Abstract
Single base pair mutations that alter the function of tumor suppressor genes and oncogenes occur frequently during oncogenesis. The guardian of the genome, p53, is inactivated by point mutation in more than 50% of human cancers. Synthetic small inhibiting RNAs (siRNAs) can suppress gene expression in mammalian cells, although their degree of selectivity might be compromised by an amplification mechanism. Here, we demonstrate that a single base difference in siRNAs discriminates between mutant and WT p53 in cells expressing both forms, resulting in the restoration of WT protein function. Therefore, siRNAs may be used to suppress expression of point-mutated genes and provide the basis for selective and personalized antitumor therapy.
The p53 transcription factor has a key role in cellular responses to stress (1–5), and is the founding member of a family of three proteins that also includes p63 and p73. In cancer cells, mutant p53 both antagonizes WT protein function (6) and has intrinsic oncogenic properties (7–11). Indeed, mutant p53 has a dominant negative effect on WT p53 (6, 11) resulting in a p53 null phenotype. In addition, although WT p53 does not seem to interact with p63 (12), mutant p53 interacts with both p63 and p73, and might interfere with the functions of these proteins (13, 14). Therapeutic strategies targeting p53 (15, 16) should be specific for the mutant without altering WT p53 function, and ideally be broadly applicable to different mutations. The demonstration that small inhibiting RNAs (siRNAs) can be used to inhibit gene expression in mammals (17–19) opens new avenues for analysis of gene function and for gene-targeted therapies. siRNAs are short double-stranded RNAs (20) that are produced in various organisms by processing of long double-stranded RNA by the enzyme DICER (21) during RNA interference (20, 22). RNA interference was first characterized in plants, after the demonstration of the “cosuppression” phenomenon (23), and was extensively used in Caenorhabditis elegans for specific gene inhibition. A long doubled-stranded RNA corresponding to the sequence of interest is produced ectopically. This double-stranded species is “diced” by a protein complex that includes the RNase III DICER, and siRNAs are used by poorly characterized protein complexes as guides to direct mRNAs of corresponding sequences toward degradation. The degradation of target mRNA is initiated with the cleavage at a position corresponding to the center of the siRNA. In addition, in C. elegans (24) and possibly in Drosophila extracts (25), the antisense strand of siRNAs is thought to serve as a primer for a putative RNA-dependent RNA polymerase that would create long double-stranded RNA sequences from the 5′ end of the target mRNA. This double-stranded sequence would serve as a substrate for the enzyme DICER, resulting in an amplification process leading to loss of target selectivity. Introduction of long double-stranded RNAs in mammalian cells activates the IFN pathway and results in a general block of protein synthesis, precluding the use of this technology in these cells. The demonstration that the first steps of the RNA interference process could be by-passed, and that siRNAs could be directly used to inhibit specific gene targets, opened the possibility of applying this approach to mammalian cells (17). siRNAs have been used for various purposes (26, 27) and shown to discriminate between point mutant mRNA targets provided that the mutation is located at the center of the molecule (17). Thus, siRNAs can theoretically be used to understand and possibly eliminate oncogenic mutant proteins. Our aim here was to determine whether siRNAs could be used to target selectively an oncogenic mutation of p53 and thus form the basis for a customized antitumor treatment.
Methods
siRNA Oligonucleotides.
The oligonucleotides used in this study were purchased from Genset Oligos (Proligo, Boulder, CO); however, similar results were observed with oligonucleotides from MWG Biotech (Ebersberg, Germany). The oligonucleotides were annealed as described (17).
Cells and Transfection.
