Abstract
Myocardin is a SAP (SAF-A/B, Acinus, PIAS) domain transcription factor that associates with serum response factor (SRF) to potently enhance SRF-dependent transcription. Here we describe two myocardin-related transcription factors (MRTFs), A and B, that also interact with SRF and stimulate its transcriptional activity. Whereas myocardin is expressed specifically in cardiac and smooth muscle cells, MRTF-A and -B are expressed in numerous embryonic and adult tissues. In SRF-deficient embryonic stem cells, myocardin and MRTFs are unable to activate SRF-dependent reporter genes, confirming their dependence on SRF. Myocardin and MRTFs comprise a previously uncharacterized family of SRF cofactors with the potential to modulate SRF target genes in a wide range of tissues.
Serum response factor (SRF) is a MADS (MCM1, Agamous, Deficiens, SRF)-box transcription factor that regulates muscle-specific and growth factor-inducible genes by binding the DNA consensus sequence CC(A/T)6GG, known as a CArG box (ref. 1; reviewed in ref. 2). The spectrum of genes activated by SRF is dictated by its differential affinity for different CArG-box sequences (3) and its association with a variety of positive and negative cofactors, many of which are cell type-specific and signal-responsive (reviewed in ref. 4).
In addition to its role in proliferation and myogenesis, targeted inactivation of the mouse Srf gene has revealed a requirement of SRF in early embryogenesis and mesoderm formation (5, 6). SRF-deficient [Srf(−/−)] embryonic stem (ES) cells retain their ability to proliferate (6, 7) and provide a powerful system for identifying transcriptional programs that depend on SRF as well as for analyzing the potential requirement of SRF for the activities of its cofactors.
Recently we described a previously uncharacterized SRF cofactor called myocardin, which is expressed specifically in cardiac and smooth muscle cells (8). Myocardin associates with the MADS box of SRF through a basic and glutamine-rich domain; this interaction brings the powerful transactivation domain (TAD) of myocardin to CArG box-containing target genes with resulting transcriptional activation. Myocardin contains an SAF-A/B, Acinus, PIAS (SAP) domain, found in a variety of nuclear proteins involved in chromatin remodeling and gene expression (9). The SAP domain of myocardin is not required for association with SRF, but it provides specificity to target gene activation and is required for the activation of some genes but not others (8). Expression in Xenopus embryos of dominant negative mutants of myocardin that associate with SRF but lack transcriptional activity prevents heart formation, revealing an essential early role for myocardin in cardiac gene expression (8).
Because SRF regulates numerous growth factor-inducible genes that are expressed in cells in which myocardin is not expressed, we investigated whether myocardin-related proteins might modulate SRF activity outside the cardiovascular system. In the present study, we describe two myocardin-related transcription factors (MRTFs), referred to as MRTF-A and MRTF-B, that differentially stimulate SRF-dependent transcription. In contrast to myocardin, MRTF-A and -B are expressed in a wide range of embryonic and adult tissues. In Srf(−/−) ES cells, myocardin and MRTFs are unable to transactivate SRF-dependent promoters, confirming the obligate role of SRF as a mediator of transcriptional activities of these factors.
Materials and Methods
Bioinformatics and cDNA Cloning.
The mouse myocardin cDNA sequence was used to search NCBI databases to identify related genes. Several human, mouse, and Xenopus cDNA clones and ESTs with homology to myocardin were identified. These sequences were used as probes to screen cDNA libraries for full-length cDNAs. The gene structures of myocardin, MRTF-A, and MRTF-B were deduced from available mouse genomic sequences.
RNA Analysis.
Adult mouse multiple-tissue Northern blots (CLONTECH) were hybridized with cDNA probes encompassing the complete ORFs of MRTF-A and -B as described (8). For in situ hybridization, 3′-untranslated regions of MRTF-A and -B were transcribed in vitro in the presence of [35S]UTP to make antisense and sense (as a control) riboprobes. In situ hybridization was performed as described (10).
Transfection Assays.
