Abstract
Using synchronized tobacco Bright Yellow-2 cells and cDNA-amplified fragment length polymorphism-based genomewide expression analysis, we built a comprehensive collection of plant cell cycle-modulated genes. Approximately 1,340 periodically expressed genes were identified, including known cell cycle control genes as well as numerous unique candidate regulatory genes. A number of plant-specific genes were found to be cell cycle modulated. Other transcript tags were derived from unknown plant genes showing homology to cell cycle-regulatory genes of other organisms. Many of the genes encode novel or uncharacterized proteins, indicating that several processes underlying cell division are still largely unknown.
Essential genes controlling cell cycle progression have been characterized in different organisms. Recently, genomewide expression analysis in yeast (1) and human cells (2) using microarrays has enlarged the collection of cell cycle-modulated genes to several hundred genes with known and unknown functions. Their transcriptional regulation is strict, and genes involved in the same biological process are most often coexpressed. In plants, the basic regulatory mechanisms controlling cell cycle progression also have been studied (3). Although the core cell cycle genes are conserved among higher eukaryotes, basic developmental differences between plants and other organisms imply that plant-specific regulatory pathways exist that control cell division (4). Especially for events occurring at mitosis, plants are thought to have developed unique mechanisms regulating karyo- and cytokinesis. A typical plant cell is surrounded by a rigid wall and cannot, as such, divide by constriction. Instead, a new cell wall between daughter nuclei is formed by a unique cytoskeletal structure called the phragmoplast, whose position is dictated by another cytoskeletal array called the preprophase band (5). Another major difference between plant and animal mitosis is found in the structure of the mitotic spindles: in animals they are tightly centered at the centrosome, whereas in plants they have a diffuse appearance (6).
To identify plant genes involved in cell division and control of cell cycle progression, we performed a genomewide expression analysis of cell cycle-modulated genes in the tobacco Bright Yellow-2 (BY2) cell line. This unique cell line can be synchronized to high levels with different types of inhibitors of cell cycle progression (7, 8). Because of the lack of extensive molecular resources such as genomic sequences, cDNA clones, or ESTs for tobacco, a microarray-based approach cannot be used for transcriptome analysis. Therefore, we used the cDNA-amplified fragment length polymorphism (AFLP) technology to identify and characterize cell cycle-modulated genes in BY2. cDNA-AFLP is a sensitive and reproducible fragment-based technology that has a number of advantages over other methods for genomewide expression analysis (9): it does not require prior sequence information, it allows the identification of novel genes, and it provides quantitative expression profiles. After a detailed analysis, we found that ≈10% of the transcripts are periodically expressed, in agreement with the results obtained in yeast (1). This comprehensive collection of plant cell cycle-modulated genes provides a basis for unraveling the basic mechanisms underlying the plant cell cycle.
Materials and Methods
Synchronization of BY2 Cells and Sampling of Material.
Synchronization, sampling of material, and evaluation of cell cycle progression and synchrony levels were performed (www.psb.rug.ac.be/papers/pebre/pnas.htm). Briefly, tobacco (Nicotiana tabacum L. cv. BY2) cultured cell suspension was synchronized by blocking cells in early S phase with aphidicolin (Sigma; 5 mg/liter). After removal of the drug, samples were taken every hour, starting from the release from the aphidicolin block (time 0) until 11 h later. The mitotic index was determined by counting the number of cells undergoing mitosis under fluorescence microscopy after the DNA had been stained with 5 mg/liter 4′,6-diamidino-2-phenylindole (Sigma). DNA content was measured by flow cytometry.
cDNA-AFLP Analysis.
RNA extraction, cDNA synthesis, and cDNA-AFLP analysis were performed (www.psb.rug.ac.be/papers/pebre/pnas.htm). Double-stranded cDNA (500 ng) was used for cDNA-AFLP analysis. The restriction enzymes used were BstYI and MseI (New England Biolabs).
