Abstract
The delivery to the plasma membrane of the general amino acid permease, Gap1p, of Saccharomyces cerevisiae is regulated by the quality of the nitrogen source in the growth medium. In an effort to define how different nitrogen sources control Gap1p sorting, we find that mutations in GDH1 and GLN1 that decrease the flux through the glutamate and glutamine synthesis pathways result in increased Gap1p sorting to the plasma membrane. Conversely, deletion of MKS1, which increases glutamate and glutamine synthesis, decreases Gap1p sorting to the plasma membrane. Glutamate and glutamine are not unusual in their ability to regulate Gap1p sorting, because the addition of all natural amino acids and many amino acid analogs to the growth medium results in increased Gap1p sorting to the vacuole. Importantly, amino acids have the capacity to signal Gap1p sorting to the vacuole regardless of whether they can be used as a source of nitrogen. Finally, we show that rapamycin does not affect Gap1p sorting, indicating that Gap1p sorting is not directly influenced by the TOR pathway. Together, these data show that amino acids are a signal for sorting Gap1p to the vacuole and imply that the nitrogen-regulated Gap1p sorting machinery responds to amino acid-like compounds rather than to the overall nutritional status associated with growth on a particular nitrogen source.
When provided with a mixture of nitrogen sources in the growth medium, the yeast Saccharomyces cerevisiae shows a preference for the utilization of particular nitrogen sources. Preferred nitrogen sources such as glutamine are used first, then nonpreferred nitrogen sources such as proline are used only after the preferred nitrogen sources have been depleted. The regulatory pathways that govern this hierarchy are collectively known as nitrogen regulation (1–3). It is unclear how the cell senses the quality of a nitrogen source, though cellular responses to nitrogen sources of varying quality have been well documented. Nitrogen source quality usually has been inferred by the cellular responses elicited by a given nitrogen source and is not correlated with the growth rate it supports (4).
To gain insight into the signals governing nitrogen regulation, we have focused our study on the nitrogen-regulated sorting of the general amino acid permease, Gap1p. Gap1p is a high-capacity permease that can transport all naturally occurring amino acids (5, 6). GAP1 transcription is positively regulated by the GATA-type transcription factors Gln3p and Gat1p/Nil1p and negatively regulated by the cytoplasmic factor Ure2p, so that GAP1 is expressed on nonpreferred nitrogen sources but repressed on preferred nitrogen sources (1). The quality of the nitrogen source also regulates the intracellular sorting of Gap1p. During growth on the poor nitrogen sources urea, proline, or ammonia (in the S288C background), Gap1p is sorted to the plasma membrane and its activity at the plasma membrane is high. During growth on glutamate, or when GAP1 is artificially transcribed during growth on glutamine (as in a ure2Δ mutant), Gap1p activity is undetectable because Gap1p is not delivered to the plasma membrane and is instead sorted to the vacuole for degradation (7, 8). One major class of mutations that influences Gap1p sorting is in ubiquitination genes (1). Because polyubiquitination of Gap1p is a signal for its sorting to the vacuole, mutation of genes required for Gap1p polyubiquitination (bul1 bul2, rsp5, or doa4) causes Gap1p to be sorted to the plasma membrane more efficiently. Gap1p sorting is thought to occur at either the endosome or trans-Golgi compartments (8, 9).
Here we show that an abundance of amino acids, resulting either from mutations that cause increased glutamate and glutamine synthesis or from the addition of amino acids to the growth media, triggers sorting of Gap1p to the vacuole.
Materials and Methods
Strains, Plasmids, and Media.
The yeast strains used in this study (listed in Table 1) are all in the S288C background. One characteristic of the S288C background is high Gap1p and Put4p activity when ammonia is used as a nitrogen source (10). Complete gene deletions of GDH1, MKS1, and END3 were constructed by gene replacement with the kanMX6 cassette by means of homologous recombination (11). The gln1-9142 strains are derived from AMP721 (gift of A. Mitchell, Columbia University, New York), which was crossed to a wild-type S288C strain until the gln1-9142 phenotype, as measured by temperature-sensitive glutamine auxotrophy and elevated Gap1p activity, segregated cleanly 2:2.
Table 1.
Strains used in this study (all are isogenic with S288C)
| Strain | Genotype | Source |
|---|---|---|
| CKY443 | MATa prototroph | Kaiser strain collection |
| CKY445 | MATα gap1Δ:LEU2 leu2 | Kaiser strain collection |
| CKY757 | MATa gdh1Δ:kanMX6 | This study |
| CKY758 | MATa mks1Δ:kanMX6 | This study |
| CKY759 | MATα PADH1-GAP1-HA | This study |
| CKY760 | MATa PADH1-GAP1-HA ura3-52 | This study |
| CKY761 | MATa pep4Δ:kanMX6 ura3-52 | This study |
| CKY762 | MATa gdh1Δ:kanMX6 PADH1-GAP1-HA | This study |
| CKY763 | MATα mks1Δ:kanMX6 PADH1-GAP1-HA | This study |
| CKY764 | MATa gln1-9142 PADH1-GAP1-HA | This study |
| CKY765 | MATa end3Δ:kanMX6 PADH1-GAP1-HA | This study |
| CKY766 | MATa end3Δ:kanMX6 mks1Δ:kanMX6 PADH1-GAP1-HA | This study |
| CKY767 | MATa gln1-9142 | This study |
Plasmids used in this study were pMS29, a PGAP1-LacZ fusion at codon 53 of GAP1 in a URA3-CEN vector (7); pPL257, GAP1 with the hemagglutinin 1 (HA1) epitope inserted at codon 62, in pRS316 (12); and pCK227, the GAP1-HA ORF and terminator fused behind the ADH1 promoter in pRS316 (9).
