Abstract
Subcellular organelle dynamics are strongly influenced by interactions with cytoskeletal filaments and their associated motor proteins, and lead to complex multiexponential relaxations that occur over a wide range of spatial and temporal scales. Here we report spatio-temporal measurements of the fluctuations of the mitochondrial reticulum in osteosarcoma cells by using Fourier imaging correlation spectroscopy, over time and distance scales of 10−2 to 103 s and 0.5–2.5 μm. We show that the method allows a more complete description of mitochondrial dynamics, through the time- and length-scale-dependent collective diffusion coefficient D(k,τ), than available by other means. Addition of either nocodazole to disrupt microtubules or cytochalasin D to disassemble microfilaments simplifies the intermediate scattering function. When both drugs are used, the reticulum morphology of mitochondria is retained even though the cytoskeletal elements have been de-polymerized. The dynamics of the organelle are then primarily diffusive and can be modeled as a collection of friction points interconnected by elastic springs. This study quantitatively characterizes organelle dynamics in terms of collective cytoskeletal interactions in living cells.
The movement and redistribution of mitochondria is an essential part of normal cell function, important to mitochondrial inheritance in the cell cycle (1, 2), transient clustering and morphogenesis in response to changing physiological conditions (3, 4), and coordinated mitochondrial behavior during key events in cell development (5, 2). Such gross, long-range movements occur over the course of minutes to days (102 to 105 s), yet the molecular basis of mitochondrial redistribution must rely on protein conformation changes that operate on relatively fast (subsecond) time scales. In most eukaryotic cells, different organelle types display unique “patterns” of motion; some fluctuate continuously over a broad range of time scales (10−3 to 102 s), whereas others exhibit so-called “saltatory motion”; rapid bursts of directed movements interrupted by pauses of variable duration (6). Mitochondria are a known cargo of microfilament (actin/myosin-based) and microtubule (tubulin/dynein/kinesin-based) cytoskeletal tracks (5, 7). They exhibit complex motility patterns that likely reflect the molecular details of a complicated regulatory system designed to rearrange mitochondria within the cytoplasmic compartment. The importance of cytoskeletal proteins as modulators of cell morphology and signaling has inspired speculation that mitochondrial dynamics and function, the aerobic production of ATP and the regulation of intracellular calcium levels, are closely related (6, 8).
An important aspect of mitochondrial dynamics is its morphology. Microscopy studies of various types now suggest that the organelle's structure can appear in one of two very different morphological states (9) at different points in the cell cycle (10). At “inter-phase” stages, mitochondria often appear as a continuously branched reticulum of connected tube-like filaments, with an approximate tube diameter of ≈0.5 μm (11). During cell division, mitochondria are partially fragmented into hundreds of ellipsoid shaped vesicles (12), thus allowing the partitioning of mitochondrial fragments (and mtDNA) into the two daughter cells. It is possible to observe intermediate morphologies between these two states. The effect of morphology on mitochondrial dynamics is significant because even in the absence of complicating factors such as crowding or the directional forces imposed on the organelle by motor proteins, the dynamics of the reticulum morphology should reflect its chain-like connectivity. In this study, we focus on the dynamics of the reticulum state.
As noted above, mitochondrial motion is complicated by the activity of motor proteins working in conjunction with cytoskeletal tracks. As a whole, the system can be viewed as a multicomponent suspension of network structures that interact through associated cytoskeletal proteins, some of which are force-generating motor proteins (8). Microtubules, microfilaments, and intermediate filaments form a gel-like entanglement network of rod-like polymers (13) that extends throughout the perinuclear region of the cytosol compartment and has a mesh size of ≈50 nm (14). Microtubules have a persistence length of ≈6 mm, whereas actin microfilaments have a persistence length of ≈15 μm (13). An unknown combination of thermal forces and those caused by motor protein cross-link junctions mediate the interactions between these filaments and the mitochondrial reticulum. Particle tracking measurements by Elbaum and coworkers (15) suggest that the effective viscosity experienced by micrometer-scale mitochondrial filaments is large enough, η0 > 0.25 poise, to dramatically slow diffusion on long time scales. Kinesin and cytoplasmic dynein are the motor proteins believed to carry mitochondria along microtubules, whereas myosin is the motor protein associated with actin-based microfilaments. Although involvement of microtubules in mitochondrial motion is well established, the roles played by microfilaments and intermediate filaments are less clear (5, 8). It is generally believed that the actin cytoskeleton plays no role in the mitochondrial transport of higher eukaryotes (16).
