Abstract
The epidermal growth factor receptor (EGFR/ErbB) family of receptor tyrosine kinases plays fundamental roles in the regulation of cell survival, proliferation, and differentiation. Here, we present evidence that ErbB3 is degraded by proteasomes, and that Nrdp1 (referred to as FLRF in mice) associates with ErbB3 and stimulates its ubiquitination and degradation by proteasomes. Nrdp1 mRNAs are expressed in a variety of human tissues. The N-terminal half of Nrdp1 possesses an atypical RING finger domain, which is required for enhancing ErbB3 degradation. Its C-terminal half by itself associates with ErbB3 and raises ErbB3 levels in cells, probably by acting as a dominant–negative form of Nrdp1. In cell-free systems, Nrdp1 has ubiquitin ligase (E3) activity and ubiquitinates ErbB3, as well as itself, in the presence of the ubiquitin-carrier protein (E2), UbcH5. These data indicate that Nrdp1 is a RING finger-type of ubiquitin ligase, which promotes degradation of ErbB3 by proteasomes and, thus, may be an important factor influencing cell growth.
The epidermal growth factor (EGF) receptor family of receptor tyrosine kinases, which includes EGFR, ErbB2 (HER2/neu), ErbB3, and ErbB4, plays fundamental roles in the regulation of cell survival, proliferation, and differentiation in response to specific growth factors. Numerous studies have documented that overexpression or amplification of ErbB receptors is associated with the development of various types of human cancer (1, 2). Consequently, blocking the activation or accelerating the degradation of ErbB proteins is an attractive approach for cancer therapy (3). In fact, herceptin, an antibody that blocks ErbB2 activation, is widely used for treatment of breast cancer.
There are more than 10 EGF-like growth factors that bind the extracellular domains of ErbB receptors and cause the formation of receptor homo- and heterodimers. Dimerization eventually stimulates autophosphorylation of specific tyrosine residues in the cytoplasmic tails of the receptors, which act as docking sites for signaling molecules (4). The unique C-terminal domain of ErbB3 binds many important signal-transducing proteins, such as phosphatidylinositol (PI) 3-kinase and Shc (5, 6). In contrast to the other members of the ErbB family, ErbB3 lacks catalytic activity and only functions by heterodimerizing with other ErbBs. In fact, dimers of ErbB3 and ErbB2, which has no direct ligands, are the most potent in causing cell transformation (7–9).
Although extensive information is available concerning the activation of ErbB receptors, their down-regulation and degradation have received little study. Ubiquitination could be an important mechanism for facilitating the degradation of these receptors. In the ubiquitin-proteasome pathway, ubiquitin (Ub) is activated by the formation of a thiol ester linkage by the ubiquitin-activating enzyme (E1) and is then transferred to the active site Cys of a ubiquitin-carrier protein (E2). Formation of isopeptide bonds between the C terminus of Ub and lysines on a substrate is catalyzed by a ubiquitin ligase (E3), which binds the substrate and catalyzes the transfer of the Ub from a specific E2 to the substrate. The formation of a chain of Ub molecules on the substrate generally targets it for degradation by the 26S proteasome (10, 11). However, for membrane proteins, ubiquitination can also be critical for their internalization and can target them to the lysosome or yeast vacuole for degradation (12, 13). For example, EGFR has been shown to be degraded in lysosomes after ubiquitination by the ubiquitin ligase, Cbl (12).
Unlike most other membrane receptors, ErbB3 does not undergo degradation by lysosomes (12, 14). This paper presents evidence that ErbB3 is degraded by proteasomes and that this process is catalyzed by a ubiquitin ligase, Nrdp1 (for neuregulin receptor degradation protein-1). In a yeast two-hybrid screen, Nrdp1 was identified as a protein interacting with the intracellular portion of bovine ErbB3 (K. L. Carraway, III, personal communication; ref. 15). The mouse ortholog of Nrdp1, FLRF, has been identified as a gene differentially expressed in fetal and adult hematopoietic stem cells and progenitors (16). While this paper was in preparation, independent evidence was presented by Carraway and coworkers (15) that Nrdp1 can negatively regulate ErbB3 function.
Materials and Methods
Expression Vectors.
For mammalian expression, the cDNA encoding full-length or truncated Nrdp1 was amplified by PCR and subcloned into pRc/CMV or pcDNA3.1 (+) (Invitrogen) from NotI to ApaI site. During the PCR, the FLAG tag (DYDDDDK) was added to the C terminus of the protein. Mutagenesis of Nrdp1 was performed by using standard technology. All mutations were evaluated by DNA sequencing. Bovine ErbB3 was subcloned in pcDNA3.1 at the EcoRI site. Human ErbB3 and EGFR in pcDNA3 were kindly provided by Yosef Yarden (The Weizmann Institute of Science, Rehovot, Israel). To generate the GST-fusion protein, the full-length or the C-terminal half of Nrdp1 was subcloned into pGEX-4T-2 (Amersham Pharmacia) at EcoRI/XhoI sites.
Cells.