Minimal media were prepared as described (8).
Assays for Amino Acid Uptake and β-Galactosidase.
Strains were cultured to 4–8 × 106 cells per ml, washed twice with nitrogen-free medium by filtration on a 0.45-μm nitrocellulose filter (Millipore), and amino acid uptake assays were performed as described (8).
β-Galactosidase activity was measured by using the permeabilized cell method (13).
Amino Acid and Analog Additions Before Uptake Assays or Density Centrifugation.
Amino acids or analogs were dissolved in ammonia medium at 4× the final concentration, and the pH of the mixture was adjusted to 4.0. One-third volume of the amino acid or analog solution was added to ammonia-growing cultures grown at 24°C at a density of 3–5 × 106 cells per ml. After 2 h, cells were collected for amino acid uptake assays or for equilibrium density centrifugation.
Equilibrium Density Centrifugation and Antibodies.
Membrane proteins were separated by equilibrium density centrifugation on continuous 20–60% sucrose gradients containing EDTA as described (8, 14).
Antibodies used were: mouse anti-HA antibody 16B12 (Babco); mouse anti-Dpm1p (Molecular Probes); rabbit anti-Pma1p (gift of S. Losko and R. Kölling, Düsseldorf, Germany); and horseradish peroxidase-coupled sheep anti-mouse and horseradish peroxidase-coupled sheep anti-rabbit (Amersham Pharmacia).
Whole-Cell Amino Acid Analysis.
Cells from an exponentially growing culture were collected by filtration on a 0.45-μm Durapore membrane filter (Millipore) and quickly washed twice. Cells were suspended in ice-cold methanol, the extract was dried in a Speed-Vac at room temperature, and pellets were stored at −80°C. Pellets were suspended in water and vortexed at 4°C for 3 min, then debris was removed by centrifugation. The supernatant was filtered with a 0.2-μm syringe filter, treated with 5% sulfosalicylic acid, and centrifuged to remove precipitated protein. Analysis was done at the University of Arizona Laboratory for Protein Sequencing and Analyses on a Beckman 7300 (postcolumn, ninhydrin method) dedicated amino acid analyzer by ion-exchange chromatography using citrate buffers of increasing ionic strength and pH at varying temperatures. Analyses were performed two to five times with similar results.
Results
A Reporter to Measure Posttranscriptional Gap1p Regulation.
Gap1p is regulated both transcriptionally and posttranslationally by the nitrogen source in the growth medium. To construct a constitutively expressed GAP1 that was insensitive to its usual transcriptional regulation, we replaced the GAP1 promoter and coding sequence with GAP1-HA under the transcriptional control of the ADH1 promoter.
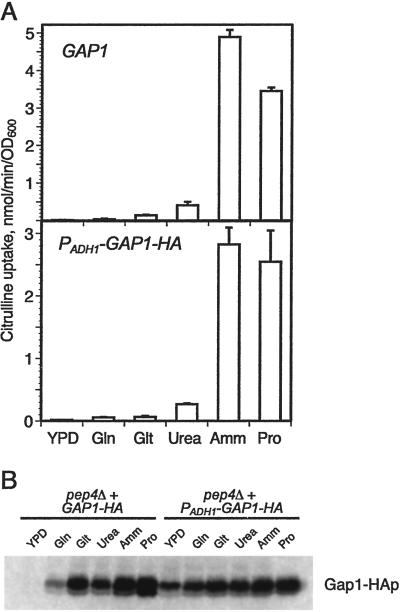
Cells containing the PADH1-GAP1-HA construct showed the same patterns of Gap1p activity on various nitrogen sources as did cells expressing GAP1 from its own promoter (Fig. 1A). In both PADH1-GAP1-HA and GAP1 strains, almost no Gap1p activity, as measured by the rate of [14C]citrulline uptake, was detected in cells grown on yeast extract/peptone/dextrose, glutamine, or glutamate, whereas Gap1p activity was seen in both strains grown on urea, ammonia, and proline. However, the steady-state levels of Gap1 protein expressed in the PADH1-GAP1-HA strain grown in different nitrogen sources were very similar (Fig. 1B). Thus, most of the regulation of Gap1p activity can be accounted for by posttranscriptional regulation of Gap1p sorting.
Fig 1.
Nitrogen regulation of Gap1p activity is largely posttranslational. (A) Gap1p activity was measured by assaying the rate of [14C]citrulline uptake in a wild-type strain (CKY443) and a strain expressing PADH1-GAP1-HA (CKY759) growing on yeast extract/peptone/dextrose or glutamine, glutamate, urea, ammonia, or proline minimal media at 24°C. (B) Gap1-HA protein levels are similar in the PADH1-GAP1-HA strain grown in different nitrogen sources. Protein extracts were prepared from pep4Δ strains (CKY761), to reduce the effect of vacuolar degradation on protein levels, containing either GAP1-HA (pPL257) or PADH1-GAP1-HA (pCK227) grown on various nitrogen sources. Gap1-HAp was detected by immunoblotting.
Mutants with Decreased Glutamate or Glutamine Synthesis Have Increased Gap1p Delivery to the Plasma Membrane.
