Abstract
RNase E, a multifunctional endoribonuclease of Escherichia coli, attacks substrates at highly specific sites. By using synthetic oligoribonucleotides containing repeats of identical target sequences protected from cleavage by 2′-O-methylated nucleotide substitutions at specific positions, we investigated how RNase E identifies its cleavage sites. We found that the RNase E catalytic domain (i.e., N-Rne) binds selectively to 5′-monophosphate RNA termini but has an inherent mode of cleavage in the 3′ to 5′ direction. Target sequences made uncleavable by the introduction of 2′-O-methyl-modified nucleotides bind to RNase E and impede cleavages at normally susceptible sites located 5′ to, but not 3′ to, the protected target. Our results indicate that RNase E can identify cleavage sites by a 3′ to 5′ “scanning” mechanism and imply that anchoring of the enzyme to the 5′-monophosphorylated end of these substrates orients the enzyme for directional cleavages that occur in a processive or quasiprocessive mode. In contrast, we find that RNase G, which has extensive structural homology with and size similarity to N-Rne, and can functionally complement RNase E gene deletions when overexpressed, has a nondirectional and distributive mode of action.
Keywords: RNA degradation, processive, RNase G
The multifunctional Escherichia coli ribonuclease, RNase E, has a demonstrated role in the processing of ribosomal RNA (1, 2), the chemical degradation of bulk cellular RNA (3–7), the decay of specific regulatory, messenger, and structural RNAs (for recent reviews, see refs. 8 and 9), the control of plasmid DNA replication (10), and the removal of poly(A) tails from transcripts (11, 12). RNase E cleaves preferentially at specific sites in single-strand RNA segments rich in A+U nucleotides (13–16). An inherent mode of action involving entry of RNase E at the 5′ end of substrates followed by a 5′ to 3′ wave of cleavages has been inferred from the greater in vivo stability of fragments at the 3′ ends of certain RNA substrates (17–20) together with evidence that (i) the enzyme prefers a free 5′ end for endonucleolytic cleavage (18, 20), (ii) RNase E degradation efficiency is affected by 5′-phosphorylation in vivo (10) and in vitro (19, 20), and (iii) 5′ regions of secondary structure can impede cleavages in vivo (21). However, RNA degradation in vivo also can occur in the opposite direction (e.g., ref. 22).
As RNase E cleavage of complex substrates potentially can be affected by differences in the affinity of the enzyme for target sites having different sequences (15, 16), by secondary structure (23), and in vivo also by the attachment of ribosomes (24–26) and/or RNA-binding proteins (27), conclusions about the inherent mode of action of RNase E from studies of long natural substrates may be problematical. We wished to elucidate the mechanism of target-site selection by RNase E in the absence of these confounding factors; to do this, we designed, and chemically synthesized, oligoribonucleotide substrates that contain repeats of identical target sequences and are devoid of regions able to engage in base paring. We found that the RNase E catalytic domain (i.e., N-Rne) binds to monophosphorylated 5′ ends of substrates but shows sequential cleavages in the 3′ to 5′ direction. We further showed that the catalytic domain binds to nucleotide sequences that define a cleavage site, even when the site is made insensitive to cleavage by 2′-O-methyl nucleotide substitutions, and that it stalls at the protected locus, preventing attack on nonmodified target sequences 5′ to that locus. In contrast to the RNase E catalytic domain, RNase G [also known as CafA protein, MreA protein, Rng (28–31)], which has extensive sequence homology and size similar to N-Rne (32), broadly similar cleavage specificity (31, 33, 34), a similar preference for 5′-monophosphate termini (31, 34), and the ability to complement Rne gene deletions (35), showed no directionality of cleavage of the same substrates.
Materials and Methods
Plasmids, Oligonucleotides, and Bacteria Strains.
Plasmids for overexpression of full-length RNase E and N-Rne have been described (23). Oligonucleotides BR13, BR13N, BR131-2M, BR13-11M, BR13-13M were synthesized by ISIS Corporation (Carlsbad, CA). Oligonucleotides BR20, F20, R20, BR30, and BR30M were purchased from Dharmacon Research, Lafayette, CO. Bacterial strain used for the His-overexpression system is BL21(DE3) (Novagen).
Protein Purification.
For the purification of N-Rne and full-length RNase E, a single bacterial colony containing corresponding plasmid was freshly inoculated to 50 ml of LB containing 50 mg/ml carbenicillin and grown to OD600 = 0.6 at 30°C before the addition of isopropyl β-d-thiogalactoside (1 mM final concentration). After isopropyl β-d-thiogalactoside was added, cells were cultured for another 20 min and collected. Cells were disrupted by French press and the 100,000 × g supernatant was used for protein purification. The His-tagged fusion proteins were purified as described (36). Recombinant RNase G protein with a C-terminal His-tag was kindly provided by K. Lee (35).
Labeling of RNA Oligonucleotides.