Although analyses of mitochondrial motility patterns (recorded, for example, by digital video microscopy) can in principle reveal complex heterogeneities in behavior, the complexity of information contained in direct microscopic observations makes straightforward interpretations difficult (17). Erratic motions occur over a spectrum of spatial and temporal scales, often encompassing several decades in time. The problem is greatly simplified by framing it in terms of the dynamics of a complex fluid. The dynamical “modes” of the system are studied through measurements of the distribution of length- and time-dependent fluctuations, which are quantitatively characterized by using space–time correlation functions (18). Space–time correlation functions describe the average regression of spontaneous microscopic density fluctuations in time (measured as a function of spatial scale). They provide the link (through theoretical models) between the microscopic dynamics and macroscopic parameters (such as diffusion coefficients) that are usually measured in the laboratory. For simple dynamical systems in which only one process is operative, the space–time correlation functions reflect a single diffusion constant for all time- and length-scales. For complex systems, such as the mitochondrial reticulum, multiple processes give rise to the overall dynamics, which can be described in terms of time- and length-scale-dependent diffusion coefficients. Furthermore, correlation functions measured over several decades contain scaling information that may reveal situations where the dynamics are not well described by a diffusion equation (e.g., sub- or superdiffusive behavior). In the present analysis we focus on the effects of time and length scales on the diffusion coefficient.
Materials and Methods
Systematic and Quantitative Characterization of Mitochondrial Dynamics by Fourier Imaging Correlation Spectroscopy (FICS).
As an initial step toward understanding the collective dynamics of the mitochondrial-specific cytoskeleton, we conducted studies of mitochondrial dynamics in live osteosarcoma cells (143B) under various intracellular conditions, where key cytoskeletal components were selectively removed. Our measurements span the full range of time (10−2 to 103 s) and distance (0.5–2.5 μm) scales necessary to characterize the dynamics of this complex system. We apply FICS, a sensitive spatially selective means to measure mitochondrial density fluctuations (17, 19, 20). In FICS experiments, modulated fluorescence signals are detected from the intersection of an excitation fringe pattern (an optically generated holographic grating) with a time-evolving configuration of fluorescently labeled mitochondrial filaments, denoted by the microscopic density C(r,t) (see Fig. 1A).
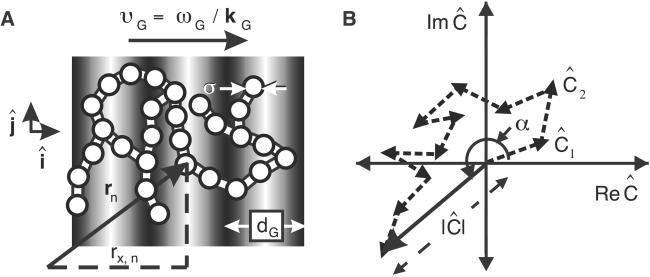
Fig 1.
(A) Schematic of the FICS experimental geometry. Fluorescently labeled filaments are represented as N interconnected disks and the excitation grating as gray-scale bars. The filament cross-sectional diameter is ≈0.5 μm, whereas the grating fringe spacing, dG, is adjusted between ≈0.56 and 2.6 μm. Signal fluctuations occur when regions of the reticulum separated by the distance dG move relative to one another. (B) At any instant, a static particle configuration is uniquely described by a sum of vectors in the complex plane whose superposition is Ĉ = (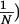 ½ ∑
½ ∑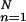 An exp[ikG⋅rn(t)] ≡ |Ĉ|exp[iα] with amplitude |Ĉ| and phase α.
An exp[ikG⋅rn(t)] ≡ |Ĉ|exp[iα] with amplitude |Ĉ| and phase α.
Fluorescence labeling was achieved by producing a stable cell line that expresses a red fluorescent homolog of the green fluorescent protein (DsRed) inside the mitochondrial matrix space (21). Cells were grown in HG-DMEM + 10% FCS, and were maintained in humidified 10% CO2 atmosphere at 37°C. To inhibit cytoskeletal components, cells were incubated for 2–4 h with 10 μM Nocodazole (Sigma) and/or 0.2 μg/ml Cytochalasin D (Sigma). To visualize the mitochondrial morphology with microfilaments or microtubules, cells were fixed for 30 min in 4% paraformaldehyde at room temperature. After permeabilizing with acetone for 20 min at −20°C, primary and secondary monoclonal antibodies conjugated to Pacific Blue (Molecular Probes) were used to label tubulin. Actin in similarly treated cells was labeled by using Alexa488 phalloidin (Molecular Probes).
Determination of a Time-Dependent Spatial Fourier Component of the Labeled Mitochondrial Density.
The experimental observable is the time-dependent spatial Fourier transform of the filament density, defined at the wave-vector specified by the optical grating. The basic principle is illustrated in Fig. 1. Mitochondrial filaments are uniformly labeled by the matrix-targeted DsRed, indicated in Fig. 1A by interconnected white disks of diameter σ. The N disks are illuminated by an excitation grating having a fringe spacing dG (adjustable between 0.25 and 4 μm).