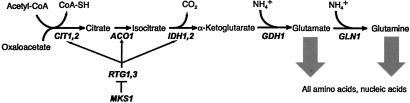
GDH1 encodes the anabolic glutamate dehydrogenase, the primary glutamate synthesizing enzyme during growth on ammonia or urea (15) (see Fig. 2). Because glutamate and glutamine serve as the nitrogen donors for all of the amino acid and nucleotide biosynthesis pathways in the cell (16), GDH1 has a major cellular role. GDH1 has been implicated in the transcriptional regulation of GAP1 and other nitrogen-regulated genes (17, 18), and in the inactivation of Gap1p when ammonia is added to cells of the Σ1278b background growing in proline medium (19).
Fig 2.
The primary routes for glutamate and glutamine synthesis during growth on ammonia or urea.
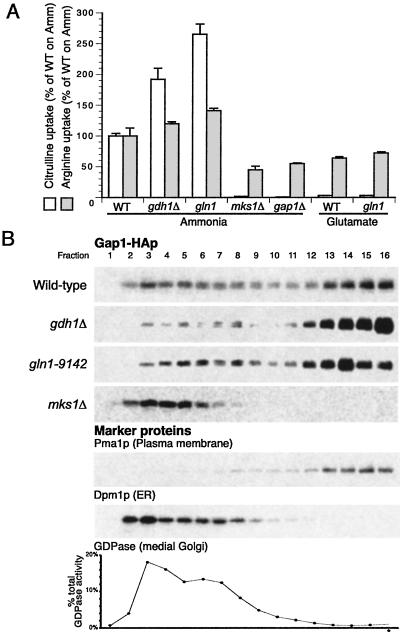
We found that gdh1Δ mutants had about twice the Gap1p activity of wild-type during growth on ammonia, both strains containing PADH1-GAP1-HA (Fig. 3A), whereas arginine uptake included as a control is largely unchanged. Fractionation of membranes of a gdh1Δ strain revealed that about two-thirds of the Gap1p localized to the plasma membrane (Fig. 3B), in contrast to wild-type, in which about two-thirds was internal, cofractionating with the ER, Golgi, and endosome. Thus, the decreased glutamate synthesis in a gdh1Δ mutant results in an increase in the amount of Gap1p sorted to the plasma membrane.
Fig 3.
Mutations that affect glutamate and glutamine synthesis pathways affect Gap1p activity and localization. (A) Gap1p activity was measured by assaying the rate of [14C]citrulline uptake in wild type (CKY759), gdh1Δ (CKY762), gln1-9142 (CKY764), mks1Δ (CKY763), and gap1Δ (CKY445), grown in ammonia or glutamate minimal medium at 24°C. All strains (except gap1Δ) contained PADH1-GAP1-HA instead of GAP1 under its own promoter. The rate of [14C]arginine uptake, which is not sensitive to nitrogen source quality, was also measured for comparison. (B) The amount of Gap1p localized to the plasma membrane is increased in gdh1Δ and gln1-9142 mutants and decreased in an mks1Δ mutant. Wild-type (CKY759), gdh1Δ (CKY762), gln1-9142 (CKY764), and mks1Δ (CKY763) cells were grown on ammonia minimal medium at 24°C, and cell extracts were subjected to isopycnic fractionation on 20–60% sucrose density gradients. Fractions were collected and proteins were separated by SDS/PAGE. Gap1-HAp was detected by immunoblotting with 16B12 anti-HA antibody. The plasma membrane marker protein Pma1p and the endoplasmic reticulum membrane protein Dpm1p were detected by immunoblotting, and the Golgi marker protein GDPase was detected by enzymatic assay. In all of the strains shown, Pma1p, Dpm1, and GDPase fractionated in the same manner. Gradients are for comparison between amounts of internal and plasma membrane-localized Gap1p in each strain and are not intended for quantitative comparison of Gap1p levels between different strains.
It has been reported in the Σ1278b background that a gdh1 mutant grown in ammonia has about a 40% decrease in total glutamate (20). By whole-cell amino acid analysis (Table 2), we verified that the gdh1Δ mutant in our background had less glutamate, glutamine, and total amino acids than the wild type. Unexpectedly, we found that the total glutamate content of gdh1Δ was only 11% lower than that of the wild type. This relatively small effect in our background may be explained by homeostatic mechanisms that decrease the rate at which glutamate is converted to other compounds or by the up-regulation of other glutamate synthesis pathways when the main pathway for glutamate synthesis has been blocked. Also, it is possible that the amino acid content of cellular compartments might show more dramatic differences than the whole cell analysis. However, because amino acid localization is thought to be highly dynamic, the technique used to separate cytoplasmic from vacuolar amino acid pools (which requires an incubation of at least 10 min in a Cu2+-containing nitrogen-free buffer) was unsatisfactory for our purposes (21). Thus, the harvest of whole cells in methanol appeared the best choice given current technology.
Table 2.
Glutamate, glutamine, glutamate + glutamine, and total amino acid content of gdh1Δ, gln1-9142, and mks1Δ
| Strain
|
Amino acid content (nmol/OD600) | |||
|---|---|---|---|---|
| Glu | Gln | Glu + Gln | Total amino acids | |
| Wild type | 19 | 23 | 42 | 96 |
| gdh1Δ | 17 | 13 | 30 | 59 |
| gln1-9142 | 24 | 10 | 34 | 96 |
| mks1Δ | 43 | 28 | 71 | 138 |
| Nitrogen-free (4 h) | 13 | 1 | 14 | 26 |
Wild type (CKY443), gdh1Δ (CKY757), gln1-9142 (CKY767), and mks1Δ (CKY758) were grown to exponential phase on minimal ammonia media at 24°C. For the nitrogen-free sample, wild-type cells were shifted from ammonia medium to nitrogen-free medium for 4 h before harvesting. Sample preparation and analysis were performed as described in Materials and Methods. One OD600 unit equals 2 × 107 cells.