293T adenovirus-transformed human embryo kidney cells, COS-7 simian virus 40-transformed monkey kidney cells, and MDA-MB-453 and –468 human breast cancer cells were maintained in DMEM (GIBCO) supplemented with 10% (vol/vol) FBS and antibiotics in 5% CO2, whereas the DMEM was supplemented with 10% (vol/vol) calf serum for NIH/3T3 mouse embryonic cells. All of the above cell lines were obtained from the American Type Culture Collection. Transfection of the different cells was carried out by using the SuperFect transfection kit according to the manual (Qiagen, Valencia, CA).
Pulse–Chase Measurements of ErbB3 Degradation.
At 48 h posttransfection, 293T cells were incubated in serum-free medium for 45 min, metabolically labeled with 35S-Met and 35S-Cys (Amersham Pharmacia) for 2 h (pulse), and then chased in nonradioactive medium for different time periods. Protein extracts (0.3 mg) were immunoprecipitated with anti-ErbB3 (Labvision, Fremont, CA), separated on SDS/6% PAGE, and analyzed with PhosphorImager (Bio-Rad).
Ubiquitination Assay.
The ubiquitination assay was carried out in buffer containing 40 mM Tris (pH 7.5), 5 mM MgCl2, 0.5 mM DTT, and 2 mM ATP. Unless indicated otherwise, the buffer (20 μl in total volume) was supplemented with 20 ng of the purified human E1 expressed in Escherichia coli (a gift of Hongtao Yu, University of Texas Southwestern Medical Center, Dallas) or 1.2 μg of bacterial lysate containing wheat germ E1, 0.1 μg of the purified UbcH5, or 1.2 μg of bacterial lysate containing UbcH5 or UbcH8 (kindly provided by Peter Howley, Harvard Medical School), 0.2 μg of 125I-Ub or its K48R mutant, and 0.1 μg of GST-Nrdp1 in the absence or presence of ErbB3. The reaction mix was incubated at room temperature for 2 h or at 30°C for 90 min, resolved by SDS/PAGE, and analyzed with PhosphorImager. To assay ubiquitination of Nrdp1 and ErbB3 immunopurified from human cell lysates, these proteins were reimmunoprecipitated at the termination of the reaction and subjected to SDS/PAGE.
Results
ErbB3 Is Rapidly Degraded in Cultured Cells but Is Stabilized by Proteasome Inhibitors.
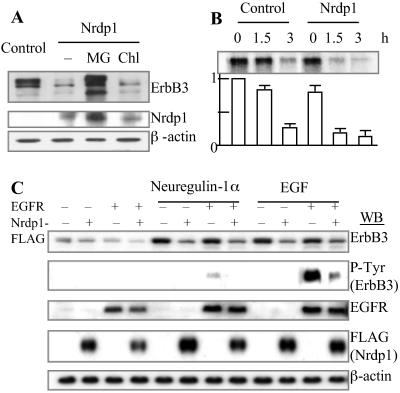
In 293T cells, the transfected ErbB3 appeared on the immunoblot primarily as a doublet (Fig. 1A), probably as a result of serine/threonine phosphorylation because the upper band disappeared after treatment with calf intestinal phosphatase (Fig. 1A Right) and was not detectable with an anti-phospho-tyrosine antibody (see below). The lysosomal inhibitor chloroquine had no significant effect on cellular levels of ErbB3 in the 293T cells, even though it did reduce in these cells the breakdown of EGFR induced by transfection of Cbl (data not shown), as previously demonstrated in Chinese hamster ovary cells (17). In contrast, the proteasome inhibitor MG132 (18) markedly increased the cellular levels of ErbB3 (Fig. 1A). Like most surface receptors, ErbB3 is a glycoprotein. It is noteworthy that MG132 dramatically elevated the levels not only of the glycosylated ErbB3, but also of a faster-migrating species (below the doublet), which appears to be a nonglycosylated form of ErbB3, because it was resistant to peptide:N-glycosidase F (PNGase F), a nonspecific N-glycosidase (Fig. 1B). In the same cells, the level of EGFR, which is degraded by lysosomes, was not affected significantly by the MG132 treatment (Fig. 1B and data not shown). Pulse–chase analysis showed that ErbB3 was degraded with a half-life of ≈2.5 h, and that MG132 or the more specific proteasome inhibitor, lactacystin (18), completely blocked ErbB3 degradation (Fig. 1C).
Fig 1.
