Abstract
Paramecium bursaria Chlorella virus type 1 (PBCV-1) is a very large, icosahedral virus containing an internal membrane enclosed within a glycoprotein coat consisting of pseudohexagonal arrays of trimeric capsomers. Each capsomer is composed of three molecules of the major capsid protein, Vp54, the 2.0-Å resolution structure of which is reported here. Four N-linked and two O-linked glycosylation sites were identified. The N-linked sites are associated with nonstandard amino acid motifs as a result of glycosylation by virus-encoded enzymes. Each monomer of the trimeric structure consists of two eight-stranded, antiparallel β-barrel, “jelly-roll” domains related by a pseudo-sixfold rotation. The fold of the monomer and the pseudo-sixfold symmetry of the capsomer resembles that of the major coat proteins in the double-stranded DNA bacteriophage PRD1 and the double-stranded DNA human adenoviruses, as well as the viral proteins VP2-VP3 of picornaviruses. The structural similarities among these diverse groups of viruses, whose hosts include bacteria, unicellular eukaryotes, plants, and mammals, make it probable that their capsid proteins have evolved from a common ancestor that had already acquired a pseudo-sixfold organization. The trimeric capsid protein structure was used to produce a quasi-atomic model of the 1,900-Å diameter PBCV-1 outer shell, based on fitting of the Vp54 crystal structure into a three-dimensional cryoelectron microscopy image reconstruction of the virus.
Chlorella viruses, isolated from freshwater sources throughout the world, are among the largest and most complex known icosahedral viruses. These viruses have a layered structure consisting of a double-stranded DNA (dsDNA) genome, surrounded by a protein core, a lipid membrane, and an outer icosahedral capsid shell (1, 2). They have amino acid sequence similarities in the major capsid protein, as well as similarities in the virion architecture, to African swine fever virus and to iridoviruses (3, 4) such as Chilo iridescent virus (1).
Paramecium bursaria Chlorella virus (PBCV), genus Chlorovirus, family Phycodnaviridae, infects certain strains of unicellular Chlorella-like green algae (2). To initiate infection, PBCV-1 attaches to its host, uses viral-encoded enzymes to digest the cell wall around the point of attachment, and injects its genome into the cell, leaving its empty capsid on the cell surface (5, 6). This infection mechanism is similar to that used by the icosahedral, dsDNA, lipid-containing bacterial viruses, such as PRD1 (7). PBCV-1 has a molecular mass of ≈1 GDa, with a linear, 330-kbp genome encoding ≈375 proteins (8), of which ≈50 are contained in the mature virion (9). The major capsid glycoprotein Vp54 accounts for ≈40% of the virion's protein mass (10). At least three other proteins may be located on the surface, including the two glycoproteins Vp280 and Vp260, each accounting for ≈1% of the mass of protein per virion, or not more than one subunit of each minor capsid protein per icosahedral asymmetric unit. One of these is probably the special “penton” protein situated at each of the 12 fivefold vertices.
A cryoelectron microscopy (cryo-EM) reconstruction of PBCV-1 (1) showed that the outer glycoprotein capsid is icosahedral and surrounds a lipid bilayer membrane. The external layer is assembled from 20 triangular units (“trisymmetrons”; refs. 11 and 12) and 12 pentagonal caps (“pentasymmetrons”) at the fivefold vertices. The trisymmetrons and pentasymmetrons are pseudohexagonal arrays of 66 and 30 trimeric “capsomers,” respectively (Fig. 1a). The outer diameter of the viral capsid is 1,650 Å measured along the two- and threefold axes and 1,900 Å along the fivefold axes. The outer capsid of PBCV-1 contains 5,040 copies of Vp54 organized into 1,680 trimers as a pseudo T = 169d quasi-equivalent lattice (13). In addition, each capsid contains pentamers of a different protein at each of the 12 icosahedral vertices.
Fig. 1.
(a) Quasi-atomic model of the PBCV-1 capsid based on fitting the crystal structure of the Vp54 trimer into the cryo-EM reconstruction. The pentasymmetrons are colored yellow to differentiate them from the variously colored trisymmetrons. (b) Symmetry elements in the PBCV-1 trisymmetron. p3 symmetry elements are shown in black and pseudo-p6 symmetry elements are shown in gray. Pseudohexameric capsomers are presented as six disks shaded alternately in black and gray. The boundary between trisymmetrons is produced by twofold axes rotating the capsomers to face north on the left and south on the right. (c) PBCV Vp54 trimers (Cα backbone in yellow) near the center of a trisymmetron fitted into the cryo-EM map (white) viewed from outside the virus (Top) and from the side (Bottom). Figures in this paper were drawn by using the programs molscript (39), raster 3d (40), and grasp (41).
