 113 ± 5 | Inactive |
| NPY | YPSKPEDMARYYSALRHYINLITRQRYa | 237 ± 30 | 3,486 ± 986 |
Data represent means ± SEM from triplicate independent determinations. Inactive, no activation was detected at concentrations up to 10 μM.
Fig 2.
Calcium signaling in HEK-MrgA1 (A–C) and HEK-MrgC11 (D–F). Cells loaded with Fura-2/AM were stimulated with each agonist, and fluorescence was recorded. Graphs represent an average plot of [Ca2+]i measurements versus time (in s) in a minimum of 8 cells from representative experiments. Individual data points represent images taken at 0.8-s intervals. (A and D) U73122 (○), the active phospholipase C inhibitor blocked agonist-induced rise in [Ca2+]i. However, U73343 (•), the inactive analogue, did not affect FLRFa or γ2-MSH-induced Ca2+ mobilization. After a 10-min pretreatment with U73122 and U73343, each agonist was added. (B and E) Extracellular [Ca2+] dependency of Ca2+ mobilization. Cells were preincubated for 2 min with 2 mM EGTA (○) or normal medium containing 1.2 mM calcium (•), and then 3 μM FLRFa or 1 μM γ2-MSH were added. (C and F) TG prevents the agonist-evoked increase of [Ca2+]i in HEK-MrgA1 (C) and HEK-MrgC11 (F). In the presence of 2 mM EGTA, TG (1 μM final concentration) was added to deplete internal Ca2+ store.
To determine whether Ca2+ influx occurs from the extracellular medium, FLRFa- or γ2-MSH-induced [Ca2+]i responses were examined in the presence of 2 mM EGTA (Fig. 2 B and E). In the presence of EGTA, the agonist-induced calcium responses were similar in amplitude to the responses obtained in medium containing the normal level of calcium (Fig. 2 B and E). However, the response rapidly returned to basal levels, suggesting that in the absence of EGTA, Ca2+ influx occurred (Fig. 2 B and E). Next, we determined the calcium source responsible for the initial peak in [Ca2+]i by depleting internal calcium stores by the application of 1 μM TG (Fig. 2 C and F). When HEK-MrgA1 or HEK-MrgC11 cells were treated with 1 μM TG, the resultant emptying of intracellular calcium stores blocked the response to FLRFa or γ2-MSH (Fig. 2 C and F). These data suggest that FLRFa or γ2-MSH can trigger the mobilization of calcium from IP3-dependent internal calcium stores, and the resultant intracellular calcium can induce the influx of extracellular calcium.
Internalization of MrgA1 and MrgC11.
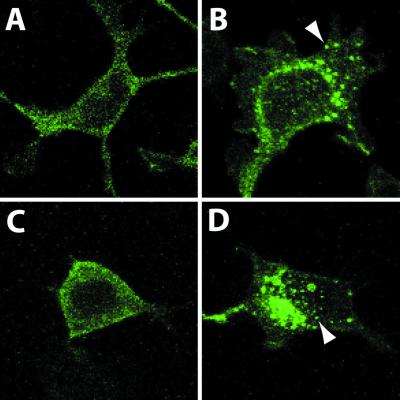
We next assessed whether the agonists can induce the internalization of MrgA1 or MrgC11. Receptor internalization is a response of GPCRs to ligand stimulation. This process indicates that agonist interacts directly with its cognate receptor. In nonstimulated conditions, MrgA1-GFP or MrgC11-GFP fusion proteins were expressed predominantly at the plasma membrane (Fig. 3 A and C). Stimulation of FLRFa or γ2-MSH induced internalization of MrgA1-GFP (Fig. 3B) or MrgC11-GFP (Fig. 3D) in >90% of cells at 37°C. However, rapid internalization was not observed at room temperature under the same conditions (data not shown).
Fig 3.
Internalization of MrgA1-GFP (A and B) and MrgC11-GFP (C and D) was induced by 3 μM FLRFa and 1 μM γ2-MSH, respectively. (A and C) Serum-starved (>4 h) HEK-MrgA1 or HEK-MrgC11 cells. (B and D) HEK-MrgA1 or HEK-MrgC11 were treated with the indicated agonists for 30 min at 37°C. Results are representative of three independent experiments, and the arrow indicates the internalization process.
MrgA1 and MrgC11 Coupling to Heterotrimeric G Proteins.