Synthetic oligonucleotide (20 pmol) was radioactively labeled with 50 μCi of [γ-32P]ATP by using 10 units of T4 polynucleotided kinase (New England Biolabs). Labeling reactions were performed at 37°C for 30 min and then stopped by heat inactivation (65°C, 15 min). Labeled products were loaded onto urea-12% acrylamide gels and electrophoresed. The major products were excised from gels, eluted in RNA elution buffer (37), and recovered by using ethanol precipitation.
RNase E and RNase G Cleavage Assays.
32P-labeled oligoribonucleotides were synthesized in vitro as described above. Endonuclease activity was assayed in reaction mixtures containing 20 mM Tris⋅HCl (pH 8.0), 10 mM MgCl2, 10 mM NaCl, 1 mM DTT, ribonuclease inhibitor RNaseOUT (Invitrogen), and 32P-labeled oligonucleotides. Reactions were incubated at 30°C and stopped at indicated time points by aliquoting 5 μl of the reaction mixture into an equal volume of loading dye containing 80% (vol/vol) formamide, 5 mM EDTA, 0.05% (wt/vol) bromophenol blue, and 0.05% (wt/vol) xylen cyanol FF. The aliquots were then denatured at 85°C for 3 min, and analyzed by electrophoresis in 15% urea-acrylamide gels.
Results
Rne Cleaves Oligoribonucleotides Containing Specific Sequence and the Cleavage Specificity Is Not Affected by 2′-O-Methyl Modification.
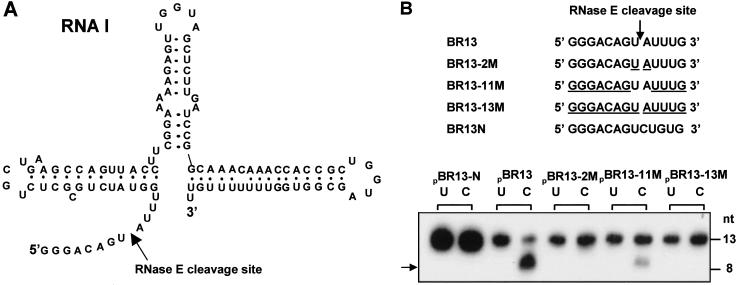
The N-terminal region of RNase E (N-Rne), which contains the catalytic domain of the enzyme, lacks both the arginine-rich major RNA-binding site of full-length RNase E (23) and the scaffold region that interacts with other protein components of degradosomes (38–40). However, N-Rne cleaves 9S RNA, a variety of messenger RNAs, and RNAI (Fig. 1A), a 108-nt antisense suppressor of ColE1 plasmid replication, at the same sites as Rne (e.g., refs. 23, 35, 41, 42), indicating that it possesses full cleavage specificity. We investigated the stringency of this specificity by determining the site(s) of cleavage of 32P-labeled monophosphorylated oligoribonucleotide substrates containing 2′-O-methyl modifications at defined positions (Fig. 1B). BR13, which includes the RNase E target sequence of RNAI, was attacked by N-Rne at its known RNase E cleavage site (23), whereas BR13-2M and BR13-13M, which are protected by 2′-O-methyl substitutions either at this cleavage site or throughout, respectively, were uncleaved. BR13N, a random-sequence oligonucleotide, was also uncleaved, consistent with dependence of RNase E cleavages on a specific order of nucleotides (15, 16). BR13-11M, which contains chemically modified nucleotides at all locations, except for the two nucleotides bracketing the bond cleaved in BR13, was attacked at the solitary nonprotected phosphodiester bond, indicating that extensive 2′-O-methyl modification does not prevent ribonucleolytic cleavage at nonmodified bonds (see ref. 43).
Fig 1.
Cleavage of chemically modified oligoribonucleotides by N-Rne. (A) Sequence and secondary structure of RNAI. The 13-nt unpaired 5′ sequence containing an RNase E cleavage site (indicated by arrow) has been reproduced in a series of chemically synthesized oligoribonucleotides. (B) A total of 0.2 pmol of 5′-labeled oligonucleotides (indicated by subscript p, e.g., pBR13) were incubated with 0.4 pmol of N-Rne at 30°C for 5 min and then the reactions were stopped by adding sequencing loading dye containing 80% formamide. RNAs in reaction mixtures were then separated in urea-15% acrylamide gels. The gel position of the labeled 8-nt cleavage product is indicated. The sequence of oligoribonucleotide is shown, and the nucleotides containing the 2′-O-methyl modification are underlined. U, uncleaved oligonucleotides (no enzyme incubation); C, cleaved oligonucleotides that had been incubated with N-Rne. The position of the cleavage products is indicated on the gel by an arrowhead.
N-Rne Binds Specifically to the Substrate and the Sequence-Specific Binding Activity Is Independent of Cleavage.