For all experiments, the cells were transfected with Lipofectamine the day after they were plated. H1299 cells (p53 null) were transfected with either WT or R248W mutant (mt) p53 expression vectors (400 ng), along with siRNAs (400 ng) as indicated. Cells were harvested 24 h later and analyzed by Western blot with the following antibodies: p53 (D01, Santa Cruz Biotechnology), actin (Sigma), tubulin (Sigma), or p21 (PharMingen). Equal loading was confirmed with Ponceau S staining. In cotransfection experiments in which different amounts of siRNA were used, the total amount of siRNA was kept constant by supplementing with control siRNA. U2OS cells (WT p53) were stably transfected with either an empty vector or a vector carrying the R248W cDNA. Cells were selected for 3 weeks in neomycin, and colonies were then pooled to avoid clonal variation. Stable transfectants were plated in 60-mm dishes (500,000 per dish) and transfected with siRNA (2 μg per dish). The next day, the cells were treated with doxorubicin (200 ng/ml) and harvested 48 h later. The cells were trypsinized and a portion was used for Western blot analysis, and the remainder for fluorescence-activated cell sorter analysis (FACS). For FACS, cells were fixed in 70% ETOH at −20°C overnight, washed with PBS and resuspended in PBS containing propidium iodide/RNase. MDAH087 were similarly transfected with mt1-siRNA or control siRNA. CasKi and SiHa cells were plated in 12-well dishes and transfected with siRNA (1.2 μg per well) targeting HPV16 E6.
Luciferase Assays.
H1299 cells were transfected with a mouse mdm2-promoter luciferase construct (28) as described (6), except that the indicated siRNA was included at a 1:1 ratio with the plasmid encoding mutant p53. All transfections also included a β-GAL expression vector as an internal transfection control. Luciferase activity was determined 24 h after transfection and normalized for β-galactosidase activity.
RT-PCR.
Quantitative RT-PCR was performed using a Light Cycler (Roche Molecular Biochemicals) according to the instructions of the manufacturer. The primers used were 5′-CCT ATG AGC CGC CTG AGG TT (forward), 5′-CGG AGA TTC TCT TCC TCT GT (reverse) for p53 and 5′-GGA CCT GAC CTG CCG TCT AGA A (forward), 5′-GGT GTC GCT GTT GAA GTC AGA G (reverse) for GAPDH. Results were calculated after standardization on GAPDH mRNA content.
Results and Discussion
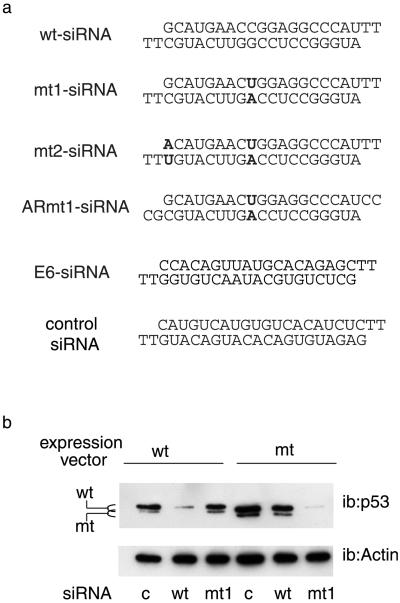
It was essential to determine the optimum conditions under which siRNAs could discriminate between WT and point-mutated molecules. Various siRNAs directed against WT p53 or the R248W mutant, a mutant that occurs at a high frequency (11), were compared for their efficiency and ability to discriminate between WT and mutant p53 (Fig. 1a; ref. 11). Because siRNAs harboring a single base pair difference in the center of the sequence were demonstrated to lack suppressive activity in vitro (17, 29), we designed the siRNAs so that the mutated base was the ninth nucleotide from the 5′ end (mt1-siRNA, Fig. 1a). An siRNA of irrelevant sequence (Fig. 1a) was used as a control in these experiments. siRNAs were transfected into p53 null cells (H1299) together with expression vectors for the corresponding proteins, and p53 expression was monitored by Western blot. WT-siRNA inhibited the expression of the WT protein, and had little effect on the mutant (Fig. 1b). Conversely, mt1-siRNA strongly inhibited the mutant protein, and showed high selectivity for the mutant in a range of doses (data not shown). At high doses, mt1-siRNA repressed the WT protein to some extent. This repression was not observed with an siRNA harboring an additional mutation at the 5′ end of the oligonucleotide (mt2-siRNA, Fig. 1a), which was slightly less efficient than mt1, but highly selective at all doses tested (data not shown). Taken together, these results demonstrate that siRNAs can be used to discriminate between the WT and mutant form of this protein.
Fig 1.