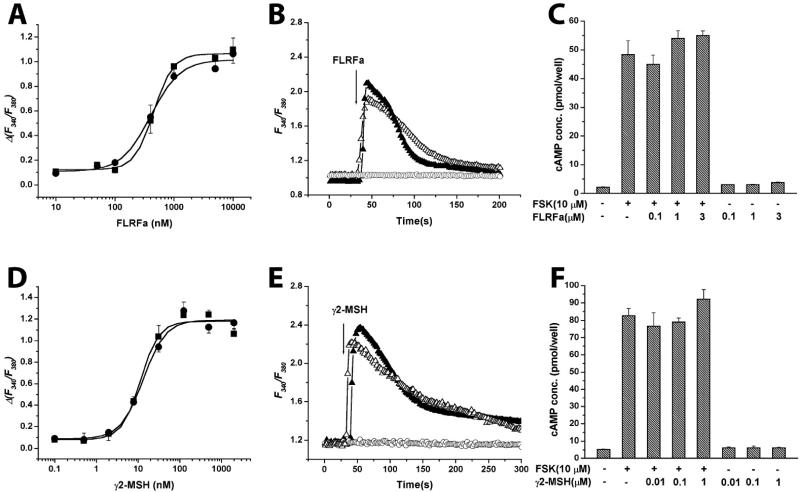
Previously, we reported that transiently overexpressed MrgA1 responded to FLRFa with high potency (EC50 ≈ 20 nM) in HEK293 cells expressing Gα15. We reexamined the dose dependence in HEK293 cells stably expressing MrgA1 (HEK-MrgA1) but not expressing exogenous Gα15. FLRFa stimulated an increase in [Ca2+]i with an EC50 of 402 ± 21 nM in this cellular system (Fig. 4A). The difference in EC50 value is possibly derived from a variety of sources such as different coupling efficiencies, different expression levels of receptor, and/or different cellular environment (21). Nonetheless, the relative ligand selectivity (FLRFa vs. NPFF) was conserved in both cellular systems.
Fig 4.
Heterotrimeric G protein coupling of MrgA1 and MrgC11. (A and D) FLRFa or γ2-MSH dose-dependently stimulates intracellular calcium mobilization in HEK-MrgA1 or HEK-MrgC11 in the absence (•) or presence (▪) of PTX (16 h, 100 ng/ml). All results shown are the mean of triplicate determination ± SEM. (B and E) Effect of Gα subunit KO on [Ca2+]i mobilization. KO MEFs were derived from KO mice at embryonic 8.5 and 9.5 days. Gα12/13 KO MEF (▴) and Gαq/11 KO MEF (○) were transfected with the cDNAs encoding the MrgA1-GFP (B) or the MrgC11-GFP (E). FLRFa or γ2-MSH evoked [Ca2+]i responses were completely ablogated in Gαq/11 KO MEF expressing MrgA1-GFP (B) or MrgC11-GFP (E). However, cotransfection (▵) of wild-type Gαq plus MrgA1-GFP or MrgC11-GFP in Gαq/11 double KO MEF restored responsiveness to FLRFa or γ2-MSH, respectively. Positively transfected cells were selected by their green fluorescence excited at 480 nm (GFP-positive cells). On the same field, cells that did not express GFP (GFP-negative cells) were selected as internal control. (C and F) cAMP production in HEK-MrgA1 (C) or HEK-MrgC11 (F). Cells were stimulated with various concentrations of FLRFa or γ2-MSH in the presence or absence of 10 μM forskolin as described in Materials and Methods. Each value represents the mean ± SEM for three independent experiments.
Heterotrimeric G proteins of the Gαi and Gαq class are involved in the propagation of signals from GPCRs leading to [Ca2+]i elevation (22). To determine whether Gαi/o proteins are involved in the [Ca2+]i response, we pretreated HEK-MrgA1 or HEK-MrgC11 cells with PTX (100 ng/ml) for 16 h. PTX blocks responses mediated by the Gαi/o system of G protein transducers but does not effect signals transmitted through Gαs, Gα12/13, or the Gαq/11 family. The dose dependency in [Ca2+]i responses for both receptors were not affected by PTX (Fig. 4 A and D). In contrast, PTX completely blocked FLRFa-induced calcium response in HEK-mNPFF2 cells (data not shown).