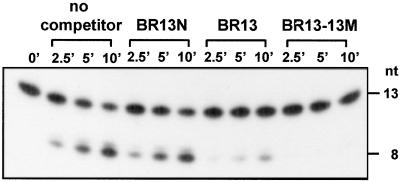
Although the RNA-cleaving ability of N-Rne implies substrate binding, interaction of this catalytic domain with RNA has not been detected under conditions that show stable binding of full-length RNase E (23, 44–46). However, during the course of our experiments, we observed that unlabeled BR13 was able to compete with cleavage of radioactively labeled BR13 by N-Rne (Fig. 2). BR13-13M, which contains an RNase E target sequence but is protected from cleavage by 2′-O-methyl nucleotide substitutions throughout, not only inhibited digestion of BR13, but also competed more efficiently on an equimolar basis than its unprotected homolog (89% and 52% inhibition by BR13-13M and BR13, respectively, at 0.5 μM concentration; Fig. 2). BR13N, which consists of unmodified nucleotides arranged in random order, lacks an N-Rne target sequence, and, like BR13-13M, is not cleaved by N-Rne (see above), had no effect on cleavage of BR13 (Fig. 2). Together, these findings provide evidence that the RNase E catalytic domain binds specifically to target sites even when they are protected from cleavage by chemical modification, and additionally implies that translocation of N-Rne may occur less readily from uncleaved target sites than from cleaved ones. Addition of a 2′-O-methyl variant of BR13N did not affect cleavage of BR13 by N-Rne (data not shown), indicating that competition by BR13-13M resulted from substrate-specific interaction with the enzyme, rather than from chemical modification of the oligonucleotide.
Fig 2.
Effect of oligonucleotide competitors on cleavage of 32P-labeled BR13 by N-Rne. Before addition of N-Rne, 10 nM 5′ labeled BR13 was mixed individually with 0.5 μM unlabeled oligonucleotides, which included BR13N, BR13, and BR13-13M. Cleavage reactions were performed as described in Materials and Methods. Reactions were stopped at the times indicated and electrophoresed as in Fig. 1. The percentage inhibition of cleavage of 32P-labeled BR13 by unlabeled BR13 and BR13-13M was 52 and 89%, respectively. Cleavage was quantified by measuring cleavage product and total substrate at the 10-min time point by using the STORM 840 PHOSPHOIMAGER (Molecular Dynamics). The percentage inhibition is defined as follows: 1 − cleavage efficiency of reaction containing competitor/cleavage efficiency of reaction without competitor × 100%.
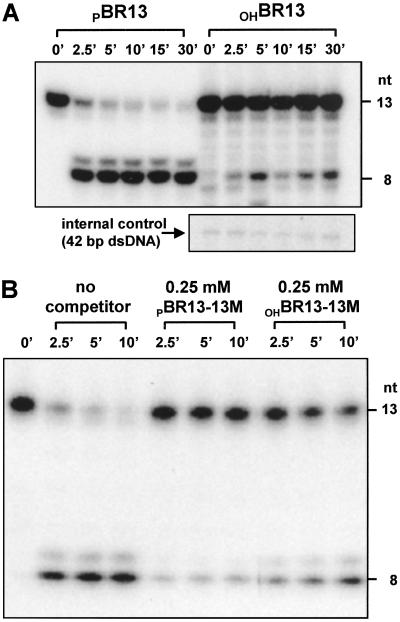
5′-Phosphorylation Modulates Cleavage by Altering the Binding of N-Rne.
Consistent with earlier evidence that efficient cleavage by full-length RNase E (20) or its catalytic domain (34) requires a 5′-monophosphate terminus, dephosphorylation of the 5′ end of BR13 sharply reduced N-Rne digestion of the substrate (compare PBR13 vs. OHBR13; Fig. 3A). We hypothesized that if the effect of 5′-phosphorylation on cleavage by N-Rne is mediated at the level of RNA binding, as has been proposed (19, 20), the extent of 5′-phosphorylation should affect the competitiveness of RNAs that contain an RNase E recognition sequence, even when these RNAs are protected from cleavage by 2′-O-methylation. We found that BR13-13M containing a 5′- monophosphorylated terminus (PBR13-13M) did in fact compete with cleavage of BR13 much more efficiently than OHBR13-13M (86% and 48% inhibition by PBR13-13M and OHBR13-13M, respectively, at 0.25 μM concentration; Fig. 3B). Because N-Rne also interacts site-specifically with a sequence of nucleotides at the cleavage site (Fig. 2), this result implies that the catalytic domain of RNase E contacts RNA substrates at two separate locations.
Fig 3.