Because glutamate is readily converted into glutamine, we wondered whether gdh1Δ affected Gap1p sorting primarily by affecting glutamate or glutamine synthesis. Glutamine synthesis requires the product of GLN1, the glutamine synthetase, an essential gene for growth on medium lacking glutamine (Fig. 2) (22). GLN1 had previously been implicated in the inactivation of Gap1p by ammonia in the Σ1278b background (23) and in the repression of the catabolic glutamate dehydrogenase GDH2 (24). The partially defective gln1-9142 mutant had more than twice the Gap1p activity of wild-type (Fig. 3A) and more Gap1p at the plasma membrane than wild-type (Fig. 3B), suggesting that intact glutamate and glutamine synthesis pathways are required for proper sorting of Gap1p to the vacuole.
To test whether impaired glutamine synthesis alone was sufficient to cause increased Gap1p trafficking to the plasma membrane, we grew the gln1-9142 mutant in glutamate medium, a condition that should result in high glutamate levels but low glutamine levels. Like wild-type grown in glutamate, the gln1-9142 mutant grown in glutamate had very low Gap1p activity (Fig. 3A) and no detectable Gap1p at the plasma membrane (not shown). This result suggests that glutamine is not the sole effector for Gap1p sorting and that the intracellular concentrations of both glutamate and glutamine can affect the amount of Gap1p sorted to the plasma membrane.
Increased Glutamate Synthesis Decreases Gap1p Sorting to the Plasma Membrane.
The MKS1 gene has been shown to be required for the negative feedback inhibition of the RTG genes by glutamate, causing increased flux through the TCA cycle to make α-ketoglutarate, the precursor of glutamate (Fig. 2) (25–27). An mks1 mutant in the Σ1278b background has been reported to have ≈60% more glutamate than a wild-type Σ1278b strain (25). By amino acid analysis, we verified that, in our background, the mks1Δ strain has abnormally high endogenous levels of glutamate, glutamine, and total amino acids (Table 2). The mks1Δ mutants containing PADH1-GAP1-HA had extremely low Gap1p activity relative to wild-type during growth on ammonia (Fig. 3A) and lacked Gap1p at the plasma membrane (Fig. 3B). These data show that increased synthesis of glutamate results in decreased sorting of Gap1p to the plasma membrane.
Nonmetabolizable Amino Acids Affect Gap1p Sorting.
We wondered whether amino acids other than glutamate and glutamine might also influence Gap1p sorting. To test the effect of other amino acids on Gap1p sorting, we added amino acids to a final concentration of 0.5–3 mM to cultures expressing PADH1-GAP1-HA in ammonia medium and measured Gap1p activity 2 h later. We found that the addition of all 19 l-α-amino acids dramatically decreased Gap1p activity (Table 3). The addition of the imino acid proline also partially decreased Gap1p activity in this assay but other compounds that can be used as a source of nitrogen such as γ-aminobutyrate (GABA) and allantoin had little effect. Proline was also able to decrease Gap1p activity in a put1Δ mutant that cannot convert proline to glutamate (not shown), suggesting that proline itself has the capacity to decrease Gap1p activity in this assay. Surprisingly, even those amino acids such as cysteine and glycine that could not support yeast growth as a sole nitrogen source were able to decrease Gap1p activity with similar efficacy as glutamate and glutamine, which are readily used by the cell (Table 3).
Table 3.
Effect of amino acid or glutamate analog additions on Gap1p activity
| Amino acid or analog | Type of compound | Gap1p activity remaining 2 h after addition, % | Doubling time as a sole nitrogen source, h |
|---|---|---|---|
| No analog | 100 | 2.9 | |
| l-glutamic acid | l-α-amino acid | 3 | 3.2 |
| l-cysteine | l-α-amino acid | 2 | >20 |
| l-glutamine | l-α-amino acid | 2 | 2.7 |
| l-glycine | l-α-amino acid | 6 | >20 |
| l-histidine | l-α-amino acid | 1 | >20 |
| l-lysine | l-α-amino acid | 18 | >20 |
| l-serine | l-α-amino acid | 2 | 3.4 |
| l-leucine | l-α-amino acid | 1 | 5.7 |
| l-alanine | l-α-amino acid | 2 | 4.7 |
| l-aspartic acid | l-α-amino acid | 2 | 3.2 |
| l-phenylalanine | l-α-amino acid | 2 | 5.2 |
| l-arginine | l-α-amino acid | 8 | 2.9 |
| l-threonine | l-α-amino acid | 2 | 6.8 |
| l-tyrosine | l-α-amino acid | 2 | 5.6 |
| l-tryptophan | l-α-amino acid | 2 | 11.7 |
| l-valine | l-α-amino acid | 2 | 4.4 |
| l-isoleucine | l-α-amino acid | 2 | 6.1 |
| l-methionine | l-α-amino acid | 2 | >20 |
| l-asparagine | l-α-amino acid | 2 | 2.8 |
| l-citrulline | l-α-amino acid | 7 | 6.9 |
| l-ornithine | l-α-amino acid | 11 | 13.4 |
| l-proline | Imino acid | 32 | 6.7 |
| γ-Methylene-d,l-glutamic acid | l-α-amino acid | 10 | 10.3 |
| l-α-aminoadipic acid | l-α-amino acid | 3 | >20 |
| l-methionine sulfoximine | l-α-amino acid | 2 | >20 |
| l-methionine sulfoxide | l-α-amino acid | 19 | 9.4 |
| l-glutamic acid γ-ethyl ester | l-α-amino acid | 7 | 7 |
| β-Glutamic acid | β-Amino acid | 24 | >20 |
| γ-Aminobutyric acid | γ-Amino acid | 55 | 4.3 |
| Allantoin | — | 80 | 3.4 |
| N-methylaspartic acid | N-methyl amino acid | 97 | >20 |
| N-methylglutamic acid | N-methyl amino acid | 89 | >20 |
| l-glutamic acid amide | Amide | 92 | >20 |
Gap1p activity was measured by [14C]citrulline uptake in a PADH1-GAP1-HA strain (CKY759) treated with the indicated amino acid or analog. All amino acids or analogs were dissolved in minimal ammonia medium, adjusted to pH 4, and added to 3 mM in minimal ammonia medium, except that those labeled with * were assayed at 0.75 mM and with ‡ was assayed at 0.5 mM. Assays were performed at least twice, in duplicate. Doubling times were measured for wild-type (CKY443) at 24°C in the presence of 6.8 mM amino acid or analog, the concentration routinely used for glutamate or glutamine minimal medium. All media were adjusted to pH 4. Those labeled with
were assayed for growth rate in the presence of 3 mM analog for solubility reasons and/or to allow comparison between the analogs shown in Figs. 4 and 5B. Also, the sample labeled with
was assayed for growth rate with 2 mM tyrosine for solubility reasons.