Two crossed laser beams with parallel polarizations create the excitation grating. The intensity of the electric field in the grating changes along each fringe, whereas its polarization is fixed in the ĵ direction (22). The grating is characterized by a wave vector, kG = (2π/dG)î, directed along the x axis. The time-dependent microscopic density is C(r, t) = Σ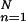 Anδ[r − rn(t)], where rn is the center-of-mass position of the nth particle, and An is proportional to that particle's excitation/emission efficiency. The phase of the grating is modulated at the angular frequency ωG (=50,000 rad/s) so that its position is swept across the sample at a velocity, υG = ωG/kG, much greater than the average speed of filament motion. The resulting fluorescence intensity has the form (19, 20)
Anδ[r − rn(t)], where rn is the center-of-mass position of the nth particle, and An is proportional to that particle's excitation/emission efficiency. The phase of the grating is modulated at the angular frequency ωG (=50,000 rad/s) so that its position is swept across the sample at a velocity, υG = ωG/kG, much greater than the average speed of filament motion. The resulting fluorescence intensity has the form (19, 20)
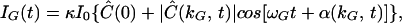 |
where κ is a proportionality factor accounting for the absorption cross-section, quantum yield, and light collection efficiency of the experimental setup, and I0 is a constant intensity level. According to Eq. 1, the signal consists of two parts; a stationary (or dc) component representing the zero-kG background fluorescence level, and a modulated (or ac) component whose amplitude, |Ĉ|, and phase, α, depend on the particle configuration
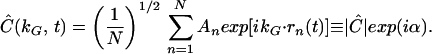 |
The amplitude of the signal modulation reflects the extent to which the structure of the N-body system contains spatial periodicity matching the spatial frequency of the excitation grating (kG = |kG| = 2π/dG). The phase of the modulation reflects the relative position (or phase) of the spatial periodicity at wave number kG of the filament configuration with respect to the phase of the excitation grating. Because filament motion occurs on a much slower time scale than the inverse modulation frequency, a lock-in detection method (20) can be used to demodulate the signal into slowly varying complex components, ReĈ = |Ĉ|cosα and ImĈ = |Ĉ|sinα. As shown in Fig. 1B, at any instant in time the experimentally detected Fourier component, Ĉ, is a vector sum of N single-particle terms. Each particle contributes an amplitude, An, and a phase, αn = kG⋅rn = kGrx,n, to the detected value of Ĉ. Because kG points in the direction of the x axis, only the x component of rn contributes to the phase.
The vector Ĉ fluctuates because of collective fluctuations of the filaments. Analysis of the time-dependent trajectory of Ĉ allows quantitative characterization of the motion. The trajectory of Ĉ can be used to construct any of its time-correlation functions. Here we focus on temporal and spatial two-point correlation functions (23),
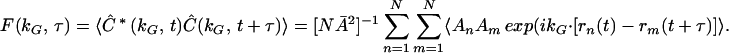 |
In Eq. 3, the angle brackets indicate an average over all starting times, t, and τ is the time interval between which successive membrane configurations are correlated. The prefactor term NĀ2 is the average fluorescence efficiency. The intermediate scattering function, F(kG,τ), is a fundamental quantity for the quantitative descriptions of liquid dynamics (24). The fluid structure is characterized by the zero-time limit, F(kG,τ = 0) = S(kG). Time- and wave-vector-dependent diffusion coefficients D(kG,τ) can be defined (25) in terms of the normalized correlation functions by D(kG,τ) = −(1/kG2) d{ln[f(kG,τ)]}/dτ, where f(kG,τ) = F(kG,τ)/S(kG). The diffusion coefficient for spherical particles in a sample at infinite dilution is given by the Stokes–Einstein formula D0 = kBT/6πη0a, where η0 is the viscosity of the solvent and a is the particle radius. For a simple fluid of non-interacting Brownian particles, f(kG,τ) = exp[−τ/τ0], where τ0 = [D0kG2]−1 is the time required for an unhindered particle to diffuse (with diffusion coefficient D0) a distance 2π kG−1. For the mitochondrial system, interactions between membrane sites and their time-evolving local environments (which depend on the structural and dynamical properties of the cytoskeleton), are expected to lead to deviations of Eq. 3 from diffusive behavior. Regions of membrane sites will collectively experience varying effective mobilities [characterized by D(kG,τ)] that reflect the relevant spatial and temporal scales involved in cytoskeletal structural relaxation. Thus, we expect the deviations of f(kG,τ) from diffusive behavior to sensitively indicate the degree of complexity of the multicomponent mitochondrial dynamics.