Transfected ErbB3 is rapidly degraded in cells but is stabilized by proteasome inhibitors. (A) Stabilization of ErbB3 by proteasome inhibitors was shown by Western blot in the 293T cells transfected with ErbB3. (Left) At 48 h posttransfection, the cells were treated with 5 μM MG132 or 100 μM chloroquine for 14 h or left untreated. β-actin was a control for loading. (Right) In addition to the conditions in Left, immunoprecipitated ErbB3 was untreated or treated with calf intestinal phosphatase. (B) MG132 increased the levels of ErbB3 in both glycosylated and nonglycosylated forms but did not affect EGFR in 293T cells cotransfected with ErbB3 and EGFR. PNGase F treatment of the cell lysates was performed according to the manual (New England Biolabs) before electrophoresis. (C) The half-life of ErbB3 in 293T cells transfected with ErbB3 was assayed by pulse–chase analysis in the absence or the presence of 5 μM MG132 or 20 μM lactacystin. Numbers under the blot represent relative amounts of ErbB3 in the samples. Lysis of cells and/or immunoprecipitation were carried out in RIPA buffer (150 mM NaCl/0.1% SDS/0.5% sodium deoxycholate/1% Nonidet P-40/50 mM Tris⋅HCl, pH 8.0/1 mM DTT/0.1 mM phenylmethanesulfonyl fluoride). (D) The levels of tyrosine-phosphorylated ErbB3 were elevated by MG132. The 293T cells were cotransfected with ErbB3 and EGFR and treated or untreated with 5 μM MG132 for 17 h. Tyrosine phosphorylation was detected by anti-P-Tyr antibodies in Western blot analysis after immunoprecipitation by anti-ErbB3 antibodies.
Functional ErbB3 on the cell surface can dimerize with EGFR and be phosphorylated on tyrosines in the presence of the ligands of either ErbB3 or EGFR. When we cotransfected the 293T cells with ErbB3 and EGFR and treated them with MG132, there was a marked increase in the levels of ErbB3 that had undergone tyrosine phosphorylation (Fig. 1D). Thus, inhibition of proteasomes increased the levels of the surface ErbB3. Like many other membrane glycoproteins, newly synthesized ErbB3 undergoes N-linked glycosylation in the endoplasmic reticulum (ER). MG132 caused accumulation of nonglycosylated form of ErbB3 in addition to the glycosylated protein (Fig. 1B). In the ER-associated degradation pathway (ERAD), a glycoprotein in the ER can be retro-translocated into the cytosol and then be degraded by the 26S proteasome after deglycosylation (19, 20). These findings raise the possibility that some of the transfected ErbB3 is degraded before it exits from the ER.
Nrdp1 Associates with ErbB3 in Mammalian Cells.
The N-terminal half of Nrdp1 (amino acids 1–133; termed Nrdp1N) possesses a RING finger domain and two zinc finger domains, and its C-terminal half (residues 134–317; termed Nrdp1C) contains a predicted coiled-coil domain (Fig. 2A). The RING finger-containing proteins represent a large family of E3s. They recruit the E2s via their RING finger domains and appear to position the substrate and the E2s optimally for ubiquitin transfer (21–23). To determine whether Nrdp1 functions as an E3 for ErbB3 degradation, we first asked whether Nrdp1 interacts with ErbB3 in human cells. When ErbB3 and FLAG-tagged Nrdp1 were cotransfected into the 293T cells, the tagged Nrdp1, which appeared as a 36-kDa protein on the immunoblot, could be coimmunoprecipitated with ErbB3 (Fig. 2B). Mutation of two residues in the RING finger of Nrdp1 (C34S and H36Q) did not prevent its association with ErbB3. ErbB3 could also be coprecipitated with the C-terminal half of Nrdp1 but not with its N-terminal half (Fig. 2B). To confirm the association between ErbB3 and Nrdp1 further, a GST pull-down assay was performed by using lysates from MDA-MB-453 human breast cancer cells, which express high levels of ErbB3 and ErbB2. ErbB3, but not ErbB2, could be pulled down by GST-fused Nrdp1C (Fig. 2C). Taken together, these data demonstrate that Nrdp1 selectively associates with ErbB3 by means of its C-terminal half.
Fig 2.
Nrdp1 associates with ErbB3 through its C-terminal half in human cells. (A) Nrdp1 domains are schematically represented. (B) Coimmunoprecipitation of ErbB3 and Nrdp1 from 293T cell lysates. After immunoprecipitation by anti-FLAG antibodies (Sigma), the presence of ErbB3 was detected by Western blot by using anti-ErbB3 antibodies. 293T cells were transfected with either FLAG-tagged Nrdp1 (wild-type, mutant C34S/H36Q, or truncated form Nrdp1C or Nrdp1N), ErbB3, or both and then were treated with 5 μM MG132 for 16 h at 24 h after transfection. Lysis of the cells and immunoprecipitation were performed in the buffer containing 20 mM Tris⋅HCl, pH 8.0, 100 mM KCl, 0.2% Nonidet P-40, 10% (vol/vol) glycerol, 1 mM ZnCl2, 10 mM β-glycerophosphate, 5 mM tetrasodium pyrophosphate, 1 mM NaF, 1 mM NaVO3, and a protease inhibitor mixture (Roche Molecular Biochemicals). The input represented 1% of proteins in the lysates used for immunoprecipitation. (C) Pull-down of ErbB3 from MDA-MB-453 cells by GST-Nrdp1C. The cell lysates were incubated with GST-Nrdp1C or GST, resolved on SDS/6% PAGE, and analyzed by immunoblotting using anti-ErbB3 antibody. The input was the ErbB3 immunoprecipitated from the same amount of the lysates used for pulldown. Because of a low affinity of the protein A beads to the anti-ErbB3 antibodies, a smaller amount of ErbB3 was immunoprecipitated in the input. In control, similar experiments were performed by using anti-ErbB2 antibody.