5′-Phosphorylation modulates cleavage by altering the binding of N-Rne. (A) Cleavage of BR13 by N-Rne. The effects of different 5′ termini on cleavages under the same conditions were examined as follows: 0.2 pmol of BR13 containing a 5′-monophosphorylated terminus (5′ PBR13), which was labeled with 32P as in Fig. 1 and purified as described in Materials and Methods, and BR13 with an unlabeled 5′-hydroxyl terminus (5′ OHBR13), were incubated with N-Rne as described. Instead of stopping reaction by addition of formamide loading dye, an equal volume of phenol/chloroform mixture was added at the indicated time points. After phenol/chloroform extraction, oligonucleotides in reaction mixtures were recovered by ethanol precipitation. For 5′ PBR13 reactions, precipitates were resuspended with formamide loading dye. For 5′ OHBR13, precipitates were resuspended in T4 kinase reaction buffer and labeled with 1 μCi of [γ-32P]ATP. A small amount of 42-bp double-stranded DNA fragment, which did not interfere with N-Rne cleavage (data not shown), was added to the reaction mixture of 5′OHBR13 before N-Rne cleavage and served as internal control for the later labeling of cleavage products. (B) Competition of BR13-13M on the cleavage of BR13 by N-Rne. Before carrying out the competition reaction, BR13-13M was incubated with T4 polynucleotide kinase in reaction buffer with or without 10 mM ATP to produce 5′ PBR13-13M and 5′ OHBR13-13M. After the kinase reaction used to label the 5′ terminus with 32P, oligonucleotides were recovered by phenol/chloroform extraction and ethanol precipitation. The recovered oligos were passed through a Microspin G-25 column (Amersham Pharmacia) to remove residual free ATP and quantified spectrophotometrically by the OD260 reading. The competition reactions were performed as in Fig. 2. The competitors, the concentrations of competitors, and time points for the reactions are as indicated. The percentage inhibition of cleavage efficiency, which is defined as in Fig. 2, was 86% and 48%, respectively, for 5′ PBR13-13M and 5′ OHBR13-13M.
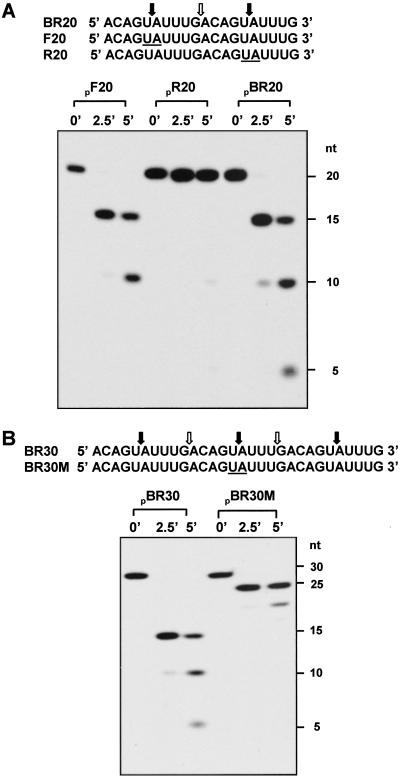
The RNase E Catalytic Domain Cleaves in the 3′ to 5′ Direction.
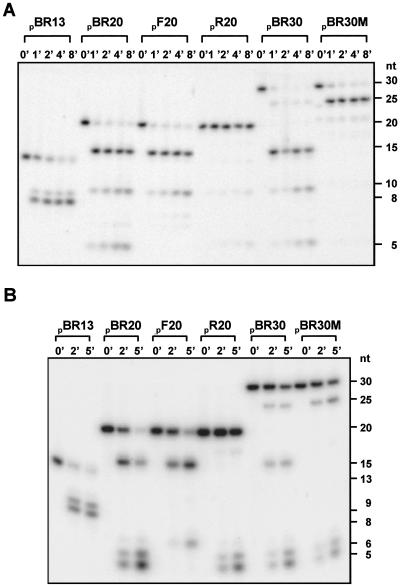
It has been proposed that RNase E enters substrates at the 5′ end and then scans toward the 3′ end for cleavage sites (8, 9, 17, 20). The ability of 2′-O-methyl substitutions to inhibit cleavage at ordinarily susceptible internucleotide bonds, together with evidence that noncleavable recognition sequences may retain RNase E, suggested an approach to test this model directly. We designed oligoribonucleotides that contain two repeats of an identical RNase E target sequence and determined the effect of a noncleavable bond in one repeat on cleavage of a bond in the other repeat (Fig. 4A). BR20, an oligoncleotide containing two repeats of a target sequence showed the expected cutting by N-Rne at internucleotide bonds 5–6 and 15–16, plus an additional cleavage at bond 10–11, a target apparently generated by the sequence combination created in BR20 (see refs. 15 and 16). Whereas a substrate (F20) containing 2′-O-methyl-substituted nucleotides in the N5 and N6 positions was cleaved at nonprotected internucleotide bonds (i.e., N15–16 and N10–11) situated 3′ to the substitutions, 2′-O-methyl substitutions at the N15-N16 (R20) bond surprisingly prevented attack at target sites located 5′ to the protected bond (Fig. 4A).
Fig 4.
N-Rne-mediated-cleavage of 32P-labeled oligonucleotides containing repeats of identical target sequence. (A) Cleavage by N-Rne of oligonucleotides containing two repeats of the RNAI single-strand region shown in Fig. 1. The sequence and the oligonucleotides having 2′-O-methyl modification (underlines) are shown. (B) Cleavage by N-Rne of oligonucleotides containing three repeats of RNAI single-strand region. The expected cleavage sites are indicated by solid arrows and adventitious cleavage sites (see text) are indicated by open arrows.