The 2.0-Å resolution structure of the trimeric capsomer described here was fitted into the previously determined cryo-EM map to obtain information of the interactions between neighboring subunits and, hence, the nature of the assembly process that forms the icosahedral particle. The structure showed a remarkable similarity of fold to the capsid proteins of the pseudo T = 25 PRD1 (14), pseudo T = 25 adenoviruses (15–17), and pseudo T = 3 picornaviruses (18), making it probable that the capsid proteins and the capsid organizations have each evolved from a common origin.
Materials and Methods
Protein Expression and Purification.
The PBCV-1 virus was propagated according to procedures described by Van Etten et al. (19) and stored in 50 mM Tris, pH 7.8, at 4°C. PBCV-1 capsid protein Vp54 was solubilized by heating the virions to 70°C for 30 min to dissociate them. Insoluble material was removed by centrifugation at 1,500 × g for 20 min. The soluble protein was placed on a DEAE-cellulose anion-exchange column and pure Vp54 protein was concentrated to ≈10 mg/ml by ultrafiltration.
Crystallization and Data Collection.
Crystals of PBCV-1 capsid protein were grown by vapor diffusion at 20°C in hanging drops with 4-μl protein samples mixed with an equal volume of reservoir solution (3.5–3.7 M sodium formate). Cube-shaped crystals grew to ≈0.25-mm diameter in 10–15 days. Two types of crystals were identified, belonging to space groups P4132 and P213, both with a = 189.0 Å and with two and four monomers per crystallographic asymmetric unit, respectively. A heavy-atom derivative was prepared by cocrystallizing the protein with 15 mM ethyl mercury phosphate. Single wavelength Hg diffraction data were collected at the Advanced Photon Source BioCARS beamlines 14BM-c (P213) and 14ID-b (P4132) by using Quantum 4 CCD detectors. The P4132 data were collected at the Hg L-III absorption edge. Intensities were integrated and scaled by using DENZO and SCALEPACK (20). Rmerge was 0.090 to 2.55-Å resolution and 0.131 to 2.0-Å resolution. The completeness of the data was 99.9% for both data sets.
Structure Determination.
The structure of the PBCV-1 major capsid protein, Vp54, was determined to 2.5-Å resolution by using the single-wavelength anomalous dispersion method with the ethyl mercury phosphate derivative data in space group P4132. The program SOLVE (21) found two Hg sites per monomer that were used to determine the initial single-wavelength anomalous dispersion phases and to establish the noncrystallographic symmetry (NCS) operators. Phases were improved by using density modification applied by the program RESOLVE (21), taking advantage of the twofold NCS redundancy for electron density averaging. The model was built and refined with the programs O (22) and CNS (23), respectively. NCS restraints were applied through all stages of refinement. Fifty-nine water molecules were included in the final stages of refinement. The final Rworking and Rfree factors were 27.1% and 30.7%, respectively (PDB ID code ). Root-mean-square deviations for the bond lengths and bond angles from idealized values were 0.0074 Å and 1.4°, respectively.
The P213 crystal form was solved because the diffraction data extended to 2.0-Å instead of only 2.5-Å resolution by using the P4132 structure as a search model for molecular replacement. This crystal form had a similar packing arrangement as the P4132 form, but with only pseudo-41 symmetry. The four molecules per crystallographic asymmetric unit were located with the program AMORE (24). Rigid body displacement from 41 symmetry was ≈1.5 Å. This solution was refined by using CNS, but the Rworking and Rfree factors failed to go below 32% and 35%, respectively. Further examination showed that the crystals were twinned with the twins related by a 90° rotation. The CNS program determined the twinning fraction to be 0.33. The structure was then refined as described above, assuming the twinning fraction and by using NCS restraints among the four independent monomers. The final Rworking and Rfree factors were 27.3% and 28.5% to 2.0-Å resolution, respectively, with root-mean-square deviations of 0.013 Å and 1.9°, respectively, for bond lengths and bond angles from idealized values (PDB ID code ). In neither structure were any main-chain dihedral angles in the disallowed region of the Ramachandran plot. The P213 diffraction data had been collected on an older crystal that had fewer ordered sugar moieties.