Phase-Stabilized FICS Measurements Are Sensitive to Nanometer Scale Mitochondrial Displacements.
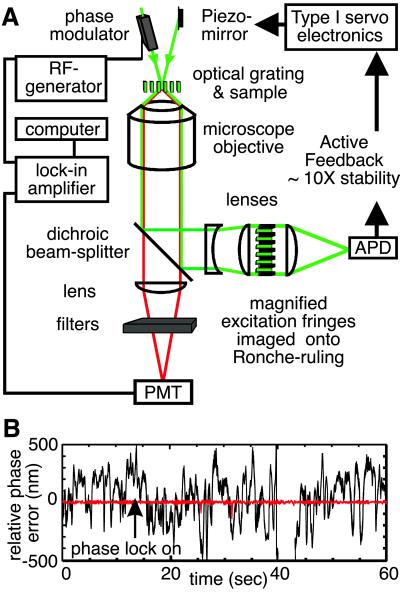
In Fig. 2 we show a schematic diagram of the FICS apparatus. The instrumentation is described in detail by Grassman et al. (20), but some modifications were made for the experiments presented below. The precision of our fluctuation measurements is limited by mechanical instabilities of the interferometer, which introduces noise in the interference pattern at the sample. To reduce mechanical noise over the time scales of interest (10−2 to 103 s), we use an active feedback system, locking the phase of the optical grating in the sample (relative to that of the reference waveform) to within ≈1/100 of the fringe spacing. As shown in Fig. 2A, the excitation beams are separated from the fluorescence signal by a dichroic beam-splitter, and the excitation pattern is magnified and imaged onto a Ronche ruling. As the optical grating is swept across the ruling, the spatially modulated signal is converted into a temporal signal that is measured by an amplified photodiode. A feedback signal generated by comparing the phase of the modulated excitation intensity (ϕE) to that of the reference waveform (ϕR) is used (by means of a Piezo-mounted optical mirror) to minimize the relative phase error (RPE = ϕE − ϕR). In Fig. 2B, we show typical time courses of the RPE for dG = 1.0 μm in the situations where the feedback signal is absent (black curve), and engaged (red curve). In passive operation, the RPE fluctuates on a range of time scales 10−2 to 101 s with root-mean-square variation, 〈RPE2〉1/2 ≈ 288 nm. When the feedback circuit is engaged, the RPE continues to fluctuate on the same time scales, but in this case 〈RPE2〉1/2 ≈ 3.9 nm. Thus, (3.9 nm/dG)⋅2π defines the precision to which our phase measurements are sensitive.
Fig 2.
(A) Schematic of the actively stabilized FICS apparatus. An interference fringe pattern is produced in the sample plane of a fluorescence microscope by two coincident laser beams. One beam passes through an electro-optics phase modulator (Conoptics, Danbury, CT). A frequency generator (Keithley) is used to modulate the phase of the excitation grating from 0 to 2π at the angular frequency ωG (=50,000 rad/s). Both the modulated fluorescence and transmitted excitation signal are collected with a fused-silica oil-immersion objective (Leica, Plan Fluotar, ×100, numerical aperture = 1.3). A dichroic beam-splitter (Chroma Technology, Brattleboro, VT) reflects the excitation (532 nm) and transmits the emission (583 nm; transmission efficiency 93%). The emission is passed through a long-pass interference filter (Omega, cutoff wavelength 570 nm, transmission efficiency 80% at 583 nm) and an excitation barrier filter. The filtered signal is imaged onto a thermoelectrically cooled photomultiplier tube (PMT, Hamamatsu, R1527) operating in current mode. The PMT output is detected by using a digital dual-phase lock-in amplifier (Stanford Research Systems, SR830) that is referenced to the RF-generator used to drive the phase-modulator. A computer, which controls an analog-to-digital data acquisition board (National Instruments), records separately the average background fluorescence intensity, κI0Ĉ(0), the complex components of the demodulated signal, ReĈ and ImĈ, and the RPE. The image of the excitation grating is passed through a Ronche-ruling, and tightly focused onto a small-area avalanche photo-diode (APD, Pacific Silicon Sensor, Westlake Village, CA). The APD output is measured by using a phase-sensitive detector referenced to the RF-generator. A type I servo is used to generate a feedback signal (delivered to a Piezo-mounted mirror) to minimize the RPE between the excitation grating and the reference waveform. (B) Typical time course for the RPE with feedback engaged (red, rms phase-error = 3.9 nm) or disengaged (black, rms phase-error = 288 nm).
Results and Discussion
Isolation of Mitochondrial-Specific Cytoskeletal Dynamics.