Additional evidence that Nrdp1 and ErbB3 interact in vivo was obtained by staining transfected COS-7 cells with a rabbit antiserum against ErbB3 and a mouse anti-FLAG antibody (Fig. 8, which is published as supporting information on the PNAS web site, www.pnas.org). In the cells transfected with either ErbB3 or FLAG-tagged Nrdp1, these proteins were found both on the membrane and in the cytosol, but with distinct distributions. However, in the cells cotransfected with both ErbB3 and Nrdp1, these proteins were colocalized in the cytosol (Fig. 8).
Nrdp1 Promotes ErbB3 Degradation.
Because Nrdp1 binds ErbB3, we examined whether Nrdp1 functions as an E3 and stimulates ErbB3 degradation. The 293T cells were cotransfected with ErbB3 and Nrdp1. As shown in Fig. 3A, Nrdp1 strongly reduced ErbB3 accumulation, and this effect of Nrdp1 was prevented by MG132 but not by chloroquine. Although the levels of both species of ErbB3 decreased dramatically upon Nrdp1 transfection, the reduction of the upper band (which probably represents Ser/Thr-phosphorylated ErbB3) seemed to be more prominent. Pulse–chase analysis demonstrated that the effect of Nrdp1 on ErbB3 levels was caused by accelerated degradation of ErbB3, whose half-life decreased from ≈2.5 h to 0.7 h on transfection of Nrdp1 (Fig. 3B). Interestingly, Nrdp1 levels also were elevated by MG132 but not by chloroquine (Fig. 3A). Thus, Nrdp1 also appears to be degraded by proteasomes.
Fig 3.
Nrdp1 promotes ErbB3 degradation. (A) Nrdp1 reduces ErbB3 accumulation but not in the presence of the proteasome inhibitor MG132. 293T cells were transfected with plasmids encoding ErbB3 and/or FLAG-tagged Nrdp1 and incubated for 24 h. The cells were then incubated in fresh medium with none, 5 μM MG132, or 100 μM chloroquine for an additional 14 h. The cell lysates were analyzed by immunoblotting by using anti-ErbB3 (Neomarker) and anti-FLAG antibody. β-actin was used as a control for loading. (B) Nrdp1 potentiates ErbB3 degradation. 293T cells were transfected with ErbB3 in the absence or presence of FLAG-tagged Nrdp1. The cells were then pulse-labeled with [35S]Met and [35S]Cys and chased for the indicated periods of time. The bar graph under the blot indicates relative amounts of ErbB3 in the samples. (C) Nrdp1-mediated ErbB3 degradation is independent of receptor heterodimerization and ligand stimulation. 293T cells were transfected with ErbB3, EGFR, and FLAG-tagged Nrdp1 in the different combinations as indicated and then were untreated or treated with 100 nM neuregulin-1α or EGF for 3 h. The cell lysates were analyzed by immunoblotting by using anti-ErbB3, anti-EGFR, anti-FLAG, or anti-β-actin antibodies, whereas the immunoprecipitates by anti-ErbB3 were used for detection of phospho-Tyr by anti-P-Tyr.
ErbB3 usually forms heterodimers with other ErbB family members, e.g., EGFR. Because the 293T cells express very low levels of EGFR and no detectable ErbB3, these cells were used to test the effect of ErbB-EGFR dimerization on ErbB3 degradation. Cotransfection of EGFR led to tyrosine-phosphorylation of ErbB3 upon stimulation by neuregulin-1α or EGF, suggesting that at least a part of the transfected receptors have folded and are functional (Fig. 3C). Interestingly, both neuregulin-1α and EGF increased ErbB3 levels. However, regardless of the presence of EGFR or the ligands, cotransfection of Nrdp1 led to a reduction in ErbB3 levels, presumably by accelerating its degradation (Fig. 3C and data not shown). Thus, the Nrdp1-mediated ErbB3 degradation appears to be independent of either receptor heterodimerization or ligand stimulation.
To test whether Nrdp1 stimulates degradation of endogenous ErbB3, MDA-MB-468 cells, which express ErbB3 and EGFR, were transfected with FLAG-tagged Nrdp1 or its C-terminal half, Nrdp1C (see Fig. 4). The endogenous ErbB3 (red) was mainly localized on the plasma membrane, whereas Nrdp1 (green) was localized both in the cytosol and on the plasma membrane. Nrdp1 bears a Met-Gly sequence at its N terminus, a putative myristoylation site, which may account for its membrane localization. However, in the cells (indicated by arrowheads in Fig. 4) that were transfected with Nrdp1, the membrane-associated ErbB3 disappeared (Fig. 4 Top), but not in those transfected with its C-terminal half (Fig. 4 Middle). In contrast, the levels of the membrane-associated EGFR were not affected by the overexpression of Nrdp1 (Fig. 4 Bottom). In addition, transfection of Nrdp1 did not influence the levels of several other signaling proteins that were transfected into the cells, e.g., AKT, TRAF2, and IKKα, as analyzed by Western blots (Fig. 9, which is published as supporting information on the PNAS web site). Thus, Nrdp1 appears to promote degradation of endogenous and transfected ErbB3 selectively.