SRF and myocardin expression constructs have been described (8). MRTF-A and -B cDNAs encoding full-length proteins or different deletion mutants were subcloned into the pcDNA3.1 expression vector (Invitrogen) in frame with a C-terminal Myc epitope tag. For GAL4 transfection experiments, full-length proteins or the TADs (residues 692–929 and 784–1080 of MRTF-A and -B, respectively) were fused in frame to the GAL4-(1–147) DNA-binding domain. Unless otherwise indicated, 100 ng of luciferase reporter and 100 ng of each activator plasmid were used. The total amount of DNA per well was kept constant by adding expression vector without a cDNA insert. Cytomegalovirus-lacZ was used as an internal control to normalize for variations in transfection efficiency. A retroviral SRF expression construct (pHeinz; D. Boos, O. Heidenreich, and A.N., unpublished data) was also used in some experiments using Srf(−/−) ES cells as indicated.
The SM22-luciferase construct contains the 1,434-bp promoter (11). The atrial natriuretic factor (ANF)-luciferase construct contains the 638-bp promoter (12). The 4xSM22 CArG-near-luciferase construct has been described (8). The (mSm)2-tk80-luciferase reporter (O. Heidenreich and A.N., unpublished data) contains the CArG-box sequence of the human c-fos promoter in which the flanking regions at the ternary complex factor (TCF) and the AP-1 binding sites were mutated by replacement with the nucleotides underlined, thereby maintaining a strong, centrally positioned SRF binding site: (CCTTACAACTAATGTCCATATTAGGACATCGTCGTACGCAT). The sequence was cloned, in tandem duplication, upstream of the tk80-luciferase reporter plasmid described previously (13). ES cell transfections used lipofectamine (Invitrogen), and associated luciferase assays were performed as described in ref. 13. The generation and maintenance of the 100 Srf(−/−) ES cell line has been described (6).
DNA-Binding Assays.
Gel-mobility shift assays were performed as described (3). An oligonucleotide corresponding to the SM22 CArG-far (11) sequences was used as a probe. Myocardin and MRTF-A and -B proteins were transcribed and translated in vitro with a TNT T7-coupled reticulocyte lysate system (Promega). The protein expression level was determined by Western blot analysis.
GST Protein-Binding Assays.
A cDNA encoding human SRF was cloned in frame to GST in the pGEX-KG vector (Amersham Pharmacia). GST-SRF fusion protein was expressed and purified as described (14). 35S-labeled myocardin and MRTF-A and -B were translated in a T7-coupled reticulocyte lysate system. For GST protein-binding assays, equal amounts of either GST-SRF or GST protein alone (as negative control) were incubated with myocardin, MRTF-A, or MRTF-B in GST binding buffer (20 mM Tris, pH 7.3/150 mM NaCl/0.5% Nonidet P-40/protease inhibitors) for 1 h at 4°C. After washing three times with GST binding buffer, proteins associated with GST-agarose beads were analyzed by 10% SDS/PAGE.
Results
A Family of MRTFs.
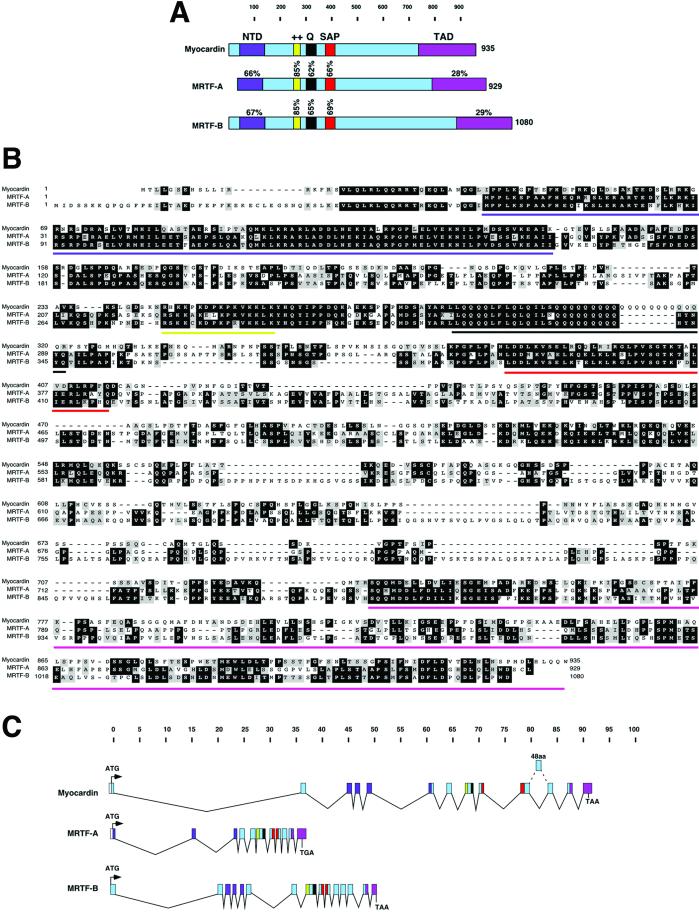
Full-length cDNAs encoding two myocardin-related proteins (MRTF-A and -B) were obtained by screening mouse embryo cDNA libraries using ESTs with homology to myocardin or applying a PCR-based cloning strategy. The protein structures of mouse myocardin and MRTF-A and -B are schematized in Fig. 1A. The overall amino acid identity between the three proteins is ≈35%, whereas they share >60% amino acid identity within the basic, glutamine (Q)-rich, and SAP domains (Fig. 1 A and B). Outside of these regions, homology among these three proteins is restricted to the N-terminal region and the C-terminal region, which functions as a TAD. It is also notable that the amino acid identity between MRTF-A and -B (42%) is greater than that between MRTFs and myocardin.