The results above imply that cleavages by N-Rne occurred with 3′ to 5′ directionality in these oligonucleotide substrates, and further suggest that dissociation of the enzyme may be impeded when it encounters and interacts with a recognition sequence protected by 2′-O-methyl modification. The use of oligoribonucleotides containing three repeats of the RNAI-derived cleavage sequence, ACAGUAUUUG, enabled us to test both of these hypotheses further.
As seen in Fig. 4B, 2′-O-methyl substitutions at nucleotides N15 and N16 of oligonucleotide BR30M not only precluded production of the 15-nt product normally generated by cleavage at this site (BR30), but also (i) impeded cleavage 5′ to the protected bond, and (ii) enhanced cleavage at susceptible sites 3′ to the protected bond. As was observed also for shorter oligonucleotide substrates (see above), the combining of multiple RNase E recognition sequences on the same RNA molecule resulted in adventitious, as well as expected, cleavages; cleavages at both expected and adventitious sites were impeded 5′ to protected bonds and enhanced 3′ to these bonds. We speculate that activity triggered by the substrate-bound stalled enzyme may account for the latter effect.
Mode of 3′ to 5′ Cleavage Site Selection Is an Inherent Property of RNase E Catalysis.
To determine whether the 3′ to 5′ directionality of cleavage we observed for N-Rne is altered by the presence of the ARRBSmaj on the endonuclease, we repeated the experiments shown in Fig. 4 by using full-length RNase E, and obtained identical results (Fig. 5A). Thus, although the capability for directional scanning of substrates and processive cleavages resides in the catalytic domain of RNase E, this ability is shared by full-length RNase E and is not affected by ARRBSmaj.
Fig 5.
Cleavages by RNase E and RNase G. on various 5′-32P-labeled oligonucleotide substrates. (A) Twenty-five nanograms of full-length RNase E was used in each reaction mixture. (B) Fifty nanograms of RNase G was used per reaction mixture. Reaction conditions and methods of analysis are indicated in Materials and Methods. Substrates are described in Figs. 1 and 4.
Recent reports indicate that the RNase G/CafA/Mre protein, which has a sequence highly similar to that of N-Rne (32) and attacks A + U-rich regions of RNA at or near the bonds cleaved by RNase E, is also affected by 5′-phosphorylation (31, 34). Moreover, Rng can functionally complement deletion of Rne when overproduced (35). However, we found that despite these similarities, the mode of action of RNase G is dramatically different from the one we observed for RNase E. As seen in Fig. 5B, the cleavage patterns observed for RNase G by using 5-32P-labeled oligonucleotides BR20, F20, R20, BR30, and BR30M show no directionally and no blockage of cleavages occurring 5′ to protected bonds, indicating that the directionality we observed for RNase E cleaves is a function of the enzyme, rather a peculiarity of the substrate used for these studies.
Discussion
It is well established that cleavages by RNase E occur in A + U-rich regions of single-strand RNA (15, 16, 47). By using chemically synthesized oligoribonucleotides modified to render specific phosphodiester bonds noncleavable by RNase E, we experimentally addressed the question of how the catalytic domain, which encodes cleavage site specificity, finds its targets. Our results rule out a 5′ to 3′ scanning mechanism intrinsic to the enzyme and instead show that the inherent mode of action of RNase E is intrinsic to the enzyme cleavage in the 3′ to 5′ direction. Our finding that cleavage sites 5′ to intranucleotide bonds made insensitive by 2′-O-methyl nucleotide substitution are not attacked, whereas targets 3′ to protected intranucleotide bonds are cleaved, argues strongly that the catalytic domain of RNase E translocates in the 3′ to 5′ direction and that during translocation the enzyme stalls at target sequences containing 2′-O-methyl nucleotides.
The results of our competition experiments support the notion that target sequences containing noncleavable 2′-O-methyl nucleotide substitutions sequester the catalytic domain of RNase E. Whether release from cleaved substitutes occurs after each cleavage or after the entire substrate has been scanned is not addressed by these studies. Earlier work has shown that the extent of 5′-phosphorylation strongly affects RNase E activity, and this phosphorylation has been hypothesized to occur through modulation of RNA binding (for review, see ref. 8). Our finding that the extent of 5′-phosphorylation affects the competitiveness of RNAs that contain an RNase E target sequence provides experimental evidence that 5′-phosphorylation does in fact alter binding of the enzyme to substrates, as has been proposed. Together, our results imply that sites on the RNase E catalytic domain make contact concurrently with two separate locations on substrates. We speculate that the S1 domain (amino acids 36–118) and the ‘minor’ arginine-rich region (amino acids 267–390) of N-Rne, both of which contain structural features known to mediate protein/RNA interactions (23, 48), may be the points of protein contact with the 5′ termini and target sequences of substrates, respectively.