We used MEF cell lines derived from Gαq/11 or Gα12/13 double gene KO mice to test whether activation of MrgA1 or MrgC11 receptors can mobilize calcium responses through the direct participation of Gαq/11. The Gαq/11 KO MEFs or Gα12/13 KO MEFs were transfected with cDNAs encoding either MrgA1-GFP or MrgC11-GFP receptor, and the ability of agonists to increase [Ca2+]i, was measured in individual cells. The GFP receptor fusion proteins were used to identify positively transfected cells, and single-cell calcium assays were performed as described in our previous studies (3). FLRFa or γ2-MSH induced robust, transient calcium responses in Gα12/13 KO cells expressing MrgA1 or MrgC11, but Gαq/11 double KO MEFs failed to respond to FLRFa or γ2-MSH (Fig. 4 B and E). The calcium responses in Gαq/11 KO cells were rescued when Gαq/11 KO cells were cotransfected with plasmids encoding wild-type Gαq and each receptor (Fig. 4 B and E). These observations demonstrated that Gαq/11 proteins are coupled to both receptors in the calcium-signaling pathway.
It is also possible that these receptors are coupled to the Gαi/o or to the Gαs family of heterotrimeric G proteins. Thus, we measured cAMP production in the presence of various concentrations of agonists and presence or absence of forskolin. No significant inhibition or activation was observed in the presence of various concentrations of FLRFa or γ2-MSH (Fig. 4 C and F). Also, FLRFa and γ2-MSH were unable to inhibit forskolin-induced cAMP accumulation in these cells (Fig. 4 C and F). Taken together, these results demonstrate that both MrgA1 and MrgC11 are coupled to Gαq/11, but not to Gαi/o or Gαs, in HEK293 cells. But, it remains to be determined how this G protein-coupling specificity is translated in DRG neurons.
Ligand Identification of MrgA1 and MrgC11.
To identify putative ligands for Mrg receptors, we screened HEK-MrgA1 or HEK-MrgC11 stable cell lines in a calcium-mobilization assay using a fluorescence-imaging plate reader (FLEXstation). We tested a panel of known peptides (≈100 peptides) at various concentrations and then measured agonist potencies (EC50) for peptides showing calcium responses. HEK293 parental cells did not respond to peptides shown in Table 1. γ2-MSH was the best agonist for MrgC11 (EC50 = 11 ± 5 nM); this neuropeptide is derived from pro-opiomelanocotin (POMC). However, MrgC11 was not activated by other POMC-derived peptides such as α-MSH, β-MSH, and endorphins (data not shown), which are largely mediated through melanocortin (MC) receptors. On the other hand, we failed to find a better agonist than FLRFa against MrgA1.
As shown in Table 1, a common feature of all activating peptides for MrgA1 and MrgC11 is the presence of RF(Y)G or RF(Y)a at the C terminus. The invertebrate neuropeptides terminating with -RP or -RN at the C terminus were inactive for both receptors up to100 μM (data not shown). However, a distinct structure–activity relationship exists between MrgA1 and MrgC11. All peptides containing RF(Y)a or RF(Y)G motif at the C terminus were able to activate MrgC11 with different potencies, but only certain peptides among them were able to activate MrgA1 (Table 1). Furthermore, either RFa or RF-OH itself was sufficient to activate MrgC11 with EC50 = 460 ± 35 nM and 632 ± 124 nM, respectively, whereas RFa or its free acid form was not able to activate MrgA1 (Table 2), suggesting that other as yet unknown structural motifs are required to activate MrgA1 in addition to the RF(Y)a or RF(Y)G motif at the C terminus. Because the amidation of RFa peptides is known to be critical for agonist activity on RFa receptors, such as GPR54 and NPFF receptors (13, 16, 17), we examined how amidation and/or deamidation of RFa peptides affect the functional affinity for both receptors. The free acid form of FMRFa resulted in about 20-fold decrease in activity for MrgA1. Also, the deamidated peptide form of YGFMRFa resulted in complete loss of activity for MrgA1, whereas deamidation rendered the peptides about only 4- to 5-fold less active for MrgC11 (Table 1). Inversely, amidation of BAM-15 peptide caused modest increase (2.7-fold) in activity for MrgA1, whereas it caused a pronounced decrease (9-fold) for MrgC11.
Table 2.