For preamplifications, a MseI primer without selective nucleotides was combined with a BstYI primer containing either a T or a C as nucleotide at the 3′ extremity. The obtained amplification mixtures were diluted 600-fold and 5 μl was used for final selective amplifications (www.psb.rug.ac.be/papers/pebre/pnas.htm). All possible primer combinations with a total of two and three additional selective nucleotides were used for transcript profiling.
Characterization of AFLP Fragments.
Bands corresponding to differentially expressed transcripts were isolated from the gel, and eluted DNA was reamplified under the same conditions as for selective amplification. Sequence information was obtained either by direct sequencing of the reamplified PCR product with the selective BstYI primer or after cloning the fragments in pGEM-T easy (Promega) and sequencing of individual clones. The obtained sequences were compared with nucleotide and protein sequences in the publicly available databases by blast sequence alignments (10). When available, tag sequences were replaced with longer EST or isolated cDNA sequences to increase the chance of finding significant homology. Based on the homology, transcript tags were classified in functional groups as shown in Table 1.
Table 1.
Functional classification of transcript tags
| Function | Tags | S, 27.7% | G2, 15.8% | M, 52.9% | G1, 3.6% |
|---|---|---|---|---|---|
| Cell cycle control | 30 | 5/8 (0.078) | 8/5 (0.068) | 14/16 (0.114) | 3/1 |
| Cell wall | 35 | 6/10 (0.047) | 4/6 (0.136) | 25/18 (7.1e−3) | 0/1 |
| Cytoskeleton | 43 | 1/12 (1.2e−5) | 4/7 (0.090) | 38/22 (2.1e−7) | 0/2 |
| Hormone response | 13 | 6/4 (0.113) | 1/2 (0.277) | 6/7 (0.185) | 0/0 |
| Kinases/phosphatases | 27 | 4/8 (0.039) | 1/4 (0.059) | 19/14 (0.025) | 1/1 |
| Protein synthesis | 50 | 15/14 (0.116) | 5/8 (0.087) | 29/26 (0.079) | 1/2 |
| Proteolysis | 21 | 2/6 (0.026) | 1/3 (0.144) | 17/11 (0.039) | 1/1 |
| Replication and modification | 74 | 57/20 (4.2e−19) | 8/12 (1.0e−5) | 8/39 (1.0e−18) | 1/3 |
| RNA processing | 20 | 1/6 (6.8e−3) | 1/3 (0.137) | 18/11 (8.1e−4) | 0/0 |
| Signal transduction | 10 | 1/3 (0.121) | 3/2 (0.201) | 6/5 (0.205) | 2/0 |
| Stress response | 20 | 6/6 (0.192) | 2/3 (0.229) | 10/10 (0.159) | 2/1 |
| Transcription factors | 27 | 4/8 (0.039) | 10/4 (3.0e−3) | 12/14 (0.112) | 1/1 |
| Transport and secretion | 31 | 5/9 (0.047) | 2/5 (0.076) | 21/16 (0.031) | 3/1 |
| Unknown function | 175 | 37/48 (0.015) | 19/28 (0.014) | 112/93 (8.3e−4) | 7/6 |
The observed/expected number of tags within the different cell cycle phases is given together with the probability values between parentheses as calculated based on the binomial distribution function, except for the G1 phase because the figures were too small. A significant enrichment of tags of a functional group within a particular cell cycle phase is indicated in bold.
Only kinases and phosphatases with unknown biological function.
Except small GTP-binding proteins, which are classified under signal transduction.
Quantitative Measurements of the Expression Profiles and Data Analysis.
Gel images were analyzed quantitatively with aflp-quantarpro image analysis software (Keygene, Wageningen, The Netherlands). All visible AFLP fragments were scored, and individual band intensities were measured per lane. The obtained data were used to determine the quantitative expression profile of each transcript. The raw data were corrected for differences in total lane intensities, after which each individual gene expression profile was variance-normalized (www.psb.rug.ac.be/papers/pebre/pnas.htm). cluster and treeview software (11) was used for hierarchical, average linkage clustering. Quality-based clustering was performed with a newly developed software program (12). This program is similar to K-means clustering, except that the number of clusters need not be defined in advance and the expression profiles that do not fit in any cluster are rejected. The minimal number of tags in a cluster and the required probability of genes belonging to a cluster were set to 10 and 0.95, respectively. With these parameters, 86% of all of the tags were grouped in 21 distinct clusters.