Fig 4.
Nrdp1 reduces levels of endogenous ErbB3. MDA-MB-468 cells were transfected with FLAG-tagged Nrdp1 or Nrdp1C. Localization of Nrdp1, Nrdp1C, ErbB3, or EGFR was visualized by confocal fluorescence microscopy after incubation with appropriate primary antibodies and FITC or Cy3 conjugates of secondary antibodies. Different images on each row, which are representative of many (>30) microscopic fields, are the same cells with different kinds of staining. The cells transfected with Nrdp1 or Nrdp1C are indicated by arrowheads. The nuclei of cells were visualized under UV light after staining with 4′,6-diamidino-2-phenylindole.
RING Finger Domain of Nrdp1 Is Critical for Degradation of ErbB3.
RING finger domains are defined by eight conserved Cys and His residues that coordinate two zinc ions, with a His in the fourth coordination site and either a Cys (C3HC4 RING) or a His (C3H2C3 RING) in the fifth position (20). The RING finger domain of Nrdp1 closely resembles those of Cbl and TRAFs, but the eighth conserved residue in the RING of Nrdp1 is Asp (D56) instead of the usual Cys (Fig. 5A). Interestingly, the RING finger of TRAF6 also has an Asp as its eighth residue. Thus, an Asp in this position may be functionally equivalent to a Cys. To explore whether the Asp in the RING finger is critical for degradation of ErbB3, D56 in Nrdp1 was mutated into V. As shown in Fig. 5B, both the D56V and C34S/H36Q mutants failed to stimulate ErbB3 degradation in the 293T cells. In NIH/3T3 cells, the mutations in the Nrdp1 RING finger (C34S-H36Q) also caused a loss of Nrdp1's capacity to promote ErbB3 degradation (Fig. 5B). Thus, the C3HC3D RING domain is essential for degradation of ErbB3. As expected, the N-terminal half of Nrdp1, which contains both the RING and zinc finger domains but does not bind ErbB3 (Fig. 1), did not stimulate ErbB3 degradation (Fig. 5B). It is noteworthy that the C-terminal half caused a dramatic accumulation of ErbB3 and was itself present at very high levels (Fig. 5B), probably by acting as a dominant–negative form of Nrdp1.
Fig 5.
The atypical RING finger domain of Nrdp1 is essential for promoting degradation of ErbB3. (A) The RING finger domains of Nrdp1, c-Cbl, hTRAF6, and hTRAF2 are aligned. The eight residues forming the RING finger domains are indicated with asterisks. Point mutations examined are indicated by arrowheads above the alignment. (B) The effect of the different mutations of Nrdp1 on degradation of ErbB3. 293T or NIH/3T3 cells were cotransfected with pcDNA3-ErbB3 and plasmids encoding different forms of FLAG-tagged Nrdp1 or empty vectors (control). The cell lysates were analyzed by immunoblotting by using anti-ErbB3 and anti-FLAG antibodies, respectively. β-actin was used as a control for loading.
Nrdp1 Is a Ubiquitin Ligase That Ubiquitinates both ErbB3 and Itself.
Protein ubiquitination requires the sequential action of E1, E2, and E3 (11). Because self-ubiquitination is a common characteristic of RING finger proteins that function as E3s (12, 20), we first studied the ability of GST-fused Nrdp1 to function both as an E3 and as a potential substrate in this assay. 125I-labeled ubiquitinated proteins were detected in a reaction mixture containing [125I]ubiquitin, GST-Nrdp1, E1, E2 (UbcH5), and ATP (Fig. 6A). All of these components were essential for this reaction. Because bacteria lack ubiquitination enzymes, these proteins were expressed in E. coli to eliminate any possible contaminating eukaryotic components. UbcH5, but not UbcH8, supported the Nrdp1-mediated ubiquitination. Other E2s tested, including UbcH7, E214K, E2F1, and Xenopus UbcX, also did not support this reaction (data not shown). In similar incubations, the 35S-labeled Nrdp1 isolated from the 293T cells also formed large conjugates with ubiquitin (Fig. 6B Left). This ability of Nrdp1 to ubiquitinate itself may account for its rapid degradation by proteasomes. To examine whether the RING finger domain is essential for this ligase activity, the RING finger mutant (C34S/H36Q) and the C-terminal half of Nrdp1 immunopurified from 293T cells were incubated in similar assays (Fig. 6B Right). Unlike the wild-type, neither mutant form could stimulate ubiquitination, indicating that the RING finger domain of Nrdp1 is required for Nrdp1 autoubiquitination.
Fig 6.