Selective inhibition of p53 by siRNAs. (a) Sequences of the siRNAs used in this study; mutated nucleotides are indicated in bold. (b) H1299 cells were transfected with expression vectors for WT (WT) or R248W mutant (mt) p53, together with 400 ng of control (c), WT (WT), or mutant siRNA (mt1), as indicated. Cells were harvested 24 h later and analyzed by Western blotting (ib, immunoblot) with the indicated antibodies.
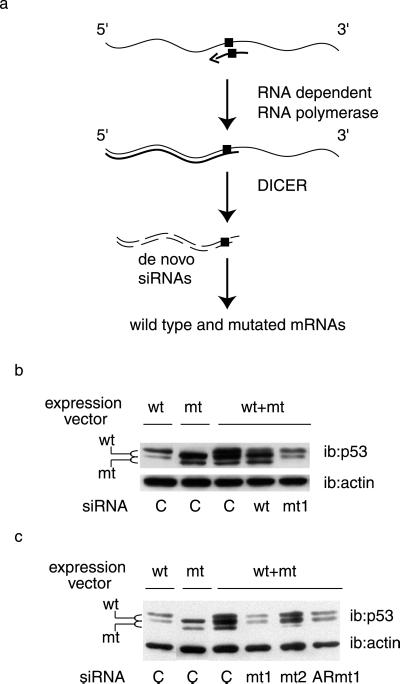
Our results in intact mammalian cells thus far confirm that synthetic siRNAs demonstrate high selectivity as demonstrated in vitro with Drosophila extracts (29), and in tissue culture cells (30). However, an amplification mechanism has been recently documented in C. elegans (24, 31). As of today, it is not known whether this mechanism is also conserved in mammals. This issue is important because, if this is the case, homologous pairing of the mutant siRNA with the mutant mRNA will result in the de novo production of siRNAs that recognize both the WT and mutant forms (Fig. 2a). Under these conditions, it would not be possible to discriminate between these two similar sequences (differing by one base) in heterozygous cells. This question can only be addressed by analyzing the effect of siRNAs when the two forms are coexpressed (Fig. 2a). We have therefore cotransfected WT and mutant p53 expression vectors with the siRNAs (Fig. 2b). The WT and mutant p53 proteins could easily be discriminated on the basis of their migration in SDS gels. Note that coexpression of the mutant protein induced a stabilization of the WT species, as documented in numerous studies (11). Transfection of WT targeting siRNA reduced the WT protein and had no effect on the mutant. We observed that in this cotransfection assay, the WT targeting siRNA did not completely suppress WT p53 protein, which might be because mutant p53 stabilizes the WT protein and thus counteracts the efficiency of the siRNA. Two siRNAs targeting the mutant protein were assayed in cotransfected cells, an siRNA entirely made of ribonucleotides (ARmt1, Fig. 1a) and an siRNA with deoxyribonucleotides (Ts) as protruding arms (mt1, Fig. 1a). Both siRNAs selectively and very efficiently suppressed the expression of the mutant protein. Reduction of the mutant protein restored WT p53 to levels equivalent to those observed on transfection of the WT protein alone (Fig. 2 b and c). These data argue against the existence of an amplification mechanism in mammalian cells, and demonstrate a good discrimination between the mutant and WT targets.
Fig 2.
Selectivity of mutant p53 inhibition. (a) Schematic model of a putative amplification process that would abolish the specificity of targeting point-mutated sequences: the antisense strand of the siRNA (arrow) is used as a primer by a putative RNA-dependent RNA polymerase for RNA strand elongation, resulting in a double-stranded 5′ portion of the target mRNA; this double-stranded sequence would in turn serve as a substrate for DICER which would result in siRNAs corresponding to the 5′ end of the target, which is common to both WT and mutant siRNAs. The square represents the single base mutation. (b and c) H1299 cells were transfected as in Fig. 1, with WT or R248W mutant (mt) expression vectors, alone or in combination, as indicated, along with the specified siRNAs, and analyzed by Western blotting 24 h later with the indicated antibodies.