Results and Discussion
Identification and Characterization of Cell Cycle-Modulated Genes.
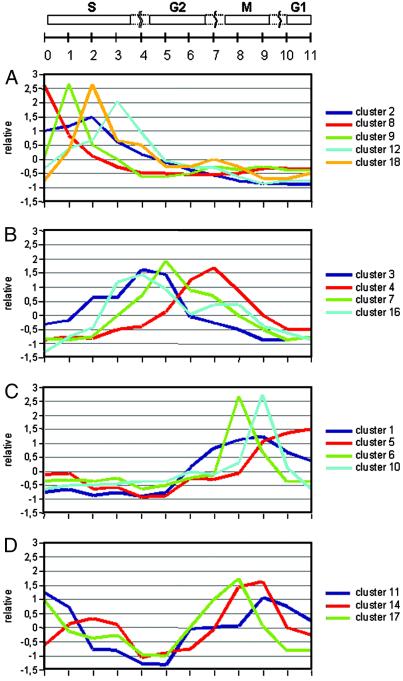
Tobacco BY2 cells were synchronized by blocking cells in early S phase with aphidicolin, an inhibitor of DNA polymerase α. After the inhibitor had been released, 12 time points at 1-h intervals, covering the cell cycle from S phase until M/G1 transition, were sampled. Flow cytometry and determination of the mitotic index showed that most cells exited S phase 4 h after release from blocking and that the peak of mitosis was reached at 8 h. mRNA extracted from each time point was subjected to cDNA-AFLP-based transcript profiling. Quantitative temporal accumulation patterns of ≈10,000 transcript tags were determined and analyzed. In total, ≈1,340 transcript tags were modulated significantly during the cell cycle. Hierarchical clustering (13) of the expression profiles resulted in four large groups with expression peaks at S, early G2, late G2, or M phase. Within each of these groups, several smaller clusters of genes with similar expression patterns were distinguished (Fig. 1). By quality-based clustering (14) (see Materials and Methods), 21 different clusters were identified (see www.psb.rug.ac.be/papers/pebre/pnas.htm). In agreement with the hierarchical clustering analysis, the four largest clusters (Fig. 2, clusters 1–4) correspond to the S, early G2, late G2, and M phases and together contain 65% of all of the tags. An additional cluster (Fig. 2C, cluster 5), not clearly separated in the hierarchical clustering, includes genes with peak expression in G1 phase and contains another 5% of the tags. The remaining clusters are much smaller and most often (e.g., clusters 6, 9, 10, and 18) include genes with narrow temporal expression patterns. In addition to these clusters, three small groups of genes displaying elevated expression during two cell cycle phases also were distinguished by quality-based clustering (Fig. 2D).
Fig 1.
Hierarchical clustering of cell cycle-modulated transcript tags. Only the tags with a significant homology to a known sequence are included. Each row represents a tag with the relative transcript accumulation patterns shown over the 12 consecutive time points (columns). Red and green reflect, respectively, transcriptional activation and repression relative to the average expression level over the time course; gray represents missing data. The dendrogram shows the relationships between clusters of genes with similar expression pattern. The main clusters, corresponding to the different cell cycle phases, are indicated on the right. The full cluster based on the entire data set can be obtained at www.psb.rug.ac.be/papers/pebre/pnas.htm.
Fig 2.
Gene expression profiles obtained by quality-based clustering of all transcript tags. Shown are the trend lines of 16 clusters containing 97% of the genes and covering the entire time course as indicated at the top. S phase-specific gene clusters are grouped in A, gene clusters with peak expression between S and M phases in B, and the M and G1 phase-specific clusters in C. (D) Three small clusters of genes with peak expression during two cell cycle phases. An overview of the complete clusters can be obtained at www.psb.rug.ac.be/papers/pebre/pnas.htm.