Cryo-EM Analysis.
The cryo-EM map of PBCV-1 reported by Yan et al. (1) was recalculated after phase-contrast transfer function correction of the EM images. The structure factors for the quasi-atomic model were calculated with the program CNS (23) by using the placement of the crystallographic atomic structure into each of the capsomers in the cryo-EM density representing the virus. The effect of the internal membrane and nucleic acid structures, of the minor structural proteins, and of the penton capsomers on the 12 fivefold vertices was neglected. Observed structure factors were derived by Fourier inversion of the cryo-EM map. These were scaled to the calculated structure factors by using the program Rstats (25) and requiring a temperature factor correction of B = 1,400 Å2. The resultant correlation coefficient was 0.85 and the R-factor was 0.50 to a resolution limit of 28 Å. There was no significant agreement at a resolution better than 28 Å. Coordinates representing the complete virus have been deposited with the Protein Data Bank (PDB ID code ).
Results and Discussion
Structure of the Vp54 Capsomer.
The structure of the PBCV-1 major capsid protein, Vp54, was determined by using single-wavelength anomalous dispersion and NCS electron density averaging. The protein crystallized as a trimer sitting on a crystallographic three-fold symmetry axis. The two independent monomers in the crystallographic asymmetric unit belong to two different trimers and are related to each other by a noncrystallographic twofold axis that is orthogonal to and intersects the crystallographic threefold axis. Thus, these six monomers (three in each trimer) are related to each other by 32-point group symmetry.
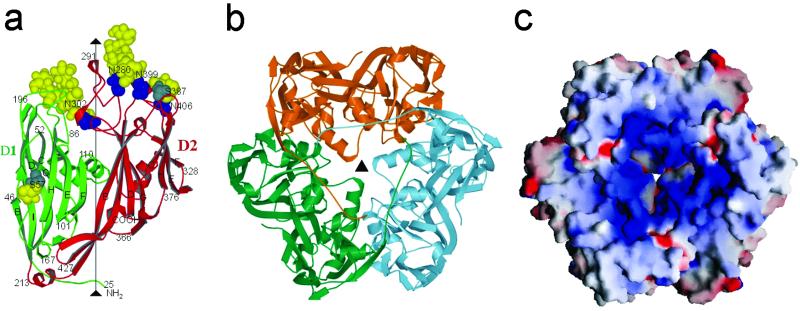
Each monomer consists of a 437-aa polypeptide chain with ≈6 kDa of carbohydrate distributed over four N-linked and two O-linked glycosylation sites (Fig. 2a). All but the first 24 amino acids could be identified in the electron density map in each of the two independent molecules. The trimers are doughnut-shaped with a block in the center of the doughnut's hole, creating a cavity on either side of the doughnut, reminiscent in shape of red blood cells (Fig. 2b). The external shape of the doughnut is triangular at one end and hexagonal at the other with a diameter of 72 Å and thickness of ≈75 Å. This shape is closely similar to that of the capsomers seen in the cryo-EM reconstruction of the mature PBCV-1 virus, as well as in iridoviruses (1). Thus, the crystallographic Vp54 trimer seems to be the biologically relevant trimer not only for PBCV-1, but apparently for all known large icosahedral viruses, including phycodnaviridae and iridiviridae. The trimer can only be dissociated into its component monomers by boiling in the presence of 0.1% SDS, but is stable at 100°C in the absence of SDS, as is also the case of the adenovirus hexone and PRD1 P3 trimers (26). The surface between monomers within the trimer shows substantial charge complementarity, which will contribute to the unusual stability of the trimeric capsomer. The stability of the capsomers may be essential for the assembly of very large icosahedral viruses.
Fig. 2.
(a) Structure of the Vp54 monomer with strategic amino acids labeled. The carbohydrate moieties (yellow) and glycosylated Asn and Ser residues (blue and gray, respectively) are shown as space-filling atoms. (b) The Vp54 trimer viewed from the inside of the virus, with each monomer in a different color. (c) Surface of the Vp54 trimer also viewed from the inside of the virus, colored according to charge distribution (positive is blue, negative is red).