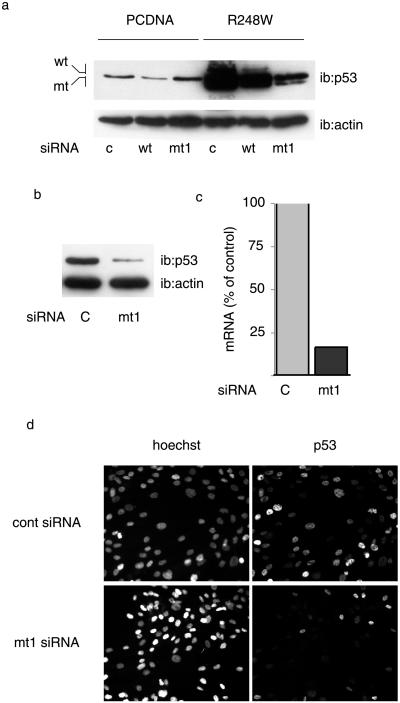
We next analyzed the suppression efficiency of siRNAs on mutant p53 expressed from an integrated genomic sequence. U2OS cells, which express WT p53, were stably transfected with an expression vector for the R248W p53 mutant or with the backbone vector (PCDNA) as a control. SiRNAs show a high level of transfectability in various cell types, and our results indicate that >90% of cells score positive when transfected with a fluorescent siRNA (data not shown). U2OS-derived cell lines were therefore transfected with siRNAs and analyzed for p53 expression (Fig. 3a). The WT-siRNA reduced, although it did not abolish, WT p53 expression in PCDNA-transfected cells, whereas mt1-siRNA had no effect. Expression of the mutant protein stabilized the expression of the WT protein (R248W cells), as we observed in transiently transfected cells and as documented (11). In these cells, transfection of mt1-siRNA strongly reduced the level of p53 protein, resulting in protein levels slightly higher than those observed in the control cells, that is, in the absence of mutant protein. Similar results were obtained with the MDAH087 cell line, derived from a Li–Fraumeni patient (32). Li–Fraumeni syndrome is due to monoallelic mutations in TP53 (33), and is characterized by a high incidence of cancer (34). MDAH087 were chosen because they express the R248W mutation, although introduction of exogenous nucleotidic sequences is difficult to achieve in this type of cells. Indeed, MDAH087 cells are poorly transfectable with plasmids and transfection with GFP reporter constructs generally results in a very low proportion of positive cells, whereas under the same conditions, >90% of the cells scored positive when transfected with an siRNA coupled to FITC. Transfection of these cells with mt1-siRNA resulted in a strong decrease in p53 expression, as shown by analysis of p53 protein (Fig. 3b) and mRNA (Fig. 3c). Analysis of p53 expression by immunofluorescence confirms that a strong suppression of mutant p53 protein exists in the majority of the cells (Fig. 3d). Thus, mt1-siRNA can be used to strongly reduce the level of endogenous mutant protein.
Fig 3.
Inhibition of endogenous mutant p53. (a) Stable transfectants were established from U2OS cells (expressing WT p53) by using a mutant expression vector (R248W) or the corresponding empty vehicle vector (PCDNA). Cells were analyzed by Western blotting with the indicated antibodies. (b–d) Fibroblasts from a Li–Fraumeni patient were transfected with siRNAs and analyzed for p53 expression by Western blotting (b), real-time RT-PCR (c), or immunofluorescence (d).