The EC50 values of FMRFa peptides chirally modified in successive single residues for HEK-MrgC11 and HEK-MrgA1 cells
| Peptides | MrgC11, nM | MrgA1, nM |
|---|---|---|
| F-M-R-Fa | 114 ± 32 | 420 ± 71 |
| (D)F-M-R-Fa | 108 ± 1 | 882 ± 55 |
| F-(D)M-R-Fa | 11 ± 4 | 1,260 ± 223 |
| F-M-(D)R-Fa | Inactive | Inactive |
| F-M-R-(D)Fa | Inactive | 643 ± 80 |
| R-Fa | 460 ± 35 | Inactive |
| R-F-OH | 632 ± 124 | Inactive |
Data represent mean ± SEM from triplicate independent determinations.
To better define the agonist specificity required for activation of both receptors, we examined the significance of the orientation of the side chains by substituting d-amino acid isomers in each position (Table 2). The change of arginine (Arg) chirality resulted in complete loss of agonist activity for both receptors, suggesting that Arg-3 is a common critical residue (Table 2). Replacement of the Met-2 residue by the d-isomer resulted in a 3-fold decrease in activity for MrgA1, whereas the change resulted in 10-fold increase in activity for MrgC11 (Table 2). This increase might be attributable to an optimization of tertiary structure for better receptor binding. Also, substitution of the Phe-4 with the d-isomer rendered the peptide inactive for MrgC11, whereas it resulted in only slight decrease in activity for MrgA1. These data provide further evidence of structure–activity differences between MrgA1 and MrgC11, though both receptors are activated by RF-amide-related peptides.
Discussion
We have further defined the Gα coupling specificity of the Mrg receptors in heterologus cellular systems. By functional and genetic studies, we demonstrated that both MrgA1 and MrgC11 are coupled to Gαq/11, but not to Gαi/o or Gαs signaling pathway. It is known that activation of hMrgX1 elicited calcium release in a PTX insensitive manner (18). This finding suggests that the main mode of signaling of Mrg receptors may be through Gαq/11 signaling pathway. However, the functional significance of the Gαq/11 pathway in DRG neurons has not been well characterized. Recently, D. Julius and coworkers (23) showed that the activation of the Gαq/11 coupled pathway modulates the ability of VR1 (TRPV1), which detects noxious heat or capsaicin in sensory neurons. VR1 belongs to the TRP superfamily that includes >20 related cation channels that play critical roles in processes ranging from sensory physiology to vasorelaxation and male fertility (24). Some of them, such as TRPV1, TRPV2, and TRPM8, are expressed in DRG neurons (24). Thus, it is conceivable that Mrg receptors may regulate the activation of a specific TRP channel, though Gαq/11 signaling pathway to modulate pain sensitivity.
Recently, Lembo and colleagues (18) proposed that BAM-22P or BAM-15 derived from preproenkephalin is the endogenous ligand for hMrgX1 (SNSR3). The human MrgX1 sequence appears most similar to the murine MrgAs and MrgC11 sequence, though it is not possible to define clear orthologous pairs (3). BAM-22P and BAM-15, weakly activate MrgA1, EC50 = 2.5 μM and 23 μM, respectively. MrgC11 is potently activated by verterbrate neuropeptides in the following order: γ2-MSH (EC50 = 11 nM) > γ1-MSH (EC50 = 17 nM) > BAM-22P (EC50 = 26 nM) ≈ Dynorphin14 > BAM-15 (EC50 = 53 nM), NPFF and Kiss (112–121). Under physiological conditions, it is not clear which peptide specifically acts as ligand for the MrgC11 receptor. Precise localization studies of receptors and peptides will be required to identify endogenous ligands.
MrgC11 is shown to be activated by all invertebrate and vertebrate neuropeptides terminating with either RF(Y)G or RF(Y)a. Previously, we found that specific RFa peptides, FLRFa, NPAF, and NPFF distinctively activate MrgA1, MrgA4, and MAS1, respectively, in HEK293-Gα15 cells (3). These results, together with the high degree of sequence homology between Mrg receptors, suggest that the cognate ligands for Mrg receptors may also terminate with the sequence RF(Y)G or RF(Y)a. We further demonstrated that the C-terminal Arg is a common critical residue for activation of both MrgA1 and MrgC11 (see Table 2). It is not surprising that the phenylalanine residue of -RFa can be replaced by tyrosine because they have very similar configuration and because of the functional similarity of the aromatic benzene (25). For example, human kisspeptin (12) ends with RFa, but the murine and rat homologues apparently end with RYa. In most cases, the glycine residue at the C terminus appears to act as amide donor. It is relatively common to find the partially processed peptide with the whole glycyl residue instead of the amide. Thus, RFa-related peptides could include peptides ending with -RFG, -RYG, -RFa, or -RYa.