The monomer's polypeptide fold consists of two consecutive “jelly-roll” domains, D1 (residues 27–212) and D2 (residues 225–437; Fig. 2a). The jelly-roll motif is a component of many virus capsid structures including single-stranded (ss) RNA plant viruses, ssRNA animal viruses (e.g., picornaviruses), ssRNA insect viruses, dsRNA plant viruses, dsRNA animal viruses (e.g., reoviruses), ssDNA bacterial viruses (e.g., microviridae), ssDNA animal viruses (e.g., parvoviruses), dsDNA bacterial viruses (e.g., PRD1), and dsDNA animal viruses (e.g., adenoviruses) (27). The jelly-roll structure is an antiparallel β-barrel (28, 29) consisting of ≈260 amino acids. The β-strands along the polypeptide are named A, B,… , I. The A-strand is frequently absent, but the remaining strands form an antiparallel ribbon with β-strands B, C, D, and E being hydrogen-bonded to I, H, G, and F, respectively. This ribbon is wound into a right-handed helix such that the sequence of β-strands that form one side of the barrel is BIDG and the other side of the barrel is CHEF (Fig. 2a; ref. 30). The loops BC (the most external), HI, DE, and FG (the most internal) form the thin, wedge-shaped end of the β-barrel that associates with five or six other barrels to form the pentameric or quasi-hexameric icosahedral vertices in simple RNA viruses, such as picornaviruses. Many viral jelly-roll domains have an α-helix A inserted between strands C and D and an α-helix B between strands E and F. The amino ends of viral capsid proteins folded into jelly-rolls are invariably internal to the virus capsid and frequently associated with the genomic nucleic acid. In contrast, the carboxy end is almost always exposed on the viral surface.
The two jelly-roll domains in Vp54 are related by a 53° rotation approximately about the central threefold axis of the trimer (Fig. 2b), giving the capsomer a pseudohexagonal symmetry. Insertions in the HI, DE, and FG loops at the wedge ends of the β-barrels separate the jelly-roll domains from each other, thus making it impossible for the barrels to approach close to the pseudo-sixfold axis, creating a central cavity in the trimer and tilting the direction of the β-strands upward roughly parallel to the central pseudo-sixfold axis. In contrast, the β-strands in jelly-roll domains of small ssRNA viruses tend to be more tangential than radial to the surface. The constriction in the center of the trimer is caused by the FG loop of domain D1. The sites of the two larger carbohydrate moieties are located in the DE and HI loops of domain D2 that form the external rim of the trimer. These insertions also contain an α-helix that is situated in the site occupied by helix B in other viral capsid proteins.
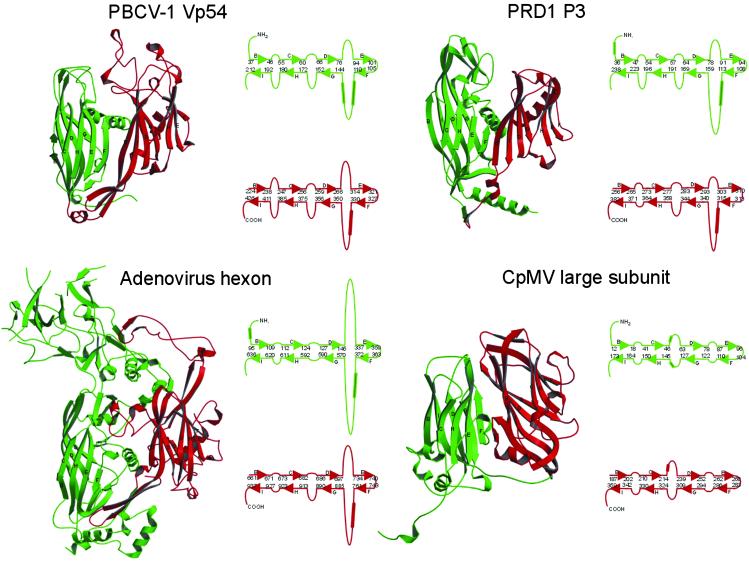
The structure of the major capsid “hexon” protein of adenovirus (Fig. 3; see ref. 31) consists of two sequential jelly-roll domains related by a 55° rotation, with large insertions in the DE and FG loops amounting to a total of 550 amino acids. Structural superposition of the jelly-roll domains of Vp54 and hexon shows that 68% and 55% of the Cα atoms can be equivalenced in domains D1 and D2, respectively. Thus, the general topology of the adenovirus hexon protein is similar to PBCV-1 Vp54, except for the very large insertions which form a “tower” on the edge of the hexon's external surface.