Nrdp1 is a ubiquitin ligase promoting ubiquitination of itself and ErbB3. (A) Nrdp1-catalyzed formation of [125I]ubiquitin conjugates was assayed in vitro. All of the components were expressed and purified from bacteria. (B) Autoubiquitination of Nrdp1 requires its RING finger domain. Wild-type, mutant (C34S/H36Q), and truncated Nrdp1 were immunopurified from the 293T cells transfected with their FLAG-tagged form and were used for in vitro ubiquitination assay. (Left) Nrdp1 was labeled in vivo with 35S. (Right) Ubiquitin was labeled with 125I. (C) Nrdp1 stimulates ErbB3 ubiquitination in vivo. The 293T cells were transfected with ErbB3, or cotransfected with ErbB3 and the wild-type or mutant (C34S/H36Q) Nrdp1-FLAG in the presence (Left) or the absence (Right) of pCMV-myc-Ub (kindly provided by Ron Kopito, Stanford University, Stanford, CA). ErbB3 ubiquitination was detected by Western blotting by using either anti-myc antibodies (Oncogene Research Products) or anti-Ub antibodies (Zymed) after immunoprecipitation by anti-ErbB3 antibodies. Arrows indicate 191-kDa markers. (D) Nrdp1 ubiquitinates endogenous ErbB3 in vitro. ErbB3 immunoprecipitated from the MDA-MB-453 cells was incubated with the lysates of 293T cells untransfected or transfected with wild-type, mutant (C34S/H36Q), or truncated Nrdp1-FLAG in the presence of [125I]ubiquitin (K48R mutant). The 125I-labeled ErbB3-ubiquitin conjugates on beads were separated on SDS/PAGE and analyzed by PhosphorImager after ubiquitination assays and precipitation from the reaction mix. Arrow indicates the 191-kDa marker.
Because Nrdp1 stimulated ErbB3 degradation, it was important to establish whether Nrdp1 causes ubiquitination of ErbB3. In the 293T cells transfected with ErbB3 and myc-tagged ubiquitin, cotransfection of Nrdp1 facilitated the formation of myc-tagged ubiquitin conjugates of ErbB3 (Fig. 6C Left). The conjugates of ErbB3 with endogenous ubiquitins could also be detected in the cells cotransfected with wild-type Nrdp1, but not with its RING finger mutant (Fig. 6C Right). Finally, we tested whether endogenous ErbB3 could also be ubiquitinated by Nrdp1. Substrates of the 26S proteasome are usually conjugated to a polyubiquitin chain in which successive ubiquitins are linked by K48-G76 isopeptide bonds (11). Because both Nrdp1 and ErbB3 could form high molecular weight ubiquitin conjugates, we took advantage of ubiquitin K48R mutant, which could not form these isopeptide bonds, to distinguish monoubiquitination of ErbB3 from Nrdp1 autoubiquitination. Thus, ErbB3 immunopurified from the MDA-MB-453 cells was incubated with 125I-labeled ubiquitin K48R mutants plus the lysates from the 293T cells transfected with Nrdp1. As shown in Fig. 6D, the wild-type Nrdp1, but not its RING finger mutant or C-terminal half, catalyzed formation of mono-ubiquitin conjugates at ≈200 kDa, suggesting that ErbB3 (180 kDa) is ubiquitinated at two or three sites. The addition of E2 (UbcH5c) stimulated this ubiquitination, suggesting that UbcH5 is an E2 for ErbB3 ubiquitination. In control, formation of the 45-kDa instead of the 200-kDa ubiquitin conjugates was detected from the FLAG-tagged Nrdp1 in a similar reaction (data not shown). Taken together, these data demonstrate that Nrdp1 is a RING finger-type of ubiquitin-protein ligase that, together with UbcH5, ubiquitinates ErbB3.
Nrdp1 mRNAs Are Expressed in Various Human Tissues.
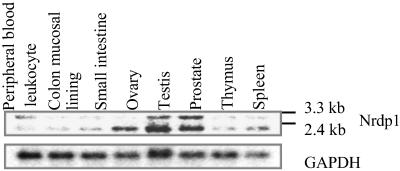
Because ErbB3 is involved in cell growth regulation, we analyzed the levels of Nrdp1 mRNAs in several human tissues with different levels of proliferative activity. In the Northern blot analysis using the cDNA encoding the full-length human Nrdp1 as the probe, two transcripts at 3.3 kb and 2.4 kb were detected in these tissues (Fig. 7). The expression of both mRNAs varied among tissues but was especially high in testis and prostate, whereas the 2.4-kb transcript was selectively expressed in ovary. Among these tissues, only the testis and colon mucosal lining are highly proliferative. Thus, the levels of Nrdp1 mRNAs do not correlate with the proliferative status of the tissues.
Fig 7.
Nrdp1 mRNAs are expressed in various human tissues. Human poly(A) RNAs from the indicated tissues on a nylon membrane (CLONTECH) were analyzed by hybridizing with 32P-labeled full-length ORF of Nrdp1 cDNA. GAPDH was a control for loading. The levels of mRNAs were detected by PhosphorImager.