Once the transcript tags were sequenced, homology searches revealed that 36.5% of the tags were significantly homologous to genes of known functions, and 13.1% of the tags matched a cDNA or genomic sequence without allocated function. In contrast, no homology with a known sequence was found for 50.4% of the tags. In agreement with findings in yeast and human (1, 2), genes of known function belong to diverse functional classes (Table 1), revealing that several biological processes are at least partially under temporal transcriptional control during the cell cycle in plants. In general, the observed transcript accumulation profiles and cell cycle specificity correlate well with the functional properties of the corresponding genes. It is interesting that the number of transcription factors with G2-phase specificity is high, which may be related to the induction of genes involved in M phase-specific processes. The overrepresentation of RNA-processing genes in the M phase might indicate that posttranscriptional regulation is involved in gene activity during mitosis. Because de novo transcription is severely reduced during mitosis (13), RNA processing (differential RNA stability, alternative splicing) or specific chromatin decondensation could be an alternative regulatory mechanism. Intriguingly, transcript tags with homology to a gene of unknown function were overrepresented in the M phase as well (Table 1). The principal differences in cell cycle events between plants and other organisms occur during mitosis; therefore, it is tempting to speculate that several of these transcripts correspond to still uncharacterized plant-specific genes triggering these events. Remarkably, several of the tags homologous to a publicly available sequence have no Arabidopsis homolog, indicating that, in addition to conserved genes, different plant species possess unique sets of cell cycle-modulated genes. Although many of these tags may be too short to significantly match an Arabidopsis sequence, analysis of longer cDNA clones corresponding to a subset of tags has revealed that ≈25% of the sequences are indeed novel (unpublished results).
The Core Cell Cycle Machinery.
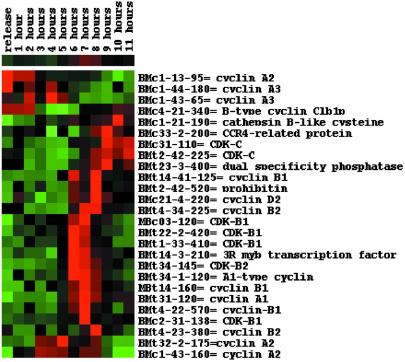
Several tags coincide with genes belonging to the core cell cycle machinery and exhibiting distinct expression profiles (Fig. 3). Transcript tags from five B1- or B2-type cyclins as well as from a D2-type cyclin show mitotic accumulation and exhibit a narrow temporal expression profile, confirming previous studies (14, 15). Based on the transcription patterns, the six A-type cyclins fall into three groups that sequentially appear during the cell cycle, adding more data to earlier observations (16). Two groups have a rather broad window of transcript accumulation: one group, homologous to A3-type cyclins, is expressed during S phase and disappears during G2 phase, whereas the other corresponds to A2-type cyclins and shows expression that increases at mid-S phase and decreases during M phase, except for one transcript, which is specific for S phase. The third group, containing an A1-type cyclin, has the same expression pattern as the B- and D2-type cyclins. Several tags derived from genes encoding the plant-specific B-type cyclin-dependent kinases (CDKs) were also identified. CDKB1 and CDKB2 peak at the G2/M transition, slightly before the mitotic cyclins, as described (17). In contrast to what has been observed in partially synchronized alfalfa cell cultures (18), the transcript levels of the tags homologous to a C-type CDK accumulated differentially during the cell cycle. The transcripts were present during late M phase and early S phase, suggesting that CDKC is active during the G1 phase.
Fig 3.
Expression profiles and hierarchical clustering of the main known and putative core cell cycle genes. The identity of each tag is given on the right.