Fig. 3.
Comparison of virus major capsid proteins. Domains D1 and D2 are green and red, respectively. The β-sheets BIDG and CHEF are indicated. To the right of each ribbon diagram are shown diagrammatically the arrangement of β-sheets with appropriate residue numbers at the end of the β-strands. Helices are indicated by cylinders. CpMV, cowpea mosaic virus.
The major capsid protein, P3, of bacteriophage PRD1 (Fig. 3) has an obvious structural similarity to the adenovirus hexon protein (14, 17) and Vp54. However, the tower structure is much smaller in P3, making it more like Vp54 than it is like the adenovirus hexon protein (Fig. 3). Superposition of the PRD1 and PBCV-1 capsid protein structures shows that 64% and 54% of the Cα atoms can be equivalenced in domains D1 and D2, respectively. All three capsid proteins, Vp54 of PBCV-1, hexon of adenovirus, and P3 of PRD1, have major insertions in loops DE and FG, with a helix in loop FG in both domains D1 and D2. As a consequence of their common folds and pseudohexameric assembly, the capsid surface of all three viruses consists of very similar hexagonal arrays of pseudohexameric capsomers.
The organization of the capsomers also resembles that of the picornaviruses and related pseudo T = 3 virus capsids. The structural viral proteins of picornaviruses are expressed as a polyprotein VP4-VP2-VP3-VP1, which is subsequently cleaved by viral proteases into the component proteins, a process that probably directs assembly (32). The three major capsid proteins (VP1, VP2, and VP3) each have a jelly-roll structure, although no detectable sequence similarity exists between them. VP2 and VP3 assemble around the icosahedral threefold axes to form hexamers with pseudo-sixfold symmetry, whereas VP1 assembles around the fivefold axes to make pentamers. In some plant viruses such as cowpea mosaic virus, the cleavage between VP2 and VP3 does not take place, resulting in a “large” subunit of VP2-VP3 and a “small” subunit of VP1 (33). Comparison of the large protein of cowpea mosaic virus (Fig. 3) with PBCV-1 Vp54 (Fig. 3) shows that the ordering of domains around the quasi-sixfold axes is the same. Thus, the capsid proteins of large dsDNA icosahedral viruses with hosts ranging from bacteria to unicellular algae to multicellular plants and animals, and the small ssRNA icosahedral plant and animal picornaviruses are likely to have evolved from a common ancestor.
The Virus Structure.
In the PBCV-1 capsid, the trisymmetrons do not correspond to the faces of an icosahedron, but instead bend gently around the icosahedral edges, leaving gaps at the fivefold vertices that are filled by the pentasymmetrons (Fig. 1a). Within each trisymmetron the capsomers have similar orientations and form a hexagonal, close-packed lattice with plane group p3, a lattice repeat of 74 Å, and a thickness of 75 Å. Similar hexagonal arrays with almost the same dimensions are also found in the capsids of Chilo iridescent virus, African swine fever virus, adenovirus, and PRD1. The pseudohexagonal structure of the trimeric capsomers gives the lattice pseudo-p6 symmetry, creating a set of pseudo-twofold axes (Fig. 1b). Boundaries between trisymmetrons are created by exact twofold operations replacing the pseudo-twofold symmetry axes, thus turning the “northward” orientation of trimers in one trisymmetron to “southward” in the neighboring trisymmetron. The pentasymmetrons are composed of five faces, each with six trimeric capsomers, giving a total of 30 mixed “northward” and “southward” orientations. The lattice repeat in the pentasymmetrons is the same as in the trisymmetrons. Hence, the entire surface of the capsid consists of a single curved two-dimensional lattice of capsomers, except for the protein at each of the 12 fivefold vertices of the icosahedron.