Certain models proposed for motion of a protein within a molecule of DNA (49, 50) seem applicable also to translocation of ribonucleases along RNA substrates. Potentially, such translocation can occur through three-dimensional space by successive cycles of dissociation/reassociation (i.e., distributive translocation). Because substrate molecules in solutions used for ribonuclease assays ordinarily are separated by large volumes of solvent, the distances between individual substrate molecules are greater than between different segments of the same oligonucleotide chain; thus, successive cycles of dissociation/reassociation are more likely to be with the same substrate chain (50, 51). Proteins can also undergo linear diffusion through one-dimensional space between sites separated by various distances along the substrate chain without dissociating from the substrate (i.e., processive translocation). The ability of target sites protected by 2′-O-methyl substitutions to impede RNase E cleavage of sites located 5′ to the protected sequence is suggestive of a mechanism of translocation by one-dimensional linear diffusion (i.e., processivity); however, our data do not rule out the possibility that RNase E molecules proceeding in the 3′ to 5′ direction may dissociate from the domain of the substrate after each cleavage and then enter the substrate domain again at or near the 3′ end of the decay intermediate it generates (i.e., quasi-processivity; ref. 52).
Sequence variation at different RNase E cleavage sites or the occurrence of regions of secondary structure within natural substrates potentially can affect the order of RNase E cleavages (14, 46, 53, 54). Attack by RNase E may be further modulated or circumvented by the presence of RNA binding proteins or ribosomes at particular sites of substrates, or by the concurrent actions of other ribonucleases, and such factors potentially may account for differences in the directionality of RNase E-mediated decay reported for different substrates in vivo (17, 46, 55). Additionally, full-length primary transcripts, which contain 5′-triphosphorylated termini, are cleaved poorly by RNase E (18, 20), and consequently may lack the 5′ end-binding activity that we have shown here for 5′-monophosphorylated termini. Primary transcripts may therefore lack an anchor to orient the enzyme for the initial RNase E cleavage, which may be determined by a mechanism different from the one used for the selection of cleavage sites on decay intermediates. The experimental design and substrates we used were intended to elucidate the inherent mode of cleavage-site selection by the RNase E catalytic domain in the absence of such modifying and/or confounding factors. Consistent with the 3′ to 5′ directionality of cleavages we observed for short oligonucleotide substrates containing multiple identical cleavage sites is kinetic evidence showing that removal of poly(A) tails from RNAI precedes, rather than follows, cleavage of a site near the 5′ end of this 108-nt-long natural substrate (11).
Like RNase E, polynucleotide phosphorylase, which exonucleolytically degrades substrates in the 3′ to 5′ direction, and poly(A) polymerase I (PAPI), which adds poly(A) 3′ tails that facilitate degradation by polynucleotide phosphorylase (56, 57), are affected by 5′-phosphorylation but initiate action at the 3′ ends of RNAs (37, 58). The ability 5′-phosphorylation to influence multiple types of transactions occurring at 3′ ends supports the notion that the termini of completed E. coli transcripts may be brought into proximity by degradosome component proteins, as suggested (11, 37, 58–60; for review, see ref. 61).
Both processive and distributive modes of action have been observed for deoxyendoribonucleases (52, 62, 63). In contrast, previously investigated proteins that attack RNA molecules endoribonucleolytically have been found normally to act only distributively (64, 65). However, it is also known that single-point mutations can change the mode of action of RNase A from distributive to processive, and concurrently can impart higher efficiency and more specificity to cleavages (66, 67). Related findings have been reported for a deoxyendoribonuclease that, when isolated from normal cells, removes damaged nucleosome DNA processively but, when isolated in mutant form from xerodema pigmentosum cells, cleaves substrates distributively and with less specific activity (68, 69). Thus, the divergent modes of action we have observed for ribonucleases E and G potentially may be determined by relatively small differences in the nonconserved regions of the catalytic domains of these proteins.
Acknowledgments
We thank Dr. Kangseok Lee of our laboratory for providing purified recombinant RNase G and Dr. Joel Belasco for helpful comments on the manuscript. These investigations were supported by National Institutes of Health Grant GM54158 (to S.N.C.).