However, ligand specificity for MrgA1 and MrgC11 was clearly different even though they are commonly activated by RFa-related peptides. The difference in ligand specificity can be summarized by the following observations. First, amidation and/or deamination of agonists differentially acts on activation of both Mrg receptors (see Table 1). Second, both MrgA1 and MrgC11 showed a different pattern of specificity for alternative chiral forms of the amino acid components of the ligand except for Arg residue (see Table 2). Moreover, the N-terminal sequences of agonists appeared to be very important for MrgA1 activation. It has been known that C-terminally amidated peptides can take a variety of conformations depending on the amino acids at the N terminus (26). Thus, assuming that the ligand for the Mrg family is a RFa-related peptide, the combination of amidation and N-terminal sequences may be sufficient to generate differences in ligand conformation unique for each Mrg receptor.
In summary, we have determined that MrgC11 is specifically expressed in a subset of nociceptive neurons in the DRG. We have also found that both MrgA1 and MrgC11 can be activated by RFa-related peptides through coupling to the Gαq/11 signaling pathway. Furthermore, we found differences in ligand specificity between MrgA1 and MrgC11, suggesting functional diversity of Mrg receptors. Most recently, adenine was proposed as an endogenous ligand for rat MrgA (27). However, we could not find any response of mouse MrgA1, MrgC11, and human MrgX1–MrgX4 to adenine (data not shown).
Acknowledgments
We thank our laboratory members for helpful discussion and cooperation, and Henry Lester and his laboratory members for help with calcium imaging. S.H., J.K., and M.I.S. are supported by National Institute for General Medical Science Grant GM-34236. X.D. is a postdoctoral fellow of the American Cancer Society, and M.J.Z. is supported by the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation Fellowship (DRG-1581).
Abbreviations
GPCR, G protein-coupled receptor
RFa, RF amide
FMRFa, FMRF amide
MEF, mouse embryonic fibroblast
KO, knockout
TG, thapsigargin
References
- 1.Baldwin J. M. (1994) Curr. Opin. Cell Biol. 6, 180-190. [DOI] [PubMed] [Google Scholar]
- 2.Strader C. D., Fong, T. M., Tota, M. R., Underwood, D. & Dixon, R. A. F. (1994) Annu. Rev. Biochem. 63, 101-132. [DOI] [PubMed] [Google Scholar]
- 3.Dong X., Han, S., Zylka, M. J., Simon, M. I. & Anderson, D. J. (2001) Cell 106, 619-632. [DOI] [PubMed] [Google Scholar]
- 4.Simonin F. & Kieffer, B. L. (2002) Nat. Neurosci. 5, 185-186. [DOI] [PubMed] [Google Scholar]
- 5.Greenberg M. J. & Price, D. A. (1992) Prog. Brain Res. 92, 25-37. [DOI] [PubMed] [Google Scholar]
- 6.Li C., Kim, K. & Nelson, L. S. (1999) Brain Res. 848, 26-34. [DOI] [PubMed] [Google Scholar]
- 7.Nelson L. S., Rosoff, M. L. & Li, C. (1998) Science 281, 1686-1690. [DOI] [PubMed] [Google Scholar]
- 8.Perry S. J., Yi-Kung Huang, E., Cronk, D., Bagust, J., Sharma, R., Walker, R. J., Wilson, S. & Burke, J. F. (1997) FEBS Lett. 409, 426-430. [DOI] [PubMed] [Google Scholar]
- 9.Vilim F. S., Aarnisalo, A. A., Nieminen, M. L., Lintunen, M., Karlstedt, K., Kontinen, V. K., Kalso, E., States, B., Panula, P. & Ziff, E. (1999) Mol. Phamacol. 55, 804-811. [PubMed] [Google Scholar]