The trisymmetron and pentasymmetron quasi-atomic structures were generated by fitting the trimeric crystal structure independently to each capsomer density within the icosahedral asymmetric unit, by using the programs SITUS (34) and EMfit (35) (Fig. 1c). The best fit of each independent capsomer into the cryo-EM density places the amino end of the polypeptide toward the virus interior, close to the lipid membrane, as has been found for all other virus capsid proteins that use a jelly-roll motif. In addition, the carbohydrate moieties (see below) are located on the exterior of the virus. The best fit of each capsomer is also consistent with placing the relative positions of domains D1 and D2 within one monomer in an anticlockwise direction around each icosahedral pseudo-sixfold axis when viewed from outside the virus (Fig. 2b), as is the case for adenoviruses (15) and PRD1 (17). The inward-pointing face of each capsomer is mostly positively charged and is in contact with the negatively charged outer surface of the viral membrane (Fig. 2c). Subtraction of the fitted x-ray structure density from the cryo-EM density shows five tube-shaped features on the inner surface of each pentasymmetron, but outside the viral membrane. Each feature has a volume roughly corresponding to the molecular mass of one of the minor capsid proteins Vp280 and Vp260.
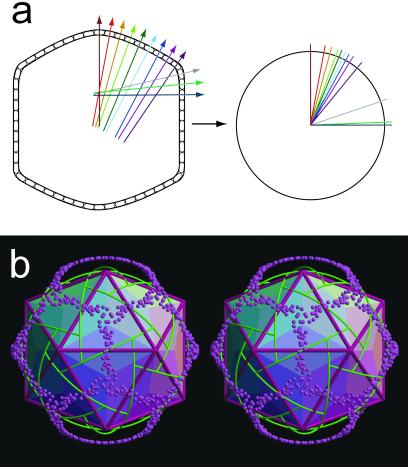
The normals of the capsomers in each trisymmetron, when shifted to a common origin, are distributed into three great circles linking the icosahedral threefold axes (Fig. 4). A uniform distribution of normals would have occurred had the array of capsomers been stretched over the surface of a sphere. In contrast, the great circles observed here show that the trisymmetron is divided into three regions related by icosahedral threefold symmetry, each of which is gradually bent about a single axis as is required to prevent stretching of the hexagonal lattice. It is probable that this subdivision of the surface is necessary for the assembly of very large icosahedral viruses.
Fig. 4.
(a Left) A diagram representing a cut through the virion showing the surface Vp54 capsomers. The direction and position of the capsomers' threefold axes (“normals”) are shown as colored lines through each capsomer. (Right) The normals are shown shifted to a common origin. The colors of the normals in the left-hand figure correspond to the colors of the transposed normals in the right-hand figure. (b) Stereo diagram showing the normals (directions of the threefold axes) for each capsomer (purple spheres) when erected from the origin of the icosahedron as shown diagrammatically above. Boundaries between trisymmetrons and pentasymmetrons are shown in green. Icosahedral faces are shaded according to regions in which the direction of the capsomers' threefold axes lie on a common great circle.
The Glycosylation Sites.
Two structures of Vp54, in space groups P4132 and P213, were determined (see Materials and Methods). These differed only in the amount of ordered carbohydrate density. The anomalously high apparent molecular weight of Vp54 suggests that it contained about 30 sugar moieties of which about 20 were ordered in the electron density. GlcNAc, which usually modifies Asn residues, fitted the carbohydrate electron density well at residues Asn-280, Asn-302, and Asn-399. There were approximately six- and seven-branched chain sugar moieties at Asn-280 and Asn-302 (Fig. 2a), respectively, as well as three and two sugar residues at Asn-399 and Asn-406, respectively, but accurate building of the polysaccharide structures was not readily possible in the density. In addition, two O-linked sugars were found at Ser-57 and Ser-387.
All of the N-linked carbohydrate linkages in Vp54 at residues 280, 302, 399, and 406, were located in a C-terminal cyanogen bromide cleavage fragment (residues 202–437), corresponding essentially to domain D2. Previous experiments (36) designed to detect Asn-linked oligosaccharides attached to the canonical NX(T/S) sequon were negative and led to the prediction that Vp54 contained only O-linked oligosaccharides (37), all within the carboxyl-terminal cyanogen bromide fragment. However, Asn-302, Asn-399, and Asn-406 occur in the amino acid sequence motif (A/G)NTXT, and Asn-280 occurs in an ANIPG sequence. None of these Asn residues occur in the NX(S/T) sequon commonly recognized by endoplasmic reticulum- and Golgi-located cellular enzymes involved in glycosylation, explaining why the previously reported (37) tests for N-glycosylation were negative.