In addition to these well-characterized cell cycle-regulatory genes, we have identified several tags derived from genes encoding transcription factors and protein kinases or phosphatases with known or putative roles in cell cycle control. One tag with a sharp peak of transcript accumulation 1 h before the B- and D-type cyclins corresponds to a 3R-MYB transcription factor. Recently, a closely related 3R-MYB has been shown to activate B-type cyclins and other genes with an M phase-specific activator domain (19). Another tag peaking in the M phase is homologous to the CCR4-associated protein CAF. CAF forms a complex with CCR4 and DBF2, resulting in a transcriptional activator involved in the regulation of diverse processes including cell wall integrity, methionine biosynthesis, and M/G1 transition (20). Most of the tags with similarity to protein kinases and phosphatases show M phase-specific accumulation (Table 1). Although the true identity and putative cell cycle-related function remains unclear for the majority, one is highly homologous to a dual-specificity phosphatase. This type of phosphatase plays a crucial role in cell cycle control in yeast and animals (21). Another M phase-specific tag is homologous to prohibitin. In the mammalian cell cycle, prohibitin represses E2F-mediated transcription via interaction with retinoblastoma, thereby blocking cellular proliferation (22).
Protein degradation by the ubiquitin-proteasome pathway also plays an important role in the control of cell cycle progression at both G1/S transition and exit from mitosis. Although there is little evidence for cell cycle-modulated expression of the genes encoding the various components of the ubiquitin-proteasome complexes, some proteins accumulate in a cell cycle-dependent way (23). In the human cell cycle study, several genes that are implicated in proteolytic control of cell cycle progression have been identified (2). Similarly, we isolated several tags from genes encoding ubiquitin-conjugating enzyme (E3), ubiquitin-protein ligase (E2), and proteasome components with an M phase-specific expression pattern. Another transcript tag that accumulates during late M phase is similar to cathepsin B-like proteins, which are proteolytically active and degrade diverse nuclear proteins, including retinoblastoma (24).
Although all core cell cycle-regulatory genes that control the G2/M transition whose expression is known to be cell cycle modulated were identified, genes encoding proteins controlling G1/S transition, such as retinoblastoma and E2F, were not found. These genes were probably missed because the G1/S transition was not included in our analysis, a hypothesis supported by the finding that the early targets of E2F, such as polymerase α and ribonucleotide reductase, are already present at high levels at the beginning of the time course.
Genes Involved in DNA Replication and Modification.
In agreement with studies performed in yeast and human fibroblasts, transcripts encoding proteins involved in DNA replication and modification accumulated during S phase and exhibited broad temporal expression profiles (Fig. 4). Different replication factors, DNA polymerase α, and histones H3 and H4 are already present at the onset of the time course, indicating that they are induced before the time point of the aphidicolin arrest. Interestingly, most of histones H1, H2A, and H2B appear somewhat later than H3 and H4, perhaps indicating that they are deposited into the nucleosomes after H3 and H4 (25, 26). As expected given that the three proteins are part of the replication-coupling assembly factor complex that mediates chromatin assembly (26), the profile of the homolog of the anti-silencing function 1 (ASF1) protein is similar to that of histones H3 and H4. Genes encoding high-mobility group proteins reach the highest accumulation during late G2, consistent with the subsequent steps involved in the chromatin folding and structuring. Tags derived from genes encoding proteins involved in DNA modification, such as S-adenosyl-l-methionine synthase and cytosine-5-methyltransferase are found in the histone cluster. Tags from methionine synthase genes, precursors for S-adenosyl-l-methionine synthase, accumulate during M phase. In contrast, these genes are expressed during late S phase in yeast (1).
Fig 4.
Hierarchical clustering of transcript tags derived from genes involved in DNA replication and modification. The identity of each tag is given on the right.
Genes involved in chromatin remodeling and transcriptional activation or repression have been identified as well. One gene is a histone deacetylase with highest transcript accumulation during G2 phase. Another gene belongs to the SNF2 family of chromodomain proteins with an M phase-specific expression pattern. Interestingly, one tag corresponds to a mammalian inhibitor of growth 1 (p33-ING1) protein. The human ING1 protein has DNA-binding activity and might be involved in chromatin-mediated transcriptional regulation (27). The protein was shown to accumulate during S phase (28), which is in agreement with the expression profile we observed. The yeast homologs of ING1 are components of the histone acetyltransferase complex and show similarity to the retinoblastoma-binding protein 2 (29). Another tag, homologous to the Arabidopsis MSI3 protein, follows a similar expression profile. MSI-like proteins are involved in the regulation of histone acetylation and deacetylation and in chromatin formation (30).