Abbreviations
N-Rne, N-terminal region of RNase E
References
- 1.Gegenheimer P., Watson, N. & Apirion, D. (1977) J. Biol. Chem. 252, 3064-3073. [PubMed] [Google Scholar]
- 2.Ghora B. K. & Apirion, D. (1978) Cell 15, 1055-1066. [DOI] [PubMed] [Google Scholar]
- 3.Ono M. & Kuwano, M. (1979) J. Mol. Biol. 129, 343-357. [DOI] [PubMed] [Google Scholar]
- 4.Mudd E. A., Carpousis, A. J. & Krisch, H. M. (1990) Genes Dev. 4, 873-881. [DOI] [PubMed] [Google Scholar]
- 5.Babitzke P. & Kushner, S. R. (1991) Proc. Natl. Acad. Sci. USA 88, 1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melefors O. & von Gabain, A. (1991) Mol. Microbiol. 5, 857-864. [DOI] [PubMed] [Google Scholar]
- 7.Taraseviciene L., Miczak, A. & Apirion, D. (1991) Mol. Microbiol. 5, 851-855. [DOI] [PubMed] [Google Scholar]
- 8.Coburn G. A. & Mackie, G. A. (1999) Prog. Nucleic Acid Res. Mol. Biol. 62, 55-108. [DOI] [PubMed] [Google Scholar]
- 9.Steege D. A. (2000) RNA 6, 1079-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin-Chao S. & Cohen, S. N. (1991) Cell 65, 1233-1242. [DOI] [PubMed] [Google Scholar]
- 11.Huang H., Liao, J. & Cohen, S. N. (1998) Nature 391, 99-102. [DOI] [PubMed] [Google Scholar]
- 12.Walsh A. P., Tock, M. R., Mallen, M. H., Kaberdin, V. R., Gabain Av, A. & McDowall, K. J. (2001) Nucleic Acids Res. 29, 1864-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehretsmann C. P., Carpousis, A. J. & Krisch, H. M. (1992) Genes Dev. 6, 149-159. [DOI] [PubMed] [Google Scholar]
- 14.Mackie G. A. & Genereaux, J. L. (1993) J. Mol. Biol. 234, 998-1012. [DOI] [PubMed] [Google Scholar]
- 15.Lin-Chao S., Wong, T. T., McDowall, K. J. & Cohen, S. N. (1994) J. Biol. Chem. 269, 10797-10803. [PubMed] [Google Scholar]
- 16.McDowall K. J., Lin-Chao, S. & Cohen, S. N. (1994) J. Biol. Chem. 269, 10790-10796. [PubMed] [Google Scholar]
- 17.Goodrich A. F. & Steege, D. A. (1999) RNA 5, 972-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mackie G. A. (2000) J. Biol. Chem. 275, 25069-25072. [DOI] [PubMed] [Google Scholar]
- 19.Spickler C., Stronge, V. & Mackie, G. A. (2001) J. Bacteriol. 183, 1106-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackie G. A. (1998) Nature 395, 720-723. [DOI] [PubMed] [Google Scholar]
- 21.Bouvet P. & Belasco, J. G. (1992) Nature 360, 488-491. [DOI] [PubMed] [Google Scholar]
- 22.von Gabain A., Belasco, J. G., Schottel, J. L., Chang, A. C. & Cohen, S. N. (1983) Proc. Natl. Acad. Sci. USA 80, 653-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDowall K. J. & Cohen, S. N. (1996) J. Mol. Biol. 255, 349-355. [DOI] [PubMed] [Google Scholar]
- 24.Braun F., Le Derout, J. & Régnier, P. (1998) EMBO J. 17, 4790-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joyce S. A. & Dreyfus, M. (1998) J. Mol. Biol. 282, 241-54. [DOI] [PubMed] [Google Scholar]
- 26.Vytvytska O., Moll, I., Kaberdin, V. R., von Gabain, A. & Blasi, U. (2000) Genes Dev. 14, 1109-1118. [PMC free article] [PubMed] [Google Scholar]
- 27.Jerome L. J., van Biesen, T. & Frost, L. S. (1999) J. Mol. Biol. 285, 1457-73. [DOI] [PubMed] [Google Scholar]
- 28.Wachi M., Umitsuki, G. & Nagai, K. (1997) Mol. Gen. Genet. 253, 515-519. [DOI] [PubMed] [Google Scholar]
- 29.Li Z., Pandit, S. & Deutscher, M. P. (1999) EMBO J. 18, 2878-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wachi M., Umitsuki, G., Shimizu, M., Takada, A. & Nagai, K. (1999) Biochem. Biophys. Res. Commun. 259, 483-488. [DOI] [PubMed] [Google Scholar]
- 31.Tock M. R., Walsh, A. P., Carroll, G. & McDowall, K. J. (2000) J. Biol. Chem. 275, 8726-8732. [DOI] [PubMed] [Google Scholar]
- 32.McDowall K. J., Hernandez, R. G., Lin-Chao, S. & Cohen, S. N. (1993) J. Bacteriol. 175, 4245-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Umitsuki G., Wachi, M., Takada, A., Hikichi, T. & Nagai, K. (2001) Genes Cells 6, 403-410. [DOI] [PubMed] [Google Scholar]
- 34.Jiang X., Diwa, A. & Belasco, J. G. (2000) J. Bacteriol. 182, 2468-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee K., Bernstein, J. A. & Cohen, S. N. (2002) Mol. Microbiol. 43, 1445-1456. [DOI] [PubMed] [Google Scholar]
- 36.McDowall K. J., Kaberdin, V. R., Wu, S. W., Cohen, S. N. & Lin-Chao, S. (1995) Nature 374, 287-290. [DOI] [PubMed] [Google Scholar]