Fig 1.
Structure of the myocardin family of transcription factors. (A) Schematic diagrams of myocardin, MRTF-A, and MRTF-B proteins. ++, basic region; NTD, N-terminal domain; Q, glutamine-rich region. The number of amino acids in each protein is shown to the right, and percent identity between the indicated domains of each MRTF and myocardin is shown. (B) Amino acid sequence homology between myocardin and MRTFs. Colored bars correspond to the conserved regions shown in A. (C) Gene organization of mouse myocardin, MRTF-A, and MRTF-B. The colors of exons correspond to the regions shown in A. Dashed lines designate an alternative exon. Kilobases of genomic DNA are shown above the gene structures. Translation initiation (ATG) and termination codons (TAA and TGA) are indicated.
The human homologue of mouse MRTF-A was identified recently in a recurrent t (1, 22) translocation of acute megakaryocytic leukemia (M7) and was named MAL1/MKL1 (15, 16). We also identified a partial human cDNA sequence (KIAA 1243) that seems to be the human homologue of MRTF-B. The alignment of myocardin, MRTF-A, and MRTF-B from human, mouse, and Xenopus are provided in Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org.
In the course of characterizing the MRTF sequences, we discovered that myocardin contains an additional 128 amino acids N-terminal to the previously published sequence (Fig. 1 A and B). This region, which we call the N-terminal domain, shares >65% amino acid identity with MRTF-A and -B (Fig. 1 A and B).
The structures of the mouse myocardin, MRTF-A, and MRTF-B genes were also determined by comparing cDNA and genomic sequences (Fig. 1C). The protein-coding regions of myocardin, MRTF-A, and MRTF-B span ≈93, 37, and 50 kb on mouse chromosomes 11, 15, and 16, respectively.
Expression Patterns of MRTFs.
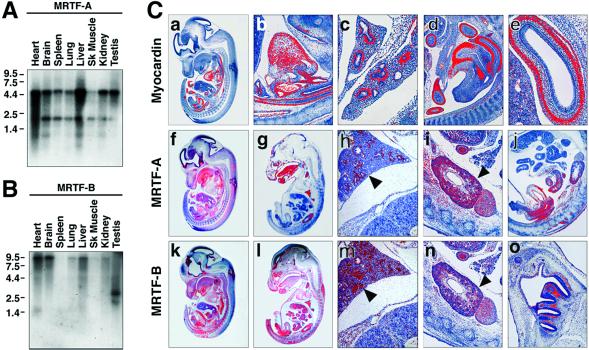
MRTF transcripts are present in a wide range of adult tissues (Fig. 2 A and B). MRTF-A has two major transcripts (≈4.5 and ≈2.5 kb) present in all tissues tested, with the most abundant expression in heart and liver. MRTF-B apparently has one major transcript of ≈9 kb with dominant expression in heart and brain. A transcript of ≈3 kb, which apparently represents an alternatively spliced form of MRTF-B, is also detected in testis.
Fig 2.