, These were at least partially toxic at ≥3 mM. However, cysteine is not toxic at the concentration shown, 0.75 mM.
This lack of correlation between the ability of a compound to act as a nitrogen source and its ability to influence Gap1p activity indicated that Gap1p activity might be regulated in response to a class of molecules, rather than to the nutritional status conferred by that nitrogen source. Accordingly, we examined the effect on Gap1p activity of a variety of glutamate analogs (28), which did not support growth or supported growth very poorly (Table 3, last column). As shown in Table 3, addition of analogs that preserve the l-α-amino acid moiety but that had modified side chains resulted in a significant decrease in Gap1p activity. In contrast, analogs such as N-methylaspartic acid, N-methyl-glutamic acid, and l-glutamic acid amide, with modifications of the amino acid moiety, had little or no effect on Gap1p, possibly because they may not be taken up by the cells.
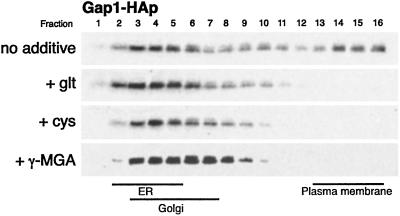
To verify that these amino acids and analogs were affecting Gap1p sorting, we examined in more detail the effects of two compounds, γ-methyleneglutamic acid (γ-MGA) and cysteine (Table 3). γ-MGA, cysteine, and glutamate also decreased proline uptake to a similar extent, suggesting that Put4p trafficking also responded to the same signals (not shown). Membrane fractionation showed that the addition of glutamate, cysteine and γ-MGA caused the loss of Gap1p at the plasma membrane (Fig. 4). Also, the total glutamate, glutamine, and amino acid levels in cells treated with cysteine or γ-MGA for 2 h were similar to the corresponding levels in untreated cells, suggesting that indeed neither cysteine nor γ-MGA was metabolized to any significant extent (not shown). These results confirmed that glutamate analogs and amino acids that cannot be used by the cell are able to signal Gap1p sorting to the vacuole, and imply that the amino acids or analogs themselves might act as signals for the the vacuolar sorting of Gap1p.
Fig 4.
Addition of the nonmetabolizable amino acid cysteine or glutamate analog γ-MGA results in less Gap1p localized at the plasma membrane. A wild-type PADH1-GAP1-HA strain (CKY759) was grown on minimal ammonia medium to exponential phase at 24°C. Glutamate, cysteine, or γ-MGA was added to 0.75 mM and, after 2 h, cells were harvested for extract preparation and fractionation on 20–60% sucrose density gradients.
Amino Acids Affect the Internal Trafficking of Gap1p, Not Its Endocytosis.
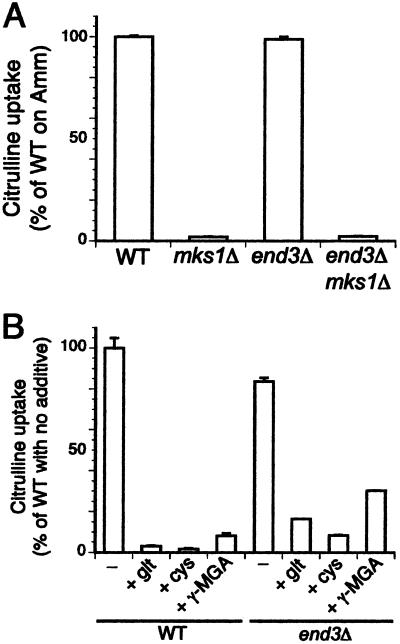
During growth on glutamate medium, Gap1p is sorted intracellularly to the vacuole (8). To verify that mks1Δ and amino acid or analog treatment affects an internal sorting event rather than endocytosis, we examined Gap1p activity in the presence and absence of end3Δ, a mutation that slows endocytosis of a variety of plasma membrane proteins (29). The end3Δ mutation did not increase the Gap1p activity of an mks1Δ mutant, indicating that in the mks1Δ strain Gap1p is sorted to the vacuole by an internal route (Fig. 5A). An end3Δ strain treated with glutamate, cysteine, or γ-MGA exhibited a significant decrease in Gap1p activity, indicating that even when endocytosis is blocked, addition of amino acids can redirect newly synthesized Gap1p to the vacuole (Fig. 5B). The end3Δ strains did exhibit higher residual Gap1p activity than wild-type after treatment with amino acids. This difference can be attributed to the expected stablization by end3Δ of Gap1p that had already been present at the plasma membrane.