- 37.Feng Y. & Cohen, S. N. (2000) Proc. Natl. Acad. Sci. USA 97, 6415-6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miczak A., Kaberdin, V. R., Wei, C. L. & Lin-Chao, S. (1996) Proc. Natl. Acad. Sci. USA 93, 3865-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaberdin V. R., Miczak, A., Jakobsen, J. S., Lin-Chao, S., McDowall, K. J. & von Gabain, A. (1998) Proc. Natl. Acad. Sci. USA 95, 11637-11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vanzo N. F., Li, Y. S., Py, B., Blum, E., Higgins, C. F., Raynal, L. C., Krisch, H. M. & Carpousis, A. J. (1998) Genes Dev. 12, 2770-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lopez P. J., Marchand, I., Joyce, S. A. & Dreyfus, M. (1999) Mol. Microbiol. 3, 188-199. [DOI] [PubMed] [Google Scholar]
- 42.Ow M. C., Liu, Q. & Kushner, S. R. (2000) Mol. Microbiol. 38, 854-866. [DOI] [PubMed] [Google Scholar]
- 43.Cummins L. L., Owens, S. R., Risen, L. M., Lesnik, E. A., Freier, S. M., McGee, D., Guinosso, C. J. & Cook, P. D. (1995) Nucleic Acids Res. 23, 2019-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carpousis A. J., Van Houwe, G., Ehretsmann, C. & Krisch, H. M. (1994) Cell 76, 889-900. [DOI] [PubMed] [Google Scholar]
- 45.Taraseviciene L., Björk, G. R. & Uhlin, B. E. (1995) J. Biol. Chem. 270, 26391-26398. [DOI] [PubMed] [Google Scholar]
- 46.Kaberdin V. R., Chao, Y. H. & Lin-Chao, S. (1996) J. Biol. Chem. 271, 13103-13109. [DOI] [PubMed] [Google Scholar]
- 47.Rapaport L. R. & Mackie, G. A. (1994) J. Bacteriol. 176, 992-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cormack R. S., Genereaux, J. L. & Mackie, G. A. (1993) Proc. Natl. Acad. Sci. USA 90, 9006-9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berg O. G., Winter, R. B. & von Hippel, P. H. (1981) Biochemistry 20, 6929-6948. [DOI] [PubMed] [Google Scholar]
- 50.Stanford N. P., Szczelkun, M. D., Marko, J. F. & Halford, S. E. (2000) EMBO J. 19, 6546-6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winters M. A. & Edmonds, M. (1973) J. Biol. Chem. 248, 4763-4768. [PubMed] [Google Scholar]
- 52.Carey D. C. & Strauss, P. R. (1999) Biochemistry 38, 16553-16560. [DOI] [PubMed] [Google Scholar]
- 53.Regnier P. & Hajnsdorf, E. (1991) J. Mol. Biol. 217, 283-292. [DOI] [PubMed] [Google Scholar]
- 54.Mackie G. A., Genereaux, J. L. & Masterman, S. K. (1997) J. Biol. Chem. 272, 609-616. [PubMed] [Google Scholar]
- 55.Hajnsdorf E., Braun, F., Haugel-Nielsen, J., Le Derout, J. & Régnier, P. (1996) Biochimie 78, 416-424. [DOI] [PubMed] [Google Scholar]
- 56.Hajnsdorf E., Braun, F., Haugel-Nielsen, J. & Régnier, P. (1995) Proc. Natl. Acad. Sci. USA 92, 3973-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O'Hara E. B., Chekanova, J. A., Ingle, C. A., Kushner, Z. R., Peters, E. & Kushner, S. R. (1995) Proc. Natl. Acad. Sci. USA 92, 1807-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu F., Lin-Chao, S. & Cohen, S. N. (1993) Proc. Natl. Acad. Sci. USA 90, 6756-6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu F. & Cohen, S. N. (1995) Nature 374, 180-183. [DOI] [PubMed] [Google Scholar]
- 60.Feng Y., Huang, H., Liao, J. & Cohen, S. N. (2001) J. Biol. Chem. 276, 31651-31656. [DOI] [PubMed] [Google Scholar]
- 61.Cohen S. N. & McDowall, K. J. (1997) Mol. Microbiol. 23, 1099-1106. [DOI] [PubMed] [Google Scholar]
- 62.Hamilton R. W. & Lloyd, R. S. (1989) J. Biol. Chem. 264, 17422-17427. [PubMed] [Google Scholar]
- 63.Nyaga S. G., Dodson, M. L. & Lloyd, R. S. (1997) Biochemistry 36, 4080-4088. [DOI] [PubMed] [Google Scholar]
- 64.Krug M. S. & Berger, S. L. (1991) Biochemistry 30, 10614-10623. [DOI] [PubMed] [Google Scholar]
- 65.Wilson H. R., Yu, D., Peters, H. K., III, Zhou, J. G. & Court, D. L. (2002) EMBO J. 21, 4154-4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.delCardayre S. B. & Raines, R. T. (1994) Biochemistry 33, 6031-6037. [DOI] [PubMed] [Google Scholar]
- 67.Kelemen B. R. & Raines, R. T. (1999) Biochemistry 38, 5302-5307. [DOI] [PubMed] [Google Scholar]
- 68.Feng S., Parrish, D. D. & Lambert, M. W. (1997) Carcinogenesis 18, 279-286. [DOI] [PubMed] [Google Scholar]
- 69.Lambert M. W. & Yang, L. (2000) Biochem. Biophys. Res. Commun. 271, 782-787. [DOI] [PubMed] [Google Scholar]