In Fig. 3, we present FICS data from live cells with DsRed labeling the mitochondrial matrix space. For all of our measurements, the laser excitation intensity at the sample is ≈13 nW μm−2. Eq. 3 is used to determine the intermediate scattering function from measurements of Ĉ(kG,t). Each curve represents an average of ≈10 individual measurements. For each experiment, ≈8.2 × 106 data points were collected at an acquisition frequency of ≈4.1 kHz. The minor influence of photo-degradation was corrected by normalizing the signal by the average background fluorescence. Additional details of our data analysis protocol are described elsewhere (17). Plots of f(kG,τ) versus D0kG2τ = τ/τ0 (with D0 = 1.8 × 10−4 μm2⋅s−1, see below) characterize the time-evolution of the collective membrane-cytoskeletal interactions over various spatial scales. This is a convenient way to plot the results because the normalized kG- and τ-dependent diffusion coefficient D(kG,τ)/D0 is equal to the inverse time constant of the correlation function. A diffusive system with D(kG,τ)/D0 independent of kG and τ exhibits exponential decays that fall on a single curve for all values of kG. Four different situations are examined; control physiological conditions (Fig. 3A), incubation of cells with the microtubule-inhibiting reagent Nocodazole (Fig. 3B), treatment with the microfilament-inhibiting reagent Cytochalasin D (Fig. 3C), and simultaneous treatment with Nocodazole and Cytochalasin D. The insets show fluorescence micrographs of corresponding specimens that have been fixed and treated with antibody or phalloidin labels (green). The insets emphasize the relationship between the mitochondrial reticulum (red) and the microtubules and/or microfilaments. We report results only for cells exhibiting the reticulum morphology.
Fig 3.
Intermediate scattering functions, f(kG,τ), for mitochondria in live osteosarcoma cells under different cytoskeletal conditions. The time-axis is plotted in reduced time units, τ0 = [kG2D0]−1 where D0 = 1.8 × 10−4 μm2⋅s−1. The Insets show typical specimens on which FICS experiments were conducted. (A) Control physiological conditions. (B) Depolymerization of microtubules using Nocodazole. (C) Depolymerization of microfilaments using Cytochalasin D. (D) Depolymerization of both microtubules and microfilaments. In all panels, the dotted curves show the expected decay of a purely diffusive system with f(kG,τ) = exp[−kG2D0τ]. The symbols (× ≡ kG = 7.8 μm−1, ⋄ ≡ kG = 3.9 μm−1) indicate the Rouse fit with N = 100, b = 5.4 μm, and Rg = b(N/6)1/2 = 22 μm.
Mitochondrial Dynamics Exhibit a Characteristic Nondiffusive Pattern That Simplifies to a Diffusive One Upon Depletion of Microtubules and Microfilaments.
The control data presented in Fig. 3A show that mitochondrial dynamics in native cells sensitively depend on spatial and temporal scales. In Fig. 3A, the dotted curve is a single exponential decay that represents the expected behavior for a freely diffusing system with D0 = 1.8 × 10−4 μm2⋅s−1. This value for the diffusion coefficient corresponds to the best fit of our data to the equation f(kG,τ) = exp[−kG2D0τ] when cells are incubated with Nocadazole and Cytochalasin D, so that both microtubules and microfilaments are depleted (Fig. 3D). Under the latter circumstances, the data fall approximately onto one universal curve for all values of kG, strongly suggesting diffusive behavior. Relative to this (diffusive) limit, the control system exhibits enhanced short- and long-time mobility that increases with increasing spatial scale. This observation supports the picture of a highly cooperative mechanism for the structural relaxation of the local environments surrounding mitochondrial filaments, which likely involves motor protein-assisted active transport.
Although the decays in Fig. 3A are generally multiexponential, they can be described from a phenomenological point of view as reporting on a system undergoing kG-dependent kinetic transitions from short- to long-time regimes. Densely crowded systems of Brownian (colloidal) particles are known to exhibit similar behavior where the transitions from short- to long-time dynamics are associated with the details of particle-particle interactions, usually mediated by long range hydrodynamic forces (23). As noted above, the complex interactions that dictate the dynamics of the mitochondrion have been thus far left uncharacterized. In Table 1, we list the short-time reduced diffusion constants, DS/D0 (established before ≈1 s) and the long-time reduced diffusion constants, DL/D0 (established ≈100 s) for each measurement of the four situations examined. These limiting values represent the diffusion coefficients before and after they are modified by the intermediate time scale relaxations. The curves appear to follow a distinct pattern, crossing over from one to another in their respective short- and long-time regimes. The curve with kG = 7.8 μm−1 is coincident with kG = 11.2 μm−1 at short time, but crosses over to the kG = 6.3 μm−1 curve on the time scale ≈0.4 τ0. Similarly, the curve with kG = 3.9 μm−1 has the same short-time diffusion coefficient as the curve with kG = 6.3 μm−1, but crosses over to the kG = 2.6 μm−1 curve on the time scale ≈0.2 τ0.