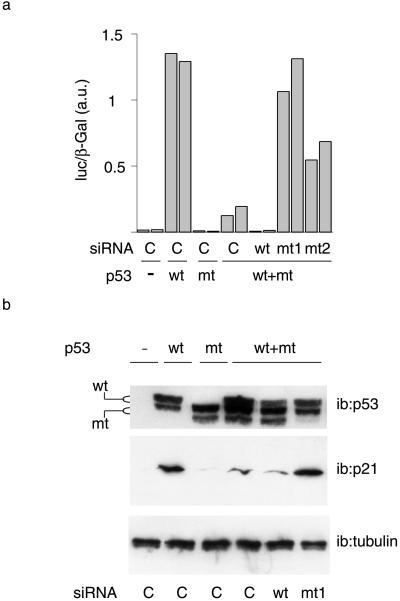
Mutant p53 has a dominant negative effect on WT p53. We examined the ability of mutant targeting siRNAs to restore WT p53 function. We first monitored the effect of siRNAs on the induction of a p53-responsive reporter construct carrying the mdm2 promoter (Fig. 4a). WT p53 strongly induced the reporter gene, and this induction was not observed with the mutant protein alone. Coexpression of WT and mutant proteins resulted in a dramatic decrease in the activation of the reporter gene, demonstrating the dominant negative effect of the mutant protein (Fig. 4a). As expected, the WT-siRNA abolished the residual reporter induction in cells cotransfected with WT and mt p53 expression vectors. In contrast, consistent with their ability to repress mutant p53 expression selectively, mt1- and mt2-siRNAs restored the induction of the reporter gene, although mt2-siRNA was slightly less efficient (Fig. 4a). Thus, mutant-targeting siRNAs restore WT activity in this model. In addition, these results confirm that the siRNAs selectively eliminate either WT or mutant protein. Similar results were observed when we monitored the expression of the proliferation inhibitor p21, an endogenous p53 target protein (35) involved in cell cycle arrest (36). Endogenous p21 was induced by WT but not mutant p53 (Fig. 4b). P21 expression was strongly decreased by cotransfection of mutant p53, demonstrating a strong transdominant negative effect in this system also. Wt-siRNA slightly decreased residual p21 induction. Most strikingly, the cotransfection of mt1siRNA resulted in a complete restoration of p21 induction. Thus, this siRNA significantly reduces or abolishes the negative transdominant effect of mutant p53 protein on the endogenous p21 promoter. Taken together, these data demonstrate that mt1-siRNA can restore WT p53 function by eliminating the mutant protein.
Fig 4.
Restoration of WT p53 function. (a) H1299 cells were transfected with both WT and mutant p53 vectors, siRNAs as indicated, and an mdm2-luciferase reporter construct along with a β-galactosidase construct as an internal control; cells were harvested 24 h later and extracts were assayed for luciferase activity. Transfections were performed in duplicate. Luciferase assays were performed in triplicate. Results are shown of two independent transfections from a representative experiment. (b) H1299 cells were transfected as in a and analyzed by Western blotting with the indicated antibodies.
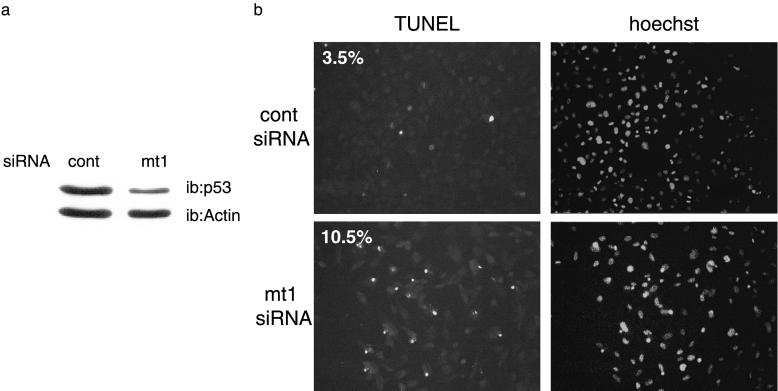
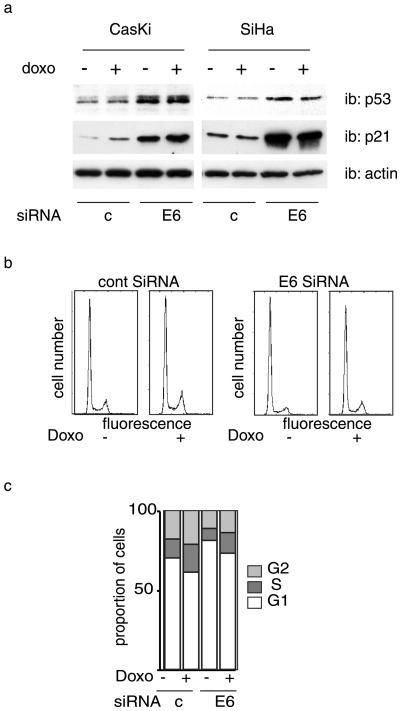
We next examined the effect of mt1-siRNA in immortalized Li–Fraumeni cells, a model that is more physiologically relevant. Whereas cell survival was not affected by the control siRNA, it was significantly decreased in cells treated with mt1-siRNA (data not shown). Furthermore, a proportion of surviving cells were undergoing apoptosis, as assessed by terminal deoxynucleotidyltransferase-mediated dUTP nick-end labeling (TUNEL) analysis (Fig. 5). P53 can be inactivated through several distinct mechanisms. In the majority of cervical cancers, p53 is inactivated by a viral protein encoded by the human papilloma virus (HPV) E6 protein (37). E6 promotes the ubiquitin-mediated degradation of p53 by the proteasome. In this case, reactivation of the p53 pathway can be achieved through inhibition of E6 protein expression. We examined whether siRNAs directed against the HPV E6 protein would be sufficient to reactivate p53 expression, and we also analyzed the phenotypic consequences. Transfection of CasKi and SiHA cells, (both HPV16 positive), with E6-targeting siRNA resulted in an increased level of p53 protein which was more pronounced for CasKI than for SiHa cells. (Fig. 6a). In both cases, the increase in p53 was accompanied by a strong