Expression patterns of myocardin and MRTFs in adult and embryonic tissues. (A and B) Northern blot analyses of MRTF-A and -B, respectively, in adult mouse tissues. (C) Expression of myocardin (a–e), MRTF-A (f–j), and MRTF-B (k–o) in mouse embryos as detected by in situ hybridization. Myocardin is expressed in cardiac and smooth muscle cells of an E13.5 embryo (a). A higher magnification of the same embryo showing myocardin expression in heart, smooth muscles of esophagus, and dorsal aorta (b), lung (c), bladder and small intestine (d), and stomach (e) is shown. MRTF-A is expressed in the tongue (f and g), lung and diaphragm (h), kidney (i), bladder (j), and colon of E13.5 (f) and E15.5 (g–j) mouse embryos. Expression of MRTF-B in E13.5 (k) and E15.5 (l–o) mouse embryos. Note higher-level expression present in the lung (m), kidney (n), and olfactory epithelium (o). Arrowheads in h and m point to the lung and in i and n to the kidney.
In contrast to the expression of myocardin, in heart and a subset of smooth muscle cells (8), MRTF-A and -B transcripts were detected throughout the embryo at embryonic day (E)10.5 (data not shown). By E13.5, MRTF-A continued to be expressed at a low level in most tissues, but higher expression was detected in a subset of neural mesenchymal cells, skeletal muscle of the tongue, and epithelial cells of the colon and small intestines (Fig. 2Cf). At E15.5, the expression of MRTF-A in the above tissues became more obvious (Fig. 2Cg). MRTF-A expression was detected also in epithelial cells of lung, kidney, bladder, and colon at this stage (Fig. 2C h–j).
Like MRTF-A, MRTF-B is expressed in epithelial cells of the lung, kidney, colon, and testis (Fig. 2C k–n). However, unlike MRTF-A, MRTF-B is expressed in the smooth muscle of the colon and small intestines. MRTF-B expression is also pronounced in mesenchymal cells adjacent to the olfactory epithelium (Fig. 2Co). Expression of MRTF-B in the developing lung differs from that of MRTF-A. Whereas the expression of MRTF-A is restricted to epithelial cells, MRTF-B seems to be expressed in both epithelial and mesenchymal cells (compare Fig. 2 Ch with Cm).
In light of the ability of myocardin to strongly induce the expression of SM22 (8), one of many smooth muscle genes that require pairs of CArG boxes for expression, we compared the expression of myocardin and MRTFs in developing smooth muscle cell lineages. At E13.5, myocardin is expressed in smooth muscle cells of the esophagus, dorsal aorta, lung, small intestine, bladder, and stomach (Fig. 2C b–e). The level of expression in the smooth muscle cells within these tissues is comparable to that in the developing heart. Myocardin expression declines in smooth muscle cells after birth, but continues to be detectable in aortic smooth muscle cells until adulthood (data not shown).
Stimulation of SRF-Dependent Transcription by MRTFs.
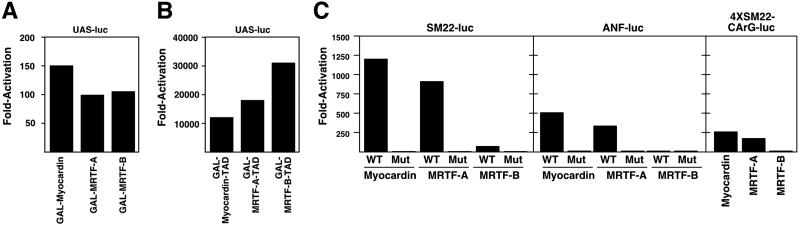
To assess the potential transcriptional activity of MRTF-A and -B, we fused their complete coding regions to the GAL4 DNA-binding domain and assayed their abilities to activate a GAL4-dependent luciferase reporter in transfected COS cells. In this assay, myocardin and both MRTFs showed an increase in transcriptional activity (Fig. 3A). The C-terminal regions of MRTF-A and -B were much more potent as transactivators than the full-length proteins (Fig. 3B), as observed with myocardin (Fig. 3B; ref. 8).
Fig 3.
Transcriptional activity of MRTFs. The complete ORFs (A) or TADs (B) of myocardin and MRTFs were fused to the GAL4 DNA-binding domain and tested for transcriptional activity by using a GAL4-dependent luciferase reporter (UAS-luc) in transfected COS cells. (C) Transactivation of luciferase reporters linked to SM22 or ANF promoters or four tandem copies of CArG-near from the SM22 promoter by myocardin and MRTFs, as indicated. WT refers to the wild-type promoter, and Mut refers to the promoters with mutations in the two CArG boxes. Values are presented as the fold activation of expression above the background level of expression of vector alone. All transfection assays were performed at least three times, and representative data are shown.