Table 1.
Short time (τ ≤ 1 s) and long time (τ ≅ 100 s) reduced diffusion constants calculated from the decays of f(kG, τ) = exp[−k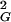 D(kG, τ)⋅τ] shown in Fig. 3
D(kG, τ)⋅τ] shown in Fig. 3
| kG, μm−1
|
dG, μm
|
τ0, sec
|
Control cells (Fig. 3A) | Nocodazole (Fig. 3B) | Cytochalasin (Fig. 3C) | Both drugs (Fig. 3D) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| DS/D0 | DL/D0 | DS/D0 | DL/D0 | DS/D0 | DL/D0 | DS/D0 | DL/D0 | |||
| 11.2 | 0.56 | 44.3 | 2.8 | 0.6 | 4.2 | 0.8 | 1.7 | 0.3 | 1.9 | 0.4 |
| 7.8 | 0.8 | 91.3 | 2.8 | 1.7 | 4.2 | 0.7 | 2.5 | 0.8 | 1.9 | 0.7 |
| 6.3 | 1.0 | 140 | 5.6 | 1.7 | 3.3 | 0.6 | 3.0 | 1.1 | 1.9 | 0.7 |
| 3.9 | 1.6 | 365 | 5.6 | 7.2 | 5.0 | 2.0 | 2.5 | 1.1 | 3.9 | 1.0 |
| 2.6 | 2.4 | 822 | 9.4 | 7.2 | 7.2 | 2.5 | 4.4 | 1.9 | 3.3 | 1.0 |
The associated relaxation time constants (in reduced units) are τX/τ0 = D0/DX where X = S or L, τ0 = [k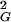 D0]−1, and D0 = 1.8 × 10−4 μm2⋅s−1. Error bars are ±0.3. The relationship between wave number and fringe spacing is kG = 2π/dG.
D0]−1, and D0 = 1.8 × 10−4 μm2⋅s−1. Error bars are ±0.3. The relationship between wave number and fringe spacing is kG = 2π/dG.
The spatial and temporal scales associated with the kinetic transitions observed in the control cells are consistent with a transport mechanism involving an ATP-dependent “hopping” process, previously seen in similarly treated osteosarcoma cells labeled with mitochondrial-specific dyes (17). In those studies, local mitochondrial displacements occurred with an average hopping distance ≈0.8 μm, corresponding to kG = 7.8 μm−1. Successive displacements of this magnitude became de-correlated over the same time scales (tens of seconds) as found for the control data here. The large dispersion of mobilities observed in Fig. 3A suggests that additional interactions are likely to be operative over the larger range of spatial scales studied here.
Depletion of Microtubules or Microfilaments Alone Leads to Partial, but Unique, Simplification of the Mitochondrial Dynamics.
In Fig. 3B, we show the effects of removing the microtubule components of the cytoskeleton by incubation of cells with Nocodazole. Although all of the curves still exhibit enhanced mobilities relative to the diffusive limit, their kG-dependences no longer follow a monotonic pattern. The short- and long-time mobilities are ordered (from smallest to largest) according to kG = 6.3, 7.8, 11.2, 3.9, and 2.6 μm−1, possibly indicating a significant structural rearrangement of the remaining intact cytoskeletal components. In comparison to the control case, the removal of microtubules has decreased the mobility on the large length scales (kG = 3.9, 2.6 μm−1). On the short length scales (e.g., kG = 11.2 μm−1) the mobility is slightly enhanced, whereas on the long length scales (e.g., kG = 2.6 μm−1) the long-time mobility is decreased by a factor of ≈3. Interestingly, for kG = 7.8 μm−1, a “cross-over” still occurs from short-time (coincident with the higher mobility curve with kG = 11.2 μm−1) to long-time (coincident with the lower mobility curve with kG = 6.3 μm−1), similar to the behavior observed in the control data but in the opposite absolute direction (see Fig. 3A). It is clear from these data that, whereas the spectrum of dynamical modes has been changed, removal of microtubules alone is not sufficient to simplify the intermediate scattering function to the diffusive limit.