induction of p21 protein (Fig. 6a), demonstrating without ambiguity the restoration of this p53-dependent pathway. Next, we assessed the effect of p53 stabilization and p21 induction on cell cycle and on response to chemotherapy. Cells were transfected with E6 siRNA and treated, or not, with the cytotoxic agent doxorubicin, a molecule that intercalates in the DNA and induces a p53-dependent response. E6 siRNA increased the proportion of cells in G1 (Fig. 6 b and c), which was also observed when the cells were treated with doxorubicin. However, despite a massive induction of p21, siRNAs did not completely arrest the cells. In addition, although the suppression of E6 with siRNA resulted in p53 protein accumulation, we did not observe any apoptosis (data not shown). Taken together, these results indicate that p53 expression can be restored in E6-expressing cells and provide an alternate means for p53 reactivation in specific cancer cells. However, elimination of HPV E6 in these cancer cells was not sufficient to induce programmed cell death. We are presently evaluating whether suppression of HPV E6 can sensitize HPV-positive cells to different types of chemotherapy.
Fig 5.
Immortalized Li–Fraumeni cells (MDAH087) were treated with control or mt1-siRNA as indicated. (a) Western blot analysis. (b) Terminal deoxynucleotidyltransferase-mediated dUTP nick-end labeling (TUNEL) analysis (proportions of TUNEL-positive cells are shown in percentages).
Fig 6.
Restoration of p53 function in E6-expressing cells. (a) CasKi and SiHa cells that express HPV E6 protein were transfected with indicated siRNAs, treated with doxorubicin, or left untreated for the controls and then analyzed by Western blotting with the indicated antibodies. (b and c) SiHA cells treated as in a were analyzed for cell cycle. The result of a typical experiment is shown.
Our results collectively demonstrate that siRNAs are powerful tools for oncogene inhibition in cancer cells. Synthetic siRNAs can first be used to analyze transformed cells and delineate the functions of mutated versions of oncogenic proteins. This aspect is of major interest in the p53 model, because mutant p53 is thought to have gain-of-function oncogenic activity that may be caused by inactivation of the p53 family members p63 and p73 (10), in addition to its interaction with cofactors that regulate the WT protein's function (7–9). Second and more importantly, siRNAs open new avenues in cancer therapy. They may provide a means for cancer prevention, or prolonging disease free survival in certain cases. For example, in Li–Fraumeni syndrome families that have one p53 allele mutated, we speculate that blocking mutant p53 would restore genome surveillance by WT p53, thereby decreasing the risk of cancer formation and decreasing the development of multiple malignancies. Furthermore, because mutant p53 is thought to promote chemotherapy resistance and angiogenesis, siRNAs targeting mutant p53 may be useful for restoring chemotherapy responsiveness, and potentially to reduce the aggressive nature of certain types of cancers (38). For this purpose, the use of synthetic siRNAs, as opposed to siRNAs expressed from a plasmid such as those recently described (39), may, in our opinion, have several advantages. In particular, the use of synthetic siRNAs that are chemically synthesized obviates the need to address the variety of technical and ethical problems raised by the use of expression vectors that, for example, must be prepared under GMP conditions. Furthermore, synthetic siRNAs should be more easy to use simultaneously in combination against various targets such as oncogenes and angiogenic factors than siRNAs derived from plasmids. Such combination therapies should prove valuable in preventing cancer cell mutations and cancer cell adaptation to treatment.