Discussion
Because E3s determine both the specificity and timing of substrate ubiquitination, the regulation of their activities or interactions with substrates is a primary mechanism governing rates of protein degradation. This paper demonstrates that Nrdp1, through its RING finger domain, promotes ErbB3 ubiquitination and degradation in cultured cells. It seems very likely that endogenous Nrdp1 functions normally in ErbB3 degradation, because the substrate-binding region (the C-terminal half) of Nrdp1 raises the levels of ErbB3, probably by acting as a dominant–negative form of Nrdp1 (Fig. 5B). Nrdp1 appears to function by itself as an E3, because it catalyzes UbcH5-dependent ubiquitination in vitro. However, additional components or covalent modifications of the substrate or of Nrdp1 may influence ErbB3 ubiquitination by Nrdp1 in vivo, just as phosphorylation of both EGFR and c-Cbl upon stimulation by EGF is required for the ubiquitination of EGFR by c-Cbl (12).
Most E3s cause ubiquitination and degradation of multiple proteins. Although Nrdp1 is conserved in Drosophila (16), there is in Drosophila only one ErbB homolog (i.e., Drosophila EGFR), which has greater similarity to ErbB2 or EGFR than to ErbB3 (data not shown). Hence, Nrdp1 might target other proteins besides ErbB3 for degradation. We examined whether the other members of ErbB family are associated with or degraded by Nrdp1. Unlike ErbB3, endogenous ErbB2 did not associate with the C-terminal half of Nrdp1 (Fig. 2C) and EGFR was not degraded by Nrdp1 (Fig. 3). During preparation of this manuscript, Diamonti et al. (15) reported that when Nrdp1 and ErbB4 were transfected into COS-7 cells, Nrdp1 also reduced the levels of ErbB4, which may also be a substrate of this E3.
Receptor degradation can play a critical role in controlling receptor signaling. In contrast to many other growth factor receptors, ErbB3 does not undergo degradation by lysosomes (12, 14). Hence, it had been proposed that ErbB3 is destined primarily to enter the recycling pathway. However, we demonstrate here that transfected ErbB3 is rapidly degraded by proteasomes (Figs. 1 and 3). In the 293T cells transfected with ErbB3, MG132 treatment led to an accumulation of nonglycosylated ErbB3 in addition to the glycosylated form (Fig. 1B). Because some substrates of the ERAD pathway undergo deglycosylation and degradation by the 26S proteasome in the cytosol after retro-translocation from the ER, these findings suggest that ErbB3 can be degraded by this pathway before exit from the ER. On the other hand, MG132 caused a buildup of functional ErbB3 on the surface, i.e., molecules that were phosphorylated on tyrosines (Fig. 1D) and overexpression of Nrdp1 reduced the levels of endogenous surface ErbB3 in MDA-MB-468 cells (Fig. 4). Thus, some ErbB3 may be degraded by proteasomes after exit from the ER. Alternatively, the ubiquitination at ER influences the levels of surface receptor. It is noteworthy that the ERAD pathway not only serves as a quality control system but also can regulate protein levels (24, 25). By either mechanism, Nrdp1-mediated degradation can markedly influence the content of functional ErbB3.
Ligand-dependent down-regulation of tyrosine kinase receptors is believed to be a critical step for modulating their activity (26). Nrdp1-mediated degradation of ErbB3 is not regulated by EGF-like ligands because Nrdp1 promotes ErbB3 degradation whether or not its ligands are present (Fig. 3D). Nevertheless, the levels of Nrdp1 in cells seem to be regulated because its mRNA levels vary widely in different human tissues. Although no simple correlation between mRNA level and proliferative capacity is evident, the high levels of mRNAs in testis, prostate, and ovary suggest possible endocrine control (Fig. 7). In addition, Nrdp1 itself seems to be degraded by proteasomes, and its ubiquitination activity seems to promote its own degradation in cells (Figs. 3 and 6). Therefore, a possible role of Nrdp1 in growth regulation or tumor suppression, through its ability to reduce levels of ErbB3 or other key proteins, seems important to study. Interestingly, the human Nrdp1 gene, like erbB3, is located at 12q13, a chromosome region frequently rearranged in human tumors (16, 27). In unpublished studies, we have found that overexpression of Nrdp1 suppresses cell growth. The molecular basis for this effect and its possible physiological significance remain to be elucidated.
Supplementary Material
Acknowledgments
We thank Drs. L. C. Cantley, T. McGarry, K.-P. Lu, and E. Cadenas for advice and critically reading an earlier version of the manuscript, Dr. K. L. Carraway, III, for sharing unpublished data, and Drs. P. Howley, Y. Yarden, R. Kopito, H. Yu, and H. Band for reagents. This work was supported in part by grants and a postdoctoral fellowship from the National Institutes of Health and by a postdoctoral fellowship from the American Heart Association, New England Affiliate.