- 10.Hinuma S., Habata, Y., Fuji, R., Kawamata, Y., Hosoya, M., Fukusumi, S., Kitada, C., Masuo, Y., Asano, T., Matsumoto, H., et al. (1998) Nature 393, 272-276. [DOI] [PubMed] [Google Scholar]
- 11.Hinuma S., Shintani, Y., Fukusumi, S., Iijima, N., Matsumoto, Y., Hosoya, M., Fujii, R., Watanabe, T., Kikuchi, K., Terao, Y., et al. (2000) Nat. Cell Biol. 2, 703-708. [DOI] [PubMed] [Google Scholar]
- 12.Kotani M., Michel, D., Vandenbogaerde, A., Communi, D., Vanderwinden, J. M., Le Poul, E., Brezillon, S., Tyldesley, R., Suarez-Huerta, N., Vandeput, F., et al. (2001) J. Biol. Chem. 276, 34631-34636. [DOI] [PubMed] [Google Scholar]
- 13.Bonini J. A., Jones, K. A., Adham, N., Forray, C., Artymyshyn, R., Durkin, M. M., Smith, K. E., Tamm, J. A., Boteju, L. W., Lakhlani, P. P., et al. (2000) J. Biol. Chem. 275, 39324-39331. [DOI] [PubMed] [Google Scholar]
- 14.Ohtaki T., Shintani, Y., Honda, S., Matsumoto, H., Hori, A., Kanehashi, K., Terao, Y., Kumano, S., Takatsu, Y., Masuda, Y., et al. (2001) Nature 411, 613-617. [DOI] [PubMed] [Google Scholar]
- 15.Panula P., Aarnisalo, A. A. & Wasowicz, K. (1996) Prog. Neurobiol. 48, 461-487. [DOI] [PubMed] [Google Scholar]
- 16.Muir A. I., Chamberlain, L., Elshourbagy, N. A., Michalovich, D., Moore, D. J., Calamari, A., Szekeres, P. G., Sarau, H. M., Chambers, J. K., Murdock, P., et al. (2001) J. Biol. Chem. 276, 28969-28975. [DOI] [PubMed] [Google Scholar]
- 17.Clements M. K., McDonald, T. P., Wang, R., Xie, G., O'Dowd, B. F., George, S. R., Austin, C. P. & Liu, Q. (2001) Biochem. Biophys. Res. Commun. 284, 1189-1193. [DOI] [PubMed] [Google Scholar]
- 18.Lembo P. M., Grazzini, E., Groblewski, T., O'Donnell, D., Roy, M. O., Zhang, J., Hoffert, C., Cao, J., Schmidt, R., Pelletier, M., et al. (2002) Nat. Neurosci. 5, 201-209. [DOI] [PubMed] [Google Scholar]
- 19.Kabarowski J. H., Feramisco, J. D., Le, L. Q., Gu, J. L., Luoh, S. W., Simon, M. I. & Witte, O. N. (2000) Proc. Natl. Acad. Sci. USA 97, 12109-12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilman A. G. (1970) Proc. Natl. Acad. Sci. USA 67, 305-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer zu Heringdorf D., Niederdraing, N., Neumann, E., Frode, R., Lass, H., Van Koppen, C. J. & Jakobs, K. H. (1998) Eur. J. Pharmacol. 354, 113-122. [DOI] [PubMed] [Google Scholar]
- 22.Gudermann T., Kalkbrenner, F. & Schultz, G. (1996) Annu. Rev. Pharmacol. Toxicol. 36, 429-459. [DOI] [PubMed] [Google Scholar]
- 23.Chuang H. H., Prescott, E. D., Kong, H., Shields, S., Jordt, S. E., Basbaum, A. I., Chao, M. V. & Julius, D. (2001) Nature 411, 957-962. [DOI] [PubMed] [Google Scholar]
- 24.Montell C., Birnbaumer, L. & Flockerzi, V. (2002) Cell 108, 595-598. [DOI] [PubMed] [Google Scholar]
- 25.Zacharias N. & Dougherty, D. A. (2002) Trends Pharmacol. Sci. 23, 281-287. [DOI] [PubMed] [Google Scholar]
- 26.Edison A. S., Espinoza, E. & Zachariah, C. (1999) J. Neurosci. 19, 6318-6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bender E., Buist, A., Jurzak, M., Langlois, X., Baggerman, G., Verhasselt, P., Ercken, M., Guo, H. Q., Wintmolders, C., Van den Wyngaert, I., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 8573-8578. [DOI] [PMC free article] [PubMed] [Google Scholar]