The expression profiles of the various ribonucleotide reductase genes are more complex. One gene is already expressed at high levels at the beginning of the time course and its expression is restricted to S phase, as described (31). In contrast, another gene is highly expressed in S phase and reappears at lower levels during M phase, whereas a third is M phase-specific. This latter expression profile has also been described for a ribonucleotide reductase gene from Xenopus where the encoded protein appears to be involved in microtubulin nucleation (32).
Numerous other transcript tags with S-phase specificity were found in addition to the ones involved in DNA replication and modification. Most interestingly, one of these tags is homologous to a mammalian gene encoding a TRAF-interacting protein (TRIP), which is a component of the tumor necrosis factor (TNF) signaling complex, and promotes cell death when complexed with TRAF (33). Another S phase-specific tag shows homology to the RING finger domain of inhibitor of apoptosis proteins that are also involved in the TNF signaling pathway. Because the TNF signaling pathway is unknown in plants, a detailed analysis will be required to determine the true functional properties of these plant homologs.
Modulated Expression of Genes Required for Mitosis and Cytokinesis.
Several paralogous genes that encode either α- or β-tubulin were highly induced and accumulated before the mitotic index peak or during early M phase (data not shown). Whereas, in yeast, tubulins fluctuate only moderately during the cell cycle (1), we found that, in BY2, tubulin genes are highly cell cycle-modulated. This transcriptional regulation is in agreement with previous demonstrations of de novo transcription of α- and β-tubulin genes during different cellular processes (34). In our analysis, no γ-tubulin genes showed up, confirming published data that the amount of γ-tubulin is constant in dividing BY2 cells (35).
Most of the kinesins we identified fall in the same cluster as the tubulins peaking before mitosis. Interestingly, two tags have a distinct transcription pattern and appear in another gene cluster. Their window of transcript accumulation is very narrow and coincides with the peak of mitosis. Most interestingly, these tags correspond to the plant-specific phragmoplast-associated type of kinesin, PAKRP1 (36). A chromokinesin not yet described in plants was identified as well. This type of motor protein uses DNA as cargo and plays a role in chromosome segregation and metaphase alignment (37).
Among the M phase-specific kinases we identified, two were unambiguously recognized to be involved in cytokinesis. One is Aurora, a protein kinase with a key role in the control of chromosome segregation, centrosome separation, and cytokinesis in yeast and animals (38), but not yet described in plants. The other is NRK1, a mitogen-activated protein kinase kinase that is phosphorylated by NPK1, a kinase involved in regulating the outward growth of the phragmoplast and cell plates (39).
Hormonal Regulation and Cell Cycle-Modulated Gene Expression.
A number of genes belonging to the class of auxin-induced genes were also differentially expressed. Cell cycle-modulated expression of auxin-induced genes has never been observed before although auxins together with cytokinins are the two major groups of plant hormones that affect cell division (3). The genes we identified fall into two groups based on their transcript accumulation profiles (data not shown). The first group displays an early S phase-specific expression pattern and consists of the parA, parB, and parC genes. Induction of the par genes is most often observed in response to stress conditions (40). The fact that the transcripts rapidly disappear after release from the cell cycle-blocking agent might indicate a stress response rather than a cell cycle-dependent auxin response.
More interesting is the second group of genes with transcripts accumulating during early M phase. This group includes auxin response factor 1 (ARF1), an auxin transporter as well as different members of the early auxin response AUX/IAA gene family. ARF1 is a transcription factor that binds to a particular auxin response element (41). Additional studies suggest that the activity of ARF1 is controlled by its dimerization with members of the AUX1/IAA family (42). The similarity in temporal expression profiles we observed supports these findings and suggests that these proteins mediate an auxin response necessary for cell cycle progression.