Glycoproteins are important structural components of many viruses (38), which are often required for guiding protein folding, protecting the virus from proteolysis, and providing a site for the specific recognition of a host. Typically, viral proteins are glycosylated by host-encoded glycosyltransferases located in the endoplasmic reticulum and Golgi (38). The glycoproteins are then transported to a host membrane where they assemble with the nucleocapsid and the virus becomes infectious by budding through the membrane as it is released from the cell. However, glycosylation of PBCV-1 Vp54 differs from this paradigm in that infectious particles accumulate inside the cell before virus release (19) and because compounds that inhibit endoplasmic reticulum-Golgi glycosylation do not affect PBCV-1 replication or the molecular weight of Vp54 (37). These differences are explained by the fact that the PBCV-1 genome encodes seven putative glycosyltransferases, at least one of which is required for Vp54 glycosylation (36). The present finding that Vp54 has Asn-linked oligosaccharides at nonstandard sequon sites is consistent with the conclusion that viral-encoded enzymes that do not normally exist in eukaryotic cells are involved in the glycosylation process. The ability of the virus to provide its own glycosylation machinery may be an advantage because of the requirement to provide stabilizing carbohydrate moieties independent of the host cell glycosylation processes. Thus, it is significant that a minimally glycosylated strain of PBCV is less stable (J.L.V.E., unpublished data).
Acknowledgments
We thank Chuan Xiao and Robert Ashmore for help in the computing of the cryo-EM maps. We also thank the personnel at the BioCARS sector of the Advanced Photon Source synchrotron for their help and encouragement. We thank Cheryl Towell and Sharon Wilder for help in preparation of the manuscript. The work was supported by a National Institutes of Health grants (to M.G.R., T.S.B., and J.L.V.E.).
Abbreviations
cryo-EM, cryoelectron microscopy
dsDNA, double-stranded DNA
ssDNA, single-stranded DNA
PBCV-1, Paramecium bursaria Chlorella virus type 1
NCS, noncrystallographic symmetry
PDB, Protein Data Bank
Data deposition: The atomic coordinates of PBCV-1 capsomer structures and whole virus structure have been deposited with the Protein Data Bank (PDB ID codes , , and , respectively).
References
- 1.Yan X., Olson, N. H., Van Etten, J. L., Bergoin, M., Rossmann, M. G. & Baker, T. S. (2000) Nat. Struct. Biol. 7, 101-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Etten J. L., Lane, L. C. & Meints, R. H. (1991) Microbiol. Rev. 55, 586-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tidona C. A., Schnitzler, P., Kehm, R. & Darai, G. (1998) Virus Genes 16, 59-66. [DOI] [PubMed] [Google Scholar]
- 4.Dixon L. K., Rock, D. & Vinuela, E. (1995) in Virus Taxonomy, eds. Murphy, F. A., Fauquet, C. M., Bishop, D. H. L., Ghabrial, S. A., Jarvis, A. W., Martelli, G. P., Mayo, M. A. & Summers, M. D. (Springer, New York), pp. 92–94.
- 5.Meints R. H., Lee, K., Burbank, D. E. & Van Etten, J. L. (1984) Virology 138, 341-346. [DOI] [PubMed] [Google Scholar]
- 6.Meints R. H., Lee, K. & Van Etten, J. L. (1986) Virology 154, 240-246. [DOI] [PubMed] [Google Scholar]
- 7.Bamford D. H., Caldentey, J. & Bamford, J. K. H. (1995) Adv. Virus Res. 45, 281-319. [DOI] [PubMed] [Google Scholar]
- 8.Rohozinski J., Girton, L. E. & Van Etten, J. L. (1989) Virology 168, 363-369. [DOI] [PubMed] [Google Scholar]
- 9.Van Etten J. L. & Meints, R. H. (1999) Annu. Rev. Microbiol. 53, 447-494. [DOI] [PubMed] [Google Scholar]