Because MRTF-A and -B share homology with the basic and Q-rich regions of myocardin, which interact with SRF (8), we tested whether they also could potentiate the activity of SRF using luciferase reporters linked to the SM22 and ANF promoters, both containing a pair of CArG boxes. MRTF-A and myocardin activated these reporters to similar levels (Fig. 3C). In contrast, MRTF-B was less effective in activating the SM22 reporter and showed almost no transcriptional activity with the ANF reporter despite the fact that MRTF-B was as potent as myocardin and MRTF-A when fused to the GAL4 DNA-binding domain. Similar to myocardin, MRTF-A and -B required the CArG boxes in the SM22 and ANF promoters for transcriptional activation, because these factors were unable to transactivate promoters with CArG box mutations (Fig. 3C). MRTF-A also activated a luciferase reporter containing the Elb minimal promoter and four tandem copies of an SM22 CArG-near to a level comparable to that of myocardin, whereas MRTF-B activated this reporter only to a minimal level (Fig. 3C). Similar to myocardin, MRTFs did not activate the c-fos promoter efficiently, which contains a single CArG box (data not shown). Myocardin, MRTF-A, and MRTF-B were expressed at comparable levels as determined by Western blot analysis of transfected cells (data not shown).
Myocardin and MRTFs Fail to Activate SRF-Dependent Transcription in Srf(−/−) ES Cells.
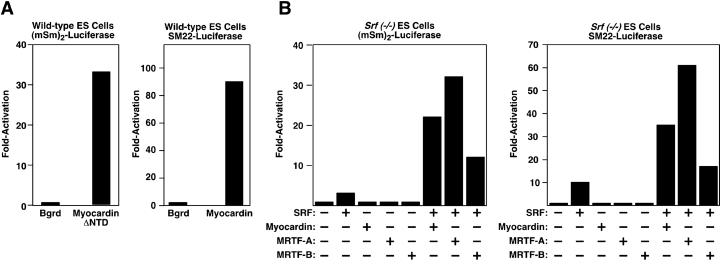
To further investigate the potential dependence of myocardin and MRTFs on SRF for transcriptional activity, we compared their abilities to transactivate SRF-dependent promoters in wild-type and Srf(−/−) ES cells. Myocardin, both full-length and the form lacking the N-terminal domain, activated expression of the SM22 promoter and an artificial reporter containing four tandem copies of the c-fos serum response element (SRE) in wild-type ES cells (Fig. 4A) but not in Srf(−/−) ES cells (Fig. 4B and data not shown). The MRTFs also were incapable of activating this reporter in Srf(−/−) ES cells, whereas introduction of SRF into Srf(−/−) ES cells with an SRF-expressing murine retroviral construct restored transcriptional activity to myocardin and MRTFs. These findings demonstrate that myocardin and MRTFs can potently activate CArG-box-dependent gene expression in a strictly SRF-dependent fashion.
Fig 4.
Lack of transcriptional activity of myocardin and MRTFs in Srf(−/−) ES cells. Wild-type (A) and Srf(−/−) (B) ES cells were transiently transfected with the SM22-luciferase and (mSm)2-luciferase reporters and expression plasmids encoding myocardin, MRTFs, and SRF as described in Materials and Methods. (A) Myocardin lacking the N-terminal domain (NTD) was used to transactivate (mSm)2-luciferase. Values are presented as the fold activation of expression above the background level of expression of vector alone. All transfection assays were performed at least three times, and representative data are shown.
Physical Interaction of SRF and MRTFs.
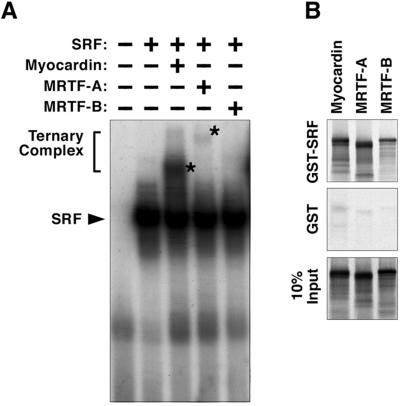
To determine whether MRTF-A and -B formed ternary complexes with SRF on DNA, we performed gel-mobility shift assays using in vitro-translated proteins and labeled probes corresponding to the SM22 CArG box. Ternary complex formation between myocardin and SRF was readily observed, whereas unexpectedly, ternary complex formation between MRTF-A and SRF was barely detectable, and complex formation between MRTF-B and SRF was undetectable under our assay conditions (Fig. 5A).