Conclusion
Using tobacco BY2 as model system together with cDNA-AFLP-based transcript profiling, we were able to build a comprehensive inventory of plant cell cycle-modulated genes. The data obtained confirm earlier results and observations, while numerous unique findings were made. Detailed characterization of the transcript tags corresponding to putative novel genes and genes of unknown function will help further unravel the control mechanisms and regulatory networks underlying the cell cycle and to identify and characterize the genes involved in plant-specific processes occurring during cell division. Very recently, a study using microarrays to analyze cell cycle-modulated gene expression in Arabidopsis has been published (43). The obtained data are in agreement with our results and also indicate that a large number of genes involved in different biological processes are differentially expressed.
Acknowledgments
We are grateful to Gerrit Beemster and Marnik Vuylsteke for comments, the sequencing group for its cloning and sequencing efforts, Rudy Vanderhaeghen for isolating and analyzing cDNA clones, and Martine De Cock for help with the manuscript. This work was funded in part by the European Union (ECCO QLG2-CT1999-00454), Génoplante (Project BI1999087), and CropDesign N.V. (0235). K.V. and L.C. are indebted to the Vlaams Instituut voor de Bevordering van het Wetenschappelijk-Technologisch Onderzoek in de Industrie for predoctoral fellowships. L.D.V. is a postdoctoral fellow of the Fund for Scientific Research (Flanders).
Abbreviations
AFLP, amplified fragment length polymorphism
BY2, Bright Yellow-2
CDK, cyclin-dependent kinase
References
- 1.Spellman P. T., Sherlock, G., Zhang, M. Q., Iyer, V. R., Anders, K., Eisen, M. B., Brown, P. O., Botstein, D. & Futcher, B. (1998) Mol. Biol. Cell 9, 3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho R. J., Huang, M., Campbell, M. J., Dong, H., Steinmetz, L., Sapinoso, L., Hampton, G., Elledge, S. J., Davis, R. W. & Lockhart, D. J. (2001) Nat. Genet. 27, 48-54. [DOI] [PubMed] [Google Scholar]
- 3.Stals H. & Inzé, D. (2001) Trends Plant Sci. 6, 359-364. [DOI] [PubMed] [Google Scholar]
- 4.den Boer B. G. W. & Murray, J. A. H. (2000) Trends Cell Biol. 10, 245-250. [DOI] [PubMed] [Google Scholar]
- 5.Smith L. G. (1999) Curr. Opin. Plant Biol. 2, 447-453. [DOI] [PubMed] [Google Scholar]
- 6.Marc J. (1997) Trends Plant Sci. 2, 223-230. [Google Scholar]
- 7.Nagata T., Nemoto, Y. & Hasezawa, S. (1992) Int. Rev. Cytol. 132, 1-30. [Google Scholar]
- 8.Planchais S., Glab, N., Inzé, D. & Bergounioux, C. (2000) FEBS Lett. 476, 78-83. [DOI] [PubMed] [Google Scholar]
- 9.Breyne P. & Zabeau, M. (2001) Curr. Opin. Plant Biol. 4, 138-142. [DOI] [PubMed] [Google Scholar]
- 10.Altschul S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisen M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Smet F., Marchal, K., Mathijs, J., Thijs, G., De Moor, B. & Moreau, Y. (2002) Bioinformatics 8, 735-746. [DOI] [PubMed] [Google Scholar]
- 13.Gottesfeld J. M. & Forbes, D. J. (1997) Trends Biochem. Sci. 22, 197-202. [DOI] [PubMed] [Google Scholar]
- 14.Mironov V., De Veylder, L., Van Montagu, M. & Inzé, D. (1999) Plant Cell 11, 509-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sorrell D. A., Combettes, B., Chaubet-Gigot, N., Gigot, C. & Murray, J. A. H. (1999) Plant Physiol. 119, 343-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reichheld J.-P., Chaubet, N., Shen, W. H., Renaudin, J.-P. & Gigot, C. (1996) Proc. Natl. Acad. Sci. USA 93, 13819-13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porceddu A., Stals, H., Reichheld, J.-P., Segers, G., De Veylder, L., De Pinho Barrôco, R., Casteels, P., Van Montagu, M., Inzé, D. & Mironov, V. (2001) J. Biol. Chem. 276, 36354-36360. [DOI] [PubMed] [Google Scholar]