Fig 5.
Deletion of mks1 and the addition of amino acids to growth media primarily affect the internal trafficking of Gap1p, not its endocytosis. (A) Wild type (CKY759), mks1Δ (CKY763), end3Δ (CKY765), and end3Δ mks1Δ (CKY766) were grown to exponential phase in minimal ammonia medium and assayed for Gap1p activity by [14C]citrulline uptake. (B) Wild type (CKY759) and end3Δ (CKY765) were grown to exponential phase and treated with 0.75 mM of the indicated amino acid or analog for 2 h before measurement of Gap1p activity by [14C]citrulline uptake. All strains contained PADH1-GAP1-HA.
We also examined the effect of a sec6–4 mutation on the stability of the Gap1p protein in a mks1Δ strain, and in the amino acid- or analog-treated wild-type strain. Because Sec6p is required for the fusion of post-Golgi secretory vesicles with the plasma membrane, any Gap1p that was en route to the plasma membrane should be trapped in secretory vesicles in a sec6 block and thus be protected from being degraded in the vacuole. Pulse–chase labeling of Gap1p in an mks1Δ mutant, and in cells treated with glutamate, cysteine, or γ-MGA, showed that a sec6 block did not change the rate of Gap1p degradation in any of these conditions (not shown). These data, taken together with the results from the experiments with end3Δ, indicate that the mks1Δ mutation and the amino acid additions primarily affect the internal sorting of Gap1p to the vacuole.
The TOR Pathway Does Not Play a Significant Role in Gap1p Sorting.
The TOR pathway has been proposed to link nutrient signals to the control of cell growth (30). Treatment of cells with rapamycin, a potent inhibitor of the TOR proteins, has been shown to activate the transcription of nitrogen-regulated genes such as GAP1 as well as many other genes (31–33), effects similar to those produced by nitrogen starvation (30).
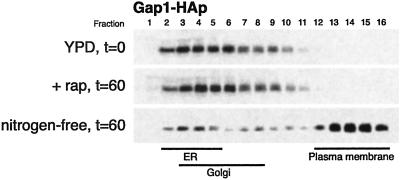
To determine whether the TOR pathway is involved in Gap1p sorting, we examined Gap1p sorting in cells containing PADH1-GAP1-HA that had been treated with rapamycin for 1 h. As a control for the effects of nitrogen starvation, we transferred a portion of the same culture to nitrogen-free media for 1 h. We found that rapamycin treatment for 1 h does not increase Gap1p activity, although GAP1 transcription, as measured by PGAP1-LacZ production, was strongly induced by rapamycin in this strain (Table 4). Similar results were seen after rapamycin treatment for 30 min (not shown). Furthermore, rapamycin treatment did not cause Gap1p to be localized to the plasma membrane (Fig. 6). In contrast, transferring the same culture to nitrogen-free media for 1 h caused a large increase in Gap1p activity, GAP1 transcription, and the amount of Gap1p localized to the plasma membrane (Table 4 and Fig. 6). Therefore, the TOR pathway does not appear to have a direct effect on Gap1p sorting. To the Gap1p trafficking machinery, rapamycin treatment is not equivalent to nitrogen starvation, and a different pathway likely regulates the nitrogen-dependent sorting of Gap1p.
Table 4.
Effect of rapamycin or nitrogen starvation on Gap1p activity and PGAP1-LacZ transcription
| Gap1p activity, pmol/min/OD600 | PGAP1-LacZ transcription, β-galactosidase units (×10−3) | |
|---|---|---|
| YPD | 14 ± 5 | 161 ± 17 |
| YPD + rap (1 h) | 5 ± 2 | 9,820 ± 322 |
| YPD → nitrogen-free (1 h) | 2,375 ± 95 | 6,175 ± 455 |
A strain containing PADH1-GAP1-HA and PGAP1-LacZ-URA3 (CKY760 + pMS29) was grown in yeast extract/peptone/dextrose (YPD) to exponential phase, then treated with either 200 μg/liter rapamycin or empty drug vehicle (DMSO) for 1 h, or shifted to nitrogen-free medium for 1 h. Gap1p activity, assayed by [14C]citrulline uptake, and β-galactosidase activity were measured at least in duplicate.
Fig 6.
Rapamycin does not mimic the effect of nitrogen starvation on Gap1p trafficking. Gap1-HAp localization was examined in the same strains used in Table 4, a wild-type PADH1-GAP1-HA strain (ECY748) containing PGAP1-LacZ (pMS29) grown to exponential phase in yeast extract/peptone/dextrose, then subjected to one of the following growth conditions for 1 h: empty drug vehicle (DMSO); 200 μg/liter rapamycin; or nitrogen-free medium. Cell membranes were fractionated on 20–60% sucrose density gradients and the fractions were subjected to SDS/PAGE.