Fig 5.
Interaction of SRF and MRTFs. (A) Gel-mobility shift assays were performed with a 32P-labeled oligonucleotide probe for SM22 CArG-far and in vitro translation products of myocardin, MRTF-A, or MRTF-B in the presence and absence of SRF. Asterisks designate the position of the ternary complexes formed between SRF and myocardin or MRTF-A. The ternary complex between SRF and MRTF-A was very weak and between SRF and MRTF-B was undetectable. (B) GST-SRF protein interaction. Myocardin, MRTF-A and -B, translated in vitro with [35S]methionine, were incubated with either GST-SRF-agarose beads or GST-agarose beads as indicated. After washing, proteins associated with beads were separated on 10% SDS/PAGE and analyzed by autoradiography. One-tenth of the in vitro-translated proteins were also separated directly on the gel as a loading control.
Using a GST protein-binding assay, we found that GST-SRF interacts with myocardin, MRTF-A, and MRTF-B (Fig. 5B). At present we cannot reconcile the difference between the gel-mobility shift assay and the GST protein-binding assay. A likely explanation for this finding is that the gel-mobility shift assay is more stringent and requires a higher affinity complex than does the GST protein-binding assay. The fact that myocardin and MRTFs have different affinities for SRF may explain (at least partially) why they transactivate SM22 and ANF reporter genes differentially despite their similar transcriptional activities when fused to the GAL4 DNA-binding domain.
Discussion
Myocardin and MRTFs comprise a previously uncharacterized family of SRF cofactors with extraordinary transcriptional potency. Whereas myocardin is expressed in a cardiac- and smooth muscle-specific manner, MRTF-A and -B are widely expressed. Their different expression patterns and differential effects on SRF activity suggest that myocardin and MRTFs participate in distinct SRF-dependent programs of gene expression.
The Myocardin Family.
Myocardin and MRTFs share a common structural organization with conserved N-terminal, basic, Q-rich, and SAP domains. These proteins also contain TADs near their C termini, which are less conserved than these other domains (see Fig. 6). The mouse myocardin protein sequence reported here has an additional 128 amino acid residues at the N terminus of the previously published sequence (8). In all the assays we have performed thus far, these two forms of myocardin are functionally identical.
Myocardin and the MRTFs constitute a subclass of SAP domain transcription factors. The SAP domain is a conserved 35-aa motif that contains two amphipathic α-helices that resemble helices 1 and 2 of the homeodomain (reviewed in ref. 9). SAP domains are found in a variety of nuclear proteins including the nuclear matrix attachment factors SAF-A and -B (17, 18), Acinus, which is a target for caspase cleavage that participates in chromatin degradation during apoptosis (19), and PIAS (protein inhibitor of activated STAT), a transcriptional repressor that associates with a variety of transcription factors (20).
The SAP domains of SAF-A and PIAS interact with matrix attachment regions, which has been proposed to stimulate transcription by forming active domains of chromatin (20, 21). Myocardin also can bind matrix attachment regions through its SAP domain (D.-Z.W., Z.W., and E.N.O., unpublished data). A myocardin mutant lacking the SAP domain retains the ability to transactivate the SM22 promoter but is unable to activate the ANF promoter (8). Whether this differential requirement of the SAP domain of myocardin for transcriptional specificity reflects a role of matrix attachment region binding or other cofactor associations remains to be determined.
The Q-rich domain of myocardin is required for association with SRF (8), and the transcriptional potency of myocardin and MRTFs correlates with the length of the Q-rich domain. Q-rich domains have been identified in a variety of other transcription factors and are presumed to mediate interactions with other components of the transcriptional machinery (22, 23).
The human MRTF-A gene was reported to be translocated to chromosome 1 in the recurrent and specific t (1, 22) translocation in acute megakaryocytic leukemia (15, 16). This translocation creates a fusion protein with the human protein One-Twenty-Two (OTT)/RBM15 (RNA-binding motif protein-15). OTT belongs to a family of nuclear proteins that share homology in a putative RNA-binding motif (24, 25). Our finding that MRTF-A is a potent transcriptional coactivator of SRF raises the possibility that OTT-MAL may induce aberrant growth via SRF.
Potentiation of SRF Activity by Myocardin and MRTFs.