- 18.Magyar Z., Mészáros, T., Miskolczi, P., Deák, M., Fehér, A., Brown, S., Kondorosi, E., Athanasiadis, A., Pongor, S., Bilgin, M., et al. (1997) Plant Cell 9, 223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito M., Araki, S., Matsunaga, S., Itoh, T., Nishihama, R., Machida, Y., Doonan, J. H. & Watanabe, A. (2001) Plant Cell 13, 1891-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu H.-Y., Toyn, J. H., Chiang, Y.-C., Draper, M. P., Johnston, L. H. & Denis, C. L. (1997) EMBO J. 16, 5289-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coleman T. R. & Dunphy, W. G. (1994) Curr. Opin. Cell Biol. 6, 877-882. [DOI] [PubMed] [Google Scholar]
- 22.Wang S., Nath, N., Adlam, M. & Chellappan, S. (1999) Oncogene 18, 3501-3510. [DOI] [PubMed] [Google Scholar]
- 23.del Pozo J. C. & Estelle, M. (2000) Plant Mol. Biol. 44, 123-128. [DOI] [PubMed] [Google Scholar]
- 24.Fu Y.-H. F., Nishinaka, T., Yokoyama, K. & Chiu, R. (1998) FEBS Lett. 421, 89-93. [DOI] [PubMed] [Google Scholar]
- 25.Luger K., Mäder, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. (1997) Nature 389, 251-260. [DOI] [PubMed] [Google Scholar]
- 26.Tyler J. K., Adams, C. R., Chen, S.-R., Kobayashi, R., Kamakaka, R. T. & Kadonaga, J. T. (1999) Nature 402, 555-560. [DOI] [PubMed] [Google Scholar]
- 27.Cheung K.-J. & Li, G. (2001) Exp. Cell Res. 268, 1-6. [DOI] [PubMed] [Google Scholar]
- 28.Garkavtsev I. & Riabowol, K. (1997) Mol. Cell. Biol. 17, 2014-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loewith R., Meijer, M., Lees-Miller, S. P., Riabowol, K. & Young, D. (2000) Mol. Cell. Biol. 20, 3807-3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ach R. A., Taranto, P. & Gruissem, W. (1997) Plant Cell 9, 1595-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chabouté M.-E., Combettes, B., Clément, B., Gigot, C. & Philipps, G. (1998) Plant Mol. Biol. 38, 797-806. [DOI] [PubMed] [Google Scholar]
- 32.Takada S., Shibata, T., Hiraoka, Y. & Masuda, H. (2000) Mol. Biol. Cell 11, 4173-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee S. Y., Lee, S. Y. & Choi, Y. (1997) J. Exp. Med. 185, 1275-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stotz H. U. & Long, S. R. (1999) Plant Mol. Biol. 41, 601-614. [DOI] [PubMed] [Google Scholar]
- 35.Stoppin-Mellet V., Peter, C. & Lambert, A. M. (2000) Plant Biol. 2, 290-296. [Google Scholar]
- 36.Lee Y. R. J. & Liu, B. (2000) Curr. Biol. 10, 797-800. [DOI] [PubMed] [Google Scholar]
- 37.Wang S.-Z. & Adler, R. (1995) J. Cell Biol. 128, 761-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bischoff J. R. & Plowman, G. D. (1999) Trends Cell Biol. 9, 454-459. [DOI] [PubMed] [Google Scholar]
- 39.Nishihama R., Ishikawa, M., Araki, S., Soyano, T., Asada, T. & Machida, Y. (2001) Genes Dev. 15, 352-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abel S. & Theologis, A. (1996) Plant Physiol. 111, 9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ulmasov T., Hagen, G. & Guilfoyle, T. J. (1997) Science 276, 1865-1868. [DOI] [PubMed] [Google Scholar]
- 42.Walker L. & Estelle, M. (1998) Curr. Opin. Plant Biol. 1, 434-439. [DOI] [PubMed] [Google Scholar]
- 43.Menges, M., Hennig, L., Gruissem, W. & Murray, J. A. (2002) J. Biol. Chem., in press. [DOI] [PubMed]