Acknowledgments
We thank Vasia Ogryzko and Linda Pritchard for critical reading of the manuscript. We also thank Christopher Neal, Jun Yang, Michael Tainsky, and Evelyne May for the kind gift of cell lines. This work was supported by grants from the Association pour la Recherche sur le Cancer, the European 5th Framework Program (QLG1-1999-00866), the National Cancer Institute (CA09212502), and the Philippe Foundation. L.A.M. was supported by a Chateaubriand fellowship.
Abbreviations
siRNAs, small inhibiting RNAs
mt, mutant
HPV, human papilloma virus
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Levine A. J. (1997) Cell 88, 323-331. [DOI] [PubMed] [Google Scholar]
- 2.Prives C. & Hall, P. A. (1999) J. Pathol. 187, 112-126. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B., Lane, D. & Levine, A. J. (2000) Nature 408, 307-310. [DOI] [PubMed] [Google Scholar]
- 4.Bargonetti J. & Manfredi, J. J. (2002) Curr. Opin. Oncol. 14, 86-91. [DOI] [PubMed] [Google Scholar]
- 5.Vousden K. H. (2002) Biochim. Biophys. Acta 1602, 47-59. [DOI] [PubMed] [Google Scholar]
- 6.Kern S. E., Pietenpol, J. A., Thiagalingam, S., Seymour, A., Kinzler, K. W. & Vogelstein, B. (1992) Science 256, 827-830. [DOI] [PubMed] [Google Scholar]
- 7.Dittmer D., Pati, S., Zambetti, G., Chu, S., Teresky, A. K., Moore, M., Finlay, C. & Levine, A. J. (1993) Nat. Genet. 4, 42-46. [DOI] [PubMed] [Google Scholar]
- 8.Harvey M., Vogel, H., Morris, D., Bradley, A., Bernstein, A. & Donehower, L. A. (1995) Nat. Genet. 9, 305-311. [DOI] [PubMed] [Google Scholar]
- 9.Joers A., Kristjuhan, A., Kadaja, L. & Maimets, T. (1998) Oncogene 17, 2351-2358. [DOI] [PubMed] [Google Scholar]
- 10.Di Como C. J., Gaiddon, C. & Prives, C. (1999) Mol. Cell. Biol. 19, 1438-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sigal A. & Rotter, V. (2000) Cancer Res. 60, 6788-6793. [PubMed] [Google Scholar]
- 12.Davison T. S., Vagner, C., Kaghad, M., Ayed, A., Caput, D. & Arrowsmith, C. H. (1999) J. Biol. Chem. 274, 18709-18714. [DOI] [PubMed] [Google Scholar]
- 13.Marin M. C., Jost, C. A., Brooks, L. A., Irwin, M. S., O'Nions, J., Tidy, J. A., James, N., McGregor, J. M., Harwood, C. A., Yulug, I. G., et al. (2000) Nat. Genet. 25, 47-54. [DOI] [PubMed] [Google Scholar]
- 14.Strano S., Munarriz, E., Rossi, M., Cristofanelli, B., Shaul, Y., Castagnoli, L., Levine, A. J., Sacchi, A., Cesareni, G., Oren, M. & Blandino, G. (2000) J. Biol. Chem. 275, 29503-29512. [DOI] [PubMed] [Google Scholar]
- 15.Bullock A. N. & Ferscht, A. R. (2001) Nat. Rev. 1, 68-76. [DOI] [PubMed] [Google Scholar]
- 16.Lane D. P. & Lain, S. (2002) Trends Mol. Med. 8, S38-S42. [DOI] [PubMed] [Google Scholar]
- 17.Elbashir S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- 18.Harborth J., Elbashir, S. M., Bechert, K., Tuschl, T. & Weber, K. (2001) J. Cell Sci. 114, 4557-4565. [DOI] [PubMed] [Google Scholar]