Our results show that myocardin and MRTFs are unable to activate SRF-dependent promoters in Srf(−/−) ES cells, confirming that SRF is an obligatory partner for these factors, at least on the SRF-dependent promoters tested, and probably others. Whether myocardin and MRTFs can cooperate with other transcription factors is an interesting question for the future.
In contrast to the high-affinity association between myocardin and SRF, the interaction of MRTFs and SRF is relatively weak and was detectable in a GST protein-binding assay but not in a DNA-binding assay. Because both MRTFs can transactivate CArG-box-dependent promoters and require SRF for this activity, we suggest that, similar to myocardin, MRTFs act through SRF to activate transcription. Consistent with this notion, dominant negative mutants of MRTFs can interfere with the activity of myocardin and vise versa (Z.W., S.L., and E.N.O., unpublished results), suggesting they may compete for the association with SRF.
Regulation of SRF Activity by Cofactor Interactions.
The activity of SRF is modulated through its interactions with a plethora of transcriptional cofactors. Among them, a family of Ets domain-containing proteins forms ternary complexes with SRF on the serum response element of the c-fos promoter (reviewed in ref. 4). This family of TCFs includes the ubiquitously expressed proteins SAP-1, SAP-2/Net (26, 27), and Elk-1 (28, 29). It is worth noting that despite their names, the latter proteins do not contain SAP domains. In addition to binding to the MADS domain of SRF, the TCF proteins can also bind directly to the Ets domain-binding consensus core motif GGA(A/T), which is adjacent to the CArG box in the c-fos promoter. Whether the association of SRF with myocardin/MRTFs and TCFs is mutually exclusive remains to be determined.
Given the tissue distribution of MRTF-A and -B expression as well as their differing affinities for SRF, it is reasonable to speculate that MRTF-A and -B may play different roles from that of myocardin. Considering that the function of TCF and SRF is signal-dependent (30, 31), it will be interesting to determine how cellular and/or extracellular stimuli regulate the functions of the myocardin family of transcriptional cofactors.
Supplementary Material
Acknowledgments
We thank R. Schwartz for reagents; J. Stark and C. Pomajzl for technical assistance; R. Bassel-Duby and D. Srivastava for comments on the manuscript; A. Tizenor for graphics; and J. Page for editorial assistance. This work was supported by grants from National Institutes of Health, Donald W. Reynolds Cardiovascular Clinical Research Center, Texas Advanced Technology Program, William G. McGowan Charitable Fund, and Robert A. Welch Foundation (to E.N.O.) and a grant from the Muscular Dystrophy Association (to D.-Z.W.) A.N. received support from Deutsche Forschungsgemeinschaft Grant SFB 446.
Abbreviations
SRF, serum response factor
ES, embryonic stem
TAD, transactivation domain
MRTF, myocardin-related transcription factor
ANF, atrial natriuretic factor
TCF, ternary complex factor
E, embryonic day
References
- 1.Norman C., Runswick, M., Pollock, R. & Treisman, R. (1988) Cell 55, 989-1003. [DOI] [PubMed] [Google Scholar]
- 2.Reecy J., Belaguli, N. & Schwartz, R. (1998) in Heart Development, eds. Harvey, R. & Rosenthal, N. (Academic, San Diego), pp. 273–290.