The effects of removing microfilaments by incubation of cells with Cytochalasin D is shown in Fig. 3C. Relative to the control, mitochondrial short- and long-time mobility in actin-depleted cells is decreased on all length scales. The smallest mobility corresponds to the shortest length scale (kG = 11.2 μm−1). In particular, the long-time mobility at this wave number is significantly decreased in comparison to the diffusive limit, possibly indicating a loss of motor-protein function and local stiffness of the membrane on this length scale. For kG = 7.8, 6.3 and 3.9 μm−1 all curves roughly superimpose, suggesting a simplified diffusive behavior in this regime that is similar to the diffusive limit. For the largest length scale investigated (kG = 2.6 μm−1) the short- and long-time mobilities are greater than those on smaller scales and the diffusive limit, although they are decreased (by a factor of ≈2 and 4, respectively) relative to the control situation. The greater mobility on the large length scale could be due to the activity (or partial activity) of intact microtubules.
In the Absence of Microtubules and Microfilaments, the Mitochondrial Reticulum Exhibits Rouse Dynamics.
As stated above, the behavior of f(kG,τ) when both microtubules and microfilaments are depleted is adequately described as diffusive on all of the time and length scales examined (Fig. 3D). Nevertheless, equivalently good agreement is obtained by modeling the mitochondrial reticulum as an interconnected chain of beads that experience a combination of forces caused by thermal fluctuations, hydrodynamic drag, and chain connectivity. The Rouse model precisely describes this situation, and provides an analytical expression for f(kG,τ) (26). The input parameters in the Rouse model are the number of beads in the chain, N, the effective bond-length (inter-bead spacing), b, the bead radius, a, and the viscosity of the medium, η0. In our fit, we have set a = 0.25 μm (consistent with the mitochondrial filament cross-section), η0 = kBT/6πD0a = 45.3 poise (consistent with the experimentally determined value D0 = 1.8 × 10−4 μm2⋅s−1), b = 5.4 μm (adjustable), and N = 100 (adjustable). The fit corresponds to a chain radius of gyration Rg = b(N/6)1/2 = 22 μm, which is a typical size for the perinuclear region of an average osteosarcoma cell. This fit is in excellent agreement with the experimental data shown in Fig. 3D. The fit is shown for two wave numbers (kG = 7.8, 3.9 μm−1) and does not significantly deviate from the diffusive limit for the range of kG investigated. This finding is consistent with observations performed in the high-kG limit (kGb ≫ 2π), in which only the diffusive motions of local subunits are observed regardless of the nature and extent of chain connectivity (27).
Concluding Remarks.
We have shown that it is possible to fully characterize the dynamics of the mitochondrial reticulum by determining the effects of collective interactions through the normalized intermediate scattering function f(kG,τ). The information contained by f(kG,τ) serves to quantify the interactions between the cytoskeletal components and the organelle, which occur over a broad range of temporal and spatial scales. The data clearly show that, under control physiological conditions, mitochondrial displacements exhibit both large and small amplitude fluctuations that occur on time scales faster than the measured diffusive limit. In the absence of microtubules, the frequency of large amplitude fluctuations is significantly decreased, whereas small amplitude displacements occur more frequently. This finding is in contrast to the situation when microfilaments are removed; then the frequencies of occurrences of both large and small amplitude displacements are significantly decreased. These results show that actin filaments and their associated myosin motors are an essential component of the mitochondrial-specific transport system, although they contribute to the overall dynamics on somewhat shorter time and distance scales than do the microtubules and their associated kinesin motors.
Part of the significance of determining f(kG,τ) for mitochondria subject to successive removal of internal constraints (i.e., individual cytoskeletal components) is that it opens the possibility of analyzing and reconstructing its complex motion in terms of well-defined theoretical models. Until now, there has been a lack of experimental data that can distinguish between different microscopic scenarios. For example, one possible model that can account for the structural relaxation shown in Fig. 3A and remains consistent with the changes observed in Fig. 3 B–D involves the combined effects of direct active transport of the mitochondrial membrane along microfilaments (by means of one class of motor proteins operating on sub-micrometer scales), and the secondary motion of microfilaments along microtubule junctions (by means of a second class of motor proteins operating on slightly larger length scales ≈1 μm). In this model, the tubulin cytoskeleton serves as a scaffold to which microfilaments are attached at certain “junction points” that evolve on a time scale comparable (but not identical) to the structural changes of mitochondria because of its contacts with the actin cytoskeleton. Depletion of microtubules then results in decreased mobility on the larger length scales, whereas depletion of microfilaments leads to decreased mobility on short length scales. This and future studies will provide a much deeper insight into the central aspects of the mitochondrial transport system.
Acknowledgments
We thank Dr. Daciana Margineantu for assistance with cell culture preparation, Clifford Dax for technical assistance with the design of the feedback electronics, and Profs. Jeffery Cina, Peter von Hippel, and Robert Mazo for insightful comments on the manuscript. This work is supported by National Science Foundation Grant CHE-9876334, American Chemical Society Petroleum Research Foundation Grant 34285-G7, and American Heart Association Beginning Grant-in-Aid 0060440Z. M.K.K. acknowledges support from the Department of Education Graduate Assistance in Areas of National Need Program Grant P200A010820.