Discussion
We have shown that yeast cells respond to elevated concentrations of amino acids, resulting either from mutations that cause increased intracellular levels of amino acids (in particular, glutamate and glutamine) or from the addition of exogenous amino acids to the growth medium, by sorting Gap1p to the vacuole. Furthermore, amino acids and amino acid analogs that cannot be metabolized by the cell as nitrogen sources can also trigger Gap1p sorting to the vacuole.
The responses of yeast to growth on different nitrogen sources have been well documented, but thus far the signals governing these responses have not been well understood. Our finding that amino acids and amino acid-related compounds can act as the signal for sorting Gap1p to the vacuole, regardless of the ability of these compounds to be used, suggests that the Gap1p trafficking machinery responds to the presence of an amino acid-related compound, rather than to the overall nitrogen nutritional status in the cell. These results anticipate that one or more components of the Gap1p sorting machinery may have a binding site for an amino acid-like molecule as an allosteric effector, determining whether the permease is to be sorted to the vacuole or to the plasma membrane.
What might be the nature of the signal detected by the Gap1p sorting apparatus? The data in Table 3 implicate l-α-amino acids as the signal recognized by the sorting machinery. Compounds that are not l-α-amino acids may also be recognized because the addition of β-glutamic acid or the imino acid proline results in some Gap1p sorting to the vacuole. However, establishing the precise nature of the interaction between the sorting machinery and the signaling molecule may not be possible until the small-molecule receptor(s) have been identified.
The finding that amino acids regulate the activity of the Gap1p sorting apparatus gives a different perspective to an old hypothesis. Grenson and colleagues proposed that Gdh1p had a role in nitrogen regulation independent of its function in glutamate synthesis (2). They found that a mutation in gdh1 eliminated the repressive effect of ammonia on Gap1p and several nitrogen-regulated catabolic enzymes in a Σ1278b strain background and that this effect was not removed by the addition of proline, which can be converted into glutamate in a GDH1-independent manner. Also, they found the cellular levels of the substrates and other products of the Gdh1p-catalyzed reaction, α-ketoglutarate, NADP, and NADPH were similar in wild-type and gdh1 growing on ammonia. The hypothesis that Gdh1p is a regulatory molecule has since been contested by others, such as Cooper, Roon, and colleagues (3). Our results provide a simple explanation for the apparent regulatory role of Gdh1p on Gap1p activity. We argue that a gdh1 mutation exerts its effect, not because Gdh1p is a regulatory molecule itself, but because all amino acids can act as regulatory molecules for the Gap1p sorting machinery, and a gdh1 mutation diminishes the net synthesis of amino acids.
Our results shed light on another old observation, the “feedback inhibition” or “transinhibition” of the Gap1p permease (2, 34–36). The phenomenon of feedback inhibition was based on the finding that cells grown in the presence of an amino acid showed a marked decrease in the activity of the corresponding permease; for example, cells grown on histidine have very low histidine permease activity. Thus, for Gap1p, which can take up all amino acids, all amino acids were hypothesized to have a posttranslational inhibitory effect, by allosteric inhibition of the permease. Our results indicate that, rather than acting as allosteric effectors of the Gap1p permease itself, the exogenous amino acids instead act as allosteric effectors of the Gap1p sorting machinery, decreasing Gap1p activity.
What is responsible for transducing amino acid signals to the Gap1p sorting machinery? We find that rapamycin does not mimic the effect of nitrogen starvation on the Gap1p sorting apparatus and thus the TOR pathway does not directly regulate Gap1p sorting. We also considered the possibility that Gap1p itself may be the relevant amino acid sensor. To test this idea we took advantage of the fact that Put4p and Gap1p are regulated coordinately, and we found that both glutamate and cysteine could decrease Put4p activity even in a gap1Δ strain (data not shown). Finally, Ssy1p, a recently identified amino acid sensor (37–39), and Ure2p, the proposed glutamine sensor for the nitrogen-regulated transcriptional apparatus (40, 41), do not appear to be involved, because both ssy1Δ and ure2Δ strains exhibited a decrease in Gap1p activity when amino acids were added to the growth medium (data not shown). Further experiments are necessary to identify the proteins that transduce the amino acid signal to the Gap1p trafficking machinery.
What might be the physiological purpose for the decrease in Gap1p activity in response to amino acids? Because Gap1p scavenges amino acids for use as nitrogen sources, it seems counterintuitive that Gap1p would be down-regulated in response to amino acids, particularly those that are nonmetabolizable. However, we have preliminary evidence that amino acids are toxic to cells that have high unregulated permease activity (data not shown). We propose that the down-regulation of Gap1p by amino acids has evolved as a mechanism to prevent unchecked uptake of high concentrations of amino acids.
Acknowledgments
We thank Aaron Mitchell for strains and the Kaiser laboratory for helpful discussions and encouragement. This work was supported by a Howard Hughes Medical Institute predoctoral fellowship (to E.J.C.) and by National Institutes of Health Grant GM56933 (to C.A.K.).
Abbreviations
γ-MGA, γ-methyleneglutamic acid
HA, hemagglutinin
References
- 1.Magasanik B. & Kaiser, C. A. (2002) Gene 290, 1-18. [DOI] [PubMed] [Google Scholar]
- 2.Wiame J. M., Grenson, M. & Arst, H. N., Jr. (1985) Adv. Microb. Physiol. 26, 1-88. [DOI] [PubMed] [Google Scholar]
- 3.Cooper T. G. (1982) in The Molecular Biology of the Yeast Saccharomyces: Metabolism and Gene Expression, eds. Strathern, J. N., Jones, E. W. & Broach, J. R. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 39–99.