- 3.Chang P. S., Li, L., McAnally, J. & Olson, E. N. (2001) J. Biol. Chem. 276, 17206-17212. [DOI] [PubMed] [Google Scholar]
- 4.Treisman R. (1994) Curr. Opin. Genet. Dev. 4, 96-101. [DOI] [PubMed] [Google Scholar]
- 5.Arsenian S., Weinhold, B., Oelgeschlager, M., Ruther, U. & Nordheim, A. (1998) EMBO J. 17, 6289-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinhold B., Schratt, G., Arsenian, S., Berger, J., Kamino, K., Schwarz, H., Ruther, U. & Nordheim, A. (2000) EMBO J. 19, 5835-5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schratt G., Weinhold, B., Lundberg, A. S., Schuck, S., Berger, J., Schwarz, H., Weinberg, R. A., Ruther, U. & Nordheim, A. (2001) Mol. Cell. Biol. 21, 2933-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang D.-Z., Chang, P. S., Wang, Z., Sutherland, L., Richardson, J. A., Small, E., Krieg, P. A. & Olson, E. N. (2001) Cell 105, 851-862. [DOI] [PubMed] [Google Scholar]
- 9.Aravind L. & Koonin, E. V. (2000) Trends Biochem. Sci. 25, 112-114. [DOI] [PubMed] [Google Scholar]
- 10.Shin C. H., Liu, Z.-P., Passier, R., Zhang, C. L., Wang, D.-Z., Harris, T. M., Yamagishi, H., Richardson, J. A., Childs, G. & Olson, E. N. (2002) Cell 110, 725-735. [DOI] [PubMed] [Google Scholar]
- 11.Li L., Liu, Z., Mercer, B., Overbeek, P. & Olson, E. N. (1997) Dev. Biol. 187, 311-321. [DOI] [PubMed] [Google Scholar]
- 12.Hiroi Y., Kudoh, S., Monzen, K., Ikeda, Y., Yazaki, Y., Nagai, R. & Komuro, I. (2001) Nat. Genet. 28, 276-280. [DOI] [PubMed] [Google Scholar]
- 13.Janknecht R., Ernst, W., Pingoud, V. & Nordheim, A. (1993) EMBO J. 12, 5097-5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guan K. L. & Dixon, J. E. (1991) Anal. Biochem. 192, 262-267. [DOI] [PubMed] [Google Scholar]
- 15.Mercher T., Coniat, M. B., Monni, R., Mauchauffe, M., Khac, F. N., Gressin, L., Mugneret, F., Leblanc, T., Dastugue, N., Berger, R. & Bernard, O. A. (2001) Proc. Natl. Acad. Sci. USA 98, 5776-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma Z., Morris, S. W., Valentine, V., Li, M., Herbrick, J. A., Cui, X., Bouman, D., Li, Y., Mehta, P. K., Nizetic, D., et al. (2001) Nat. Genet. 28, 220-221. [DOI] [PubMed] [Google Scholar]
- 17.Gohring F., Schwab, B. L., Nicotera, P., Leist, M. & Fackelmayer, F. O. (1997) EMBO J. 16, 7361-7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kipp M., Gohring, F., Ostendorp, T., van Drunen, C. M., van Driel, R., Przybylski, M. & Fackelmayer, F. O. (2000) Mol. Cell. Biol. 20, 7480-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahara S., Aoto, M., Eguchi, Y., Imamoto, N., Yoneda, Y. & Tsujimoto, Y. (1999) Nature 401, 168-173. [DOI] [PubMed] [Google Scholar]
- 20.Liu B., Liao, J., Rao, X., Kushner, S. A., Chung, C. D., Chang, D. D. & Shuai, K. (1998) Proc. Natl. Acad. Sci. USA 95, 10626-10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sachdev S., Bruhn, L., Sieber, H., Pichler, A., Melchior, F. & Grosschedl, R. (2001) Genes Dev. 15, 3088-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saluja D., Vassallo, M. F. & Tanese, N. (1998) Mol. Cell. Biol. 18, 5734-5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Escher D., Bodmer-Glavas, M., Barberis, A. & Schaffner, W. (2000) Mol. Cell. Biol. 20, 2774-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiellettte E. L., Harding, K. W., Mace, K. A., Ronshaugen, M. R., Wang, F. Y. & McGinnis, W. (1999) Development (Cambridge, U.K.) 126, 5373-5385. [DOI] [PubMed] [Google Scholar]
- 25.Newberry E. P., Latifi, T. & Towler, D. A. (1999) Biochemistry 17, 10678-10690. [DOI] [PubMed] [Google Scholar]
- 26.Dalton S. & Treisman, R. (1992) Cell 68, 597-612. [DOI] [PubMed] [Google Scholar]
- 27.Price M. A., Rogers, A. E. & Treisman, R. (1995) EMBO J. 14, 2589-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rao V. N., Huebner, K., Isobe, M., ar-Rushdi, A., Croce, C. M. & Reddy, E. S. (1989) Science 244, 66-70. [DOI] [PubMed] [Google Scholar]
- 29.Hipskind R. A., Rao, V. N., Mueller, C. G., Reddy, E. S. & Nordheim, A. (1991) Nature 354, 531-534. [DOI] [PubMed] [Google Scholar]
- 30.Hill C. S., Marais, R., John, S., Wynne, J., Dalton, S. & Treisman, R. (1993) Cell 73, 395-406. [DOI] [PubMed] [Google Scholar]
- 31.Hill C. S., Wynne, J. & Treisman, R. (1995) Cell 81, 1159-1170. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.