Abbreviations
EGF, epidermal growth factor
PI, phosphatidylinositol
Ub, ubiquitin
ER, endoplasmic reticulum
References
- 1.Slamon D. J., Clark, G. M., Wong, S. G., Levin, W. J., Ullrich, A. & McGuire, W. L. (1987) Science 235, 177-182. [DOI] [PubMed] [Google Scholar]
- 2.Gullick W. J. (1996) Cancer Surv. 27, 339-349. [PubMed] [Google Scholar]
- 3.Mendelsohn J. & Baselga, J. (2000) Oncogene 19, 6550-6565. [DOI] [PubMed] [Google Scholar]
- 4.Weiss F. U., Daub, H. & Ullrich, A. (1997) Curr. Biol. Gen. Dev. 7, 80-86. [DOI] [PubMed] [Google Scholar]
- 5.Soltoff S. P., Carraway, K. L., III, Prigent, S. A., Gullick, W. G. & Cantley, L. C. (1994) Mol. Cell. Biol. 14, 3550-3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prigent S. A. & Gullick, W. J. (1994) EMBO J. 13, 2831-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alimandi M., Romano, A., Curia, M. C., Muraro, R., Fedi, P., Aaronson, S. A., Di Fiore, P. P. & Kraus, M. H. (1995) Oncogene 10, 1813-1821. [PubMed] [Google Scholar]
- 8.Pinkas-Kramarski R., Soussan, L., Waterman, H., Levkowitz, G., Alroy, I., Klapper, L., Lavi, S., Seger, R., Ratzkin, B. J., Sela, M. & Yarden, Y. (1996) EMBO J. 15, 2452-2467. [PMC free article] [PubMed] [Google Scholar]
- 9.Waterman H., Alroy, I., Strano, S., Seger, R. & Yarden, Y. (1999) EMBO J. 18, 3348-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coux O., Tanaka, K. & Goldberg, A. L. (1996) Annu. Rev. Biochem. 65, 801-847. [DOI] [PubMed] [Google Scholar]
- 11.Hershko A. & Ciechanover, A. (1998) Annu. Rev. Biochem. 67, 425-479. [DOI] [PubMed] [Google Scholar]
- 12.Levkowitz G., Waterman, H., Ettenberg, S. A., Katz, M., Tsygankov, A. Y., Alroy, I., Lavi, S., Iwai, K., Reiss, Y., Ciechanover, A., et al. (1999) Mol. Cell 4, 1029-1040. [DOI] [PubMed] [Google Scholar]
- 13.Hicke L. & Riezman, H. (1996) Cell 84, 277-287. [DOI] [PubMed] [Google Scholar]
- 14.Waterman H., Sabanai, I., Geiger, B. & Yarden, Y. (1998) J. Biol. Chem. 273, 13819-13827. [DOI] [PubMed] [Google Scholar]
- 15.Diamonti A. J., Guy, P. M., Ivanof, C., Wong, K., Sweeney, C. & Carraway, K. L., III (2002) Proc. Natl. Acad. Sci. USA 99, 2866-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdullah J. M., Li, X., Nachtman, R. G. & Jurecic, R. (2001) Blood Cells Mol. Dis. 27, 320-333. [DOI] [PubMed] [Google Scholar]
- 17.Levkowitz G., Waterman, H., Zamir, E., Kam, Z., Oved, S., Langdon, W. Y., Beguinot, L., Geiger, B. & Yarden, Y. (1998) Genes Dev. 12, 3663-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kisselev A. F. & Goldberg, A. L. (2001) Chem. Biol. 8, 739-758. [DOI] [PubMed] [Google Scholar]
- 19.Tsai B., Ye, Y. & Rapoport, T. A. (2002) Nat. Rev. 3, 246-255. [DOI] [PubMed] [Google Scholar]
- 20.Wiertz E. J., Jones, T. R., Sun, L., Bogyo, M., Geuze, H. J. & Ploegh, H. L. (1996) Cell 84, 769-779. [DOI] [PubMed] [Google Scholar]
- 21.Lorick K. L., Jensen, J. P., Fang, S., Ong, A. M., Hatakeyama, S. & Weissman, A. M. (1999) Proc. Natl. Acad. Sci. USA 96, 11364-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng N., Wang, P., Jeffrey, P. D. & Pavletich, N. P. (2000) Cell 102, 533-539. [DOI] [PubMed] [Google Scholar]
- 23.Joazeiro C. A. P. & Weissman, A. M. (2000) Cell 102, 549-552. [DOI] [PubMed] [Google Scholar]
- 24.Wilhovsky S., Gardner, R. & Hampton, R. (2000) Mol. Biol. Cell 11, 1697-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fisher E. A., Zhou, M., Mitchell, D. M., Wu, X., Omura, S., Wang, H., Goldberg, A. L. & Ginsberg, H. N. (1997) J. Biol. Chem. 272, 20427-20434. [DOI] [PubMed] [Google Scholar]
- 26.Petrelli A., Gilestro, G., Lanzardo, S., Comoglio, P. M., Migone, N. & Giordano, S. (2002) Nature 416, 187-190. [DOI] [PubMed] [Google Scholar]
- 27.Zimonjic D. B., Rezanka, J., DiPaolo, J. A. & Popescu, N. C. (1995) Cancer Genet. Cytogenet. 80, 100-102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.