- 4.Bossinger J., Lawther, R. P. & Cooper, T. G. (1974) J. Bacteriol. 118, 821-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grenson M., Hou, C. & Crabeel, M. (1970) J. Bacteriol. 103, 770-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jauniaux J. C. & Grenson, M. (1990) Eur. J. Biochem. 190, 39-44. [DOI] [PubMed] [Google Scholar]
- 7.Stanbrough M. & Magasanik, B. (1995) J. Bacteriol. 177, 94-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberg K. J., Rowley, N. & Kaiser, C. A. (1997) J. Cell Biol. 137, 1469-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helliwell S. B., Losko, S. & Kaiser, C. A. (2001) J. Cell Biol. 153, 649-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Courchesne W. E. & Magasanik, B. (1983) Mol. Cell. Biol. 3, 672-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wach A., Brachat, A., Pohlmann, R. & Philippsen, P. (1994) Yeast 10, 1793-1808. [DOI] [PubMed] [Google Scholar]
- 12.Ljungdahl P. O., Gimeno, C. J., Styles, C. A. & Fink, G. R. (1992) Cell 71, 463-478. [DOI] [PubMed] [Google Scholar]
- 13.Adams A., Gottschling, D. & Kaiser, C., (1996) Methods in Yeast Genetics: A Laboratory Course Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 14.Kaiser C. A., Chen, E. J. & Losko, S. (2002) Methods Enzymol. 351, 325-338. [DOI] [PubMed] [Google Scholar]
- 15.Grenson M., Dubois, E., Piotrowska, M., Drillien, R. & Aigle, M. (1974) Mol. Gen. Genet. 128, 73-85. [DOI] [PubMed] [Google Scholar]
- 16.Magasanik B. (1992) in The Molecular Biology of the Yeast Saccharomyces Cerevisiae: Metabolism and Gene Expression, eds. Strathern, J. N., Jones, E. W. & Broach, J. R. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 283–317.
- 17.Dubois E., Grenson, M. & Wiame, J. M. (1973) Biochem. Biophys. Res. Commun. 50, 967-972. [DOI] [PubMed] [Google Scholar]
- 18.Dunlop P. C., Meyer, G. M. & Roon, R. J. (1980) J. Bacteriol. 143, 422-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grenson M. & Hou, C. (1972) Biochem. Biophys. Res. Commun. 48, 749-756. [DOI] [PubMed] [Google Scholar]
- 20.Messenguy F., Colin, D. & Ten Have, J.-P. (1980) Eur. J. Biochem. 108, 439-447. [DOI] [PubMed] [Google Scholar]
- 21.Ohsumi Y., Kitamoto, K. & Anraku, Y. (1988) J. Bacteriol. 170, 2676-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell A. P. (1985) Genetics 111, 243-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grenson M. (1983) Eur. J. Biochem. 133, 135-139. [DOI] [PubMed] [Google Scholar]
- 24.Mitchell A. P. & Magasanik, B. (1984) Mol. Cell. Biol. 4, 2758-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feller A., Ramos, F., Pierard, A. & Dubois, E. (1997) Yeast 13, 1337-1346. [DOI] [PubMed] [Google Scholar]
- 26.Dilova I., Chen, C. Y. & Powers, T. (2002) Curr. Biol. 12, 389-395. [DOI] [PubMed] [Google Scholar]
- 27.Sekito T., Liu, Z., Thornton, J. & Butow, R. A. (2002) Mol. Biol. Cell. 13, 795-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meister A., (1965) Biochemistry of the Amino Acids (Academic, New York).
- 29.Benedetti H., Raths, S., Crausaz, F. & Riezman, H. (1994) Mol. Biol. Cell. 5, 1023-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmelzle T. & Hall, M. N. (2000) Cell 103, 253-262. [DOI] [PubMed] [Google Scholar]
- 31.Hardwick J. S., Kuruvilla, F. G., Tong, J. K., Shamji, A. F. & Schreiber, S. L. (1999) Proc. Natl. Acad. Sci. USA 96, 14866-14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cardenas M. E., Cutler, N. S., Lorenz, M. C., Di Como, C. J. & Heitman, J. (1999) Genes Dev. 13, 3271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beck T. & Hall, M. N. (1999) Nature 402, 689-692. [DOI] [PubMed] [Google Scholar]
- 34.Crabeel M. & Grenson, M. (1970) Eur. J. Biochem. 14, 197-204. [DOI] [PubMed] [Google Scholar]
- 35.Woodward J. R. & Cirillo, V. P. (1977) J. Bacteriol. 130, 714-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woodward J. R. & Kornberg, H. L. (1981) Biochem. J. 196, 531-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iraqui I., Vissers, S., Bernard, F., de Craene, J. O., Boles, E., Urrestarazu, A. & Andre, B. (1999) Mol. Cell. Biol. 19, 989-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klasson H., Fink, G. R. & Ljungdahl, P. O. (1999) Mol. Cell. Biol. 19, 5405-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Didion T., Regenberg, B., Jorgensen, M. U., Kielland-Brandt, M. C. & Andersen, H. A. (1998) Mol. Microbiol. 27, 643-650. [DOI] [PubMed] [Google Scholar]
- 40.Courchesne W. E. & Magasanik, B. (1988) J. Bacteriol. 170, 708-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coschigano P. W. & Magasanik, B. (1991) Mol. Cell. Biol. 11, 822-832. [DOI] [PMC free article] [PubMed] [Google Scholar]