- 19.Caplen N. J., Parrish, S., Imani, F., Fire, A. & Morgan, R. A. (2001) Proc. Natl. Acad. Sci. USA 98, 9742-9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elbashir S. M., Lendeckel, W. & Tuschl, T. (2001) Genes Dev. 15, 188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernstein E., Caudy, A. A., Hammond, S. M. & Hannon, G. J. (2001) Nature 409, 363-366. [DOI] [PubMed] [Google Scholar]
- 22.Sharp P. A. (2001) Genes Dev. 15, 485-490. [DOI] [PubMed] [Google Scholar]
- 23.Vaucheret H. & Fagard, M. (2001) Trends Genet. 17, 29-35. [DOI] [PubMed] [Google Scholar]
- 24.Sijen T., Fleenor, J., Simmer, F., Thijssen, K. L., Parrish, S., Timmons, L., Plasterk, R. H. & Fire, A. (2001) Cell 107, 465-476. [DOI] [PubMed] [Google Scholar]
- 25.Lipardi C., Wei, Q. & Paterson, B. M. (2001) Cell 107, 297-307. [DOI] [PubMed] [Google Scholar]
- 26.Garrus J. E., von Schwedler, U. K., Pornillos, O. W., Morham, S. G., Zavitz, K. H., Wang, H. E., Wettstein, D. A., Stray, K. M., Cote, M., Rich, R. L., et al. (2001) Cell 107, 55-65. [DOI] [PubMed] [Google Scholar]
- 27.Novina C. D., Murray, M. F., Dykxhoorn, D. M., Beresford, P. J., Riess, J., Lee, S. K., Collman, R. G., Lieberman, J., Shankar, P. & Sharp, P. A. (2002) Nat. Med. 8, 681-686. [DOI] [PubMed] [Google Scholar]
- 28.Deffie A., Hao, M., Montes de Oca Luna, R., Hulboy, D. L. & Lozano, G. (1995) Mol. Cell. Biol. 15, 3926-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elbashir S. M., Martinez, J., Patkaniowska, A., Lendeckel, W. & Tuschl, T. (2001) EMBO J. 20, 6877-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kisielow M., Kleiner, S., Nagasawa, M., Faisal, A. & Nagamine, Y. (2002) Biochem. J. 363, 1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishikura K. (2001) Cell 107, 415-418. [DOI] [PubMed] [Google Scholar]
- 32.Bischoff F. Z., Yim, S. O., Pathak, S., Grant, G., Siciliano, M. J., Giovanella, B. C., Strong, L. C. & Tainsky, M. A. (1990) Cancer Res. 50, 7979-7984. [PubMed] [Google Scholar]
- 33.Malkin D., Li, F. P., Strong, L. C., Fraumeni, J. F., Jr., Nelson, C. E., Kim, D. H., Kassel, J., Gryka, M. A., Bischoff, F. Z., Tainsky, M. A., et al. (1990) Science 250, 1233-1238. [DOI] [PubMed] [Google Scholar]
- 34.Birch J. M. (1994) Eur. J. Cancer 13, 1935-1941. [DOI] [PubMed] [Google Scholar]
- 35.el-Deiry W. S., Tokino, T., Velculescu, V. E., Levy, D. B., Parsons, R., Trent, J. M., Lin, D., Mercer, W. E., Kinzler, K. W. & Vogelstein, B. (1993) Cell 75, 817-825. [DOI] [PubMed] [Google Scholar]
- 36.Waldman T., Kinzler, K. W. & Vogelstein, B. (1995) Cancer Res. 55, 5187-5190. [PubMed] [Google Scholar]
- 37.Mantovani F. & Banks, L. (2001) Oncogene 20, 7874-7887. [DOI] [PubMed] [Google Scholar]
- 38.Aas T., Borresen, A. L., Geisler, S., Smith-Sorensen, B., Johnsen, H., Varhaug, J. E., Akslen, L. A. & Lonning, P. E. (1996) Nat. Med. 2, 811-814. [DOI] [PubMed] [Google Scholar]
- 39.Brummelkamp T. R., Bernards, R. & Agami, R. (2002) Science 296, 550-553. [DOI] [PubMed] [Google Scholar]