- 10.Skrdla M. P., Burbank, D. E., Xia, Y., Meints, R. H. & Van Etten, J. L. (1984) Virology 135, 308-315. [DOI] [PubMed] [Google Scholar]
- 11.Wrigley N. G. (1969) J. Gen. Virol. 5, 123-134. [DOI] [PubMed] [Google Scholar]
- 12.Wrigley N. G. (1970) J. Gen. Virol. 6, 169-173. [DOI] [PubMed] [Google Scholar]
- 13.Caspar D. L. D. & Klug, A. (1962) Cold Spring Harbor Symp. Quant. Biol. 27, 1-24. [DOI] [PubMed] [Google Scholar]
- 14.Benson S. D., Bamford, J. K. H., Bamford, D. H. & Burnett, R. M. (1999) Cell 98, 825-833. [DOI] [PubMed] [Google Scholar]
- 15.Stewart P. L., Fuller, S. D. & Burnett, R. M. (1993) EMBO J. 12, 2589-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butcher S. J., Bamford, D. H. & Fuller, S. D. (1995) EMBO J. 14, 6078-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.San Martin C., Burnett, R. M., de Haas, F., Heinkel, R., Rutten, T., Fuller, S. D., Butcher, S. J. & Bamford, D. H. (2001) Structure (London) 9, 917-930. [DOI] [PubMed] [Google Scholar]
- 18.Rossmann M. G., Arnold, E., Erickson, J. W., Frankenberger, E. A., Griffith, J. P., Hecht, H. J., Johnson, J. E., Kamer, G., Luo, M., Mosser, A. G., et al. (1985) Nature 317, 145-153. [DOI] [PubMed] [Google Scholar]
- 19.Van Etten J. L., Burbank, D. E., Xia, Y. & Meints, R. H. (1983) Virology 126, 117-125. [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 21.Terwilliger T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 23.Brünger A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 24.Navaza J. (1994) Acta Crystallogr. A 50, 157-163. [Google Scholar]
- 25.Collaborative Computational Project Number 4 (1994) Acta Crystallogr. D 50, 760-763.15299374 [Google Scholar]
- 26.Benson S. D., Bamford, J. K. H., Bamford, D. H. & Burnett, R. M. (2002) Acta Crystallogr. D 58, 39-59. [DOI] [PubMed] [Google Scholar]
- 27.Rossmann M. G. & Johnson, J. E. (1989) Annu. Rev. Biochem. 58, 533-573. [DOI] [PubMed] [Google Scholar]
- 28.Harrison S. C., Olson, A. J., Schutt, C. E., Winkler, F. K. & Bricogne, G. (1978) Nature 276, 368-373. [DOI] [PubMed] [Google Scholar]
- 29.Abad-Zapatero C., Abdel-Meguid, S. S., Johnson, J. E., Leslie, A. G. W., Rayment, I., Rossmann, M. G., Suck, D. & Tsukihara, T. (1980) Nature 286, 33-39. [DOI] [PubMed] [Google Scholar]
- 30.Branden C. & Tooze, J., (1991) Introduction to Protein Structure (Garland, New York).
- 31.Roberts M. M., White, J. L., Grütter, M. G. & Burnett, R. M. (1986) Science 232, 1148-1151. [DOI] [PubMed] [Google Scholar]
- 32.Arnold E., Luo, M., Vriend, G., Rossmann, M. G., Palmenberg, A. C., Parks, G. D., Nicklin, M. J. H. & Wimmer, E. (1987) Proc. Natl. Acad. Sci. USA 84, 21-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin T., Chen, Z., Usha, R., Stauffacher, C. V., Dai, J.-B., Schmidt, T. & Johnson, J. E. (1999) Virology 265, 20-34. [DOI] [PubMed] [Google Scholar]
- 34.Wriggers W., Milligan, R. A. & McCammon, J. A. (1999) J. Struct. Biol. 125, 185-189. [DOI] [PubMed] [Google Scholar]
- 35.Rossmann M. G., Bernal, R. & Pletnev, S. V. (2001) J. Struct. Biol. 136, 190-200. [DOI] [PubMed] [Google Scholar]
- 36.Graves M. V., Bernadt, C. T., Cerny, R. & Van Etten, J. L. (2001) Virology 285, 332-345. [DOI] [PubMed] [Google Scholar]
- 37.Que Q., Li, Y., Wang, I.-N., Lane, L. C., Chaney, W. G. & Van Etten, J. L. (1994) Virology 203, 320-327. [DOI] [PubMed] [Google Scholar]
- 38.Knipe D. M. (1996) in Fields Virology, eds. Fields, B. N., Knipe, D. M. & Howley, P. M. (Lippincott, Philadelphia), pp. 273–299.
- 39.Kraulis P. (1991) J. Appl. Crystallogr. 24, 946-950. [Google Scholar]
- 40.Merritt E. A. & Bacon, D. J. (1997) Methods Enzymol. 277, 505-524. [DOI] [PubMed] [Google Scholar]
- 41.Nicholls A., Bharadwaj, R. & Honig, B. (1993) Biophys. J. 64, 166-170. [Google Scholar]