Abbreviations
FICS, Fourier imaging correlation spectroscopy
RPE, relative phase error
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Nunnari J., Marshall, W. F., Straight, A., Murray, A., Sedat, J. W. & Walter, P. (1997) Mol. Biol. Cell 8, 1233-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okamoto K., Perlman, P. S. & Butow, R. A. (1998) J. Cell Biol. 142, 613-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rojo G., Chamorro, M., Salas, R. L., Vinuela, E., Cuezva, J. M. & Salas, J. (1998) J. Virol. 72, 7583-7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dittmann F., Weiss, D. G. & Munz, A. (1987) Roux's Arch. Dev. Biol. 196, 407-413. [DOI] [PubMed] [Google Scholar]
- 5.Yaffe M. P. (1999) Science 283, 1493-1497. [DOI] [PubMed] [Google Scholar]
- 6.Hollenbeck P. J. (1996) Front. Biosci. 1, 91-116. [DOI] [PubMed] [Google Scholar]
- 7.Karbowski M., Spodnik, J. H., Teranishi, M., Wozniak, M., Nishizawa, Y., Usukura, J. & Wakabayashi, T. (2000) J. Cell Sci. 114, 281-291. [DOI] [PubMed] [Google Scholar]
- 8.Rappaport L., Oliviero, P. & Samuel, J. L. (1998) Mol. Cell. Biochem. 184, 101-105. [PubMed] [Google Scholar]
- 9.Bereiter-Hahn J. (1990) Int. Rev. Cytol. 122, 1-63. [DOI] [PubMed] [Google Scholar]
- 10.Margineantu D. H., Cox, W. G., Sudell, L., Sherwood, S. W., Beechem, J. M. & Capaldi, R. A. (2002) Mitochondrion 1, 425-435. [DOI] [PubMed] [Google Scholar]
- 11.Rizzuto R., Pinton, P., Carrington, W., Fay, F. S., Fogarty, K. E., Lifshitz, L. M., Tuft, R. A. & Pozzan, T. (1998) Science 280, 1763-1766. [DOI] [PubMed] [Google Scholar]
- 12.Perkins G., Renken, C., Martone, M. E., Young, S. J., Ellisman, M. & Frey, T. (1997) J. Struct. Biol. 119, 260-272. [DOI] [PubMed] [Google Scholar]
- 13.Howard J., (2001) Mechanics of Motor Proteins and the Cytoskeleton (Sinauer, Sunderland, MA).
- 14.Luby-Phelps K. (1994) Curr. Opin. Cell. Biol. 6, 3-9. [DOI] [PubMed] [Google Scholar]
- 15.Caspi A., Granek, R. & Elbaum, M. (2000) Phys. Rev. Lett. 85, 5655-5658. [DOI] [PubMed] [Google Scholar]
- 16.Griparic L. & van der Bliek, A. M. (2001) Traffic 2, 235-244. [DOI] [PubMed] [Google Scholar]
- 17.Margineantu D., Capaldi, R. A. & Marcus, A. H. (2000) Biophys. J. 79, 1833-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandler D., (1987) Introduction to Modern Statistical Mechanics (Oxford Univ. Press, New York), pp. 234–242.
- 19.Knowles M. K., Grassman, T. J. & Marcus, A. H. (2000) Phys. Rev. Lett. 85, 2837-2840. [DOI] [PubMed] [Google Scholar]
- 20.Grassman T. J., Knowles, M. K. & Marcus, A. H. (2000) Phys. Rev. E 62, 8245. [DOI] [PubMed] [Google Scholar]
- 21.Margineantu D. H., Brown, R. M., Brown, G. K., Marcus, A. H. & Capaldi, R. A. (2002) Mitochondrion 1, 327-338. [DOI] [PubMed] [Google Scholar]
- 22.Fayer M. D. (1982) Ann. Rev. Phys. Chem. 33, 63-87. [Google Scholar]
- 23.Pusey P. N. & Tough, R. J. A. (1985) in Dynamic Light Scattering: Applications of Photon Correlation Spectroscopy, ed. Pecora, R. (Plenum, New York), pp. 85–179.
- 24.Hansen J.-P. & McDonald, I. R., (1986) Theory of Simple Liquids (Academic, San Diego).
- 25.Segrè P. N. & Pusey, P. N. (1997) Pysica A. 235, 9-18. [Google Scholar]
- 26.Doi M. & Edwards, S. F., (1986) The Theory of Polymer Dynamics (Oxford Univ. Press, New York), pp. 133.
- 27.Lin S.-C. & Schurr, J. M. (1978) Biopolymers 17, 425-461. [Google Scholar]