Abstract
The Dnmt1o form of the Dnmt1 (cytosine-5)-methyltransferase enzyme is synthesized and stored in the cytoplasm of the oocyte and is used after fertilization to maintain methylation patterns on imprinted genes. After implantation of the blastocyst, Dnmt1o is replaced by the Dnmt1 form, which has an additional 118 aa at its amino terminus. To investigate functional differences between Dnmt1o and Dnmt1, mice were generated with a mutant allele, Dnmt1V, which synthesized Dnmt1o instead of Dnmt1 in all somatic cells. Homozygous Dnmt1V mice were phenotypically normal, and had normal levels of genomic methylation, indicating that Dnmt1o adopts the maintenance methyltransferase function of Dnmt1. Despite the apparent equivalence of Dnmt1o and Dnmt1 maintenance methyltransferase function in somatic cells, the Dnmt1o protein was found at high levels (with a corresponding high enzymatic activity) in Dnmt1V mice. In heterozygous Dnmt1V/+ embryonic stem cells and early embryos, equal steady-state levels of Dnmt1o and Dnmt1 proteins were produced from the Dnmt1V and the WT Dnmt1 alleles, respectively. However, in older embryos and adults, the Dnmt1V allele produced five times the steady-state level of protein of the WT Dnmt1 allele. The difference in Dnmt1o and Dnmt1 levels is due to a developmentally regulated mechanism that degrades the Dnmt1 protein. The intrinsic stability of the Dnmt1o protein is the most likely reason for its use as a maternal-effect protein; stable ooplasmic stores of Dnmt1o would be available to traffick into the nuclei of the eight-cell stage embryo and maintain methylation patterns on alleles of imprinted genes during the fourth embryonic S phase.
The Dnmt1 (cytosine-5)-methyltransferase catalyzes the addition of methyl groups to cytosine bases in DNA, and it is found in most, if not all, cells of the mammalian organism (1). Based on in vitro studies, Dnmt1 has a 5- to 30-fold preference for hemimethylated DNA substrates over unmethylated substrates, indicating that the main function of Dnmt1 is to maintain methylation patterns (2). There are two isoforms of Dnmt1, which are expressed in a sequential pattern during development (3, 4). The Dnmt1o protein, which has a relative molecular mass (Mr) of 175,000, is expressed during oocyte growth and maturation, and also during preimplantation development. The Mr 190,000 Dnmt1 form of the enzyme replaces Dnmt1o after implantation of the embryo (1). Dnmt1 has the same primary structure as Dnmt1o, except for the addition of a unique 118-aa domain at its amino terminus. Homozygous mutant offspring of mice that are heterozygous for Dnmt1 hypomorphic alleles have reduced levels of Dnmt1 protein and exhibit marked reductions in the level of genomic methylation, including reduction in the methylation associated with imprinted genes (5–7).
Dnmt1o is a maternal-effect protein that is synthesized in the growing oocyte, stored in the ooplasm of the mature M2 oocyte, and functions after fertilization to maintain DNA methylation patterns on alleles of imprinted genes (4, 8, 9). Dnmt1o is synthesized from an alternative oocyte-specific Dnmt1 promoter. This promoter is turned off after fertilization because Dnmt1o transcripts are not found in the embryo after the zygote stage (9). The oocytes of females that are homozygous for a targeted deletion of the oocyte-specific promoter lack Dnmt1o. Offspring of these homozygous Dnmt1Δ1o females exhibit a 50% reduction in the number of normally methylated alleles of imprinted genes, and most of them die during the later stages of fetal development (8). This observation is consistent with a role for Dnmt1o in maintaining methylation patterns at just one embryonic S phase. Moreover, it is consistent with the observation that oocyte-derived Dnmt1o trafficks to nuclei at just one cleavage stage of preimplantation development, namely the eight-cell stage.
When analyzed in vitro, there are no known functional differences between the essential maintenance methylating functions of the Mr 190,000 Dnmt1 protein and the Mr 175,000 Dnmt1o protein (3). That is, the Dnmt1o and Dnmt1 proteins appear to have equivalent maintenance methylation activities. The only difference in primary structure between Dnmt1o and Dnmt1 is in the 118-aa domain that is unique to the Dnmt1 protein. We might expect a functional difference between the two proteins, should there be one, to be mediated through this Dnmt1-specific domain. In this regard, the amino terminal domain of the human DNMT1 protein is known to interact with a transcriptional repressor protein DMAP1 (10). A specific interaction between the human DNMT1 and DMAP1 proteins (or between the mouse Dnmt1 and Dmap1 proteins) could possibly affect the maintenance methylation function of Dnmt1, but this is not known. To determine whether there are functional differences between Dnmt1o and Dnmt1, we generated mice with a mutant Dnmt1 allele, Dnmt1V, which synthesized Dnmt1o instead of Dnmt1 in all somatic cells.
Materials and Methods
Dnmt1 Gene Targeted Mutation.
Standard gene disruption methods were used to assemble the construct of Fig. 1 and to induce homologous recombination in mouse W9.5 embryonic stem (ES) cells (gift of C. Stewart). Excision of the HSV-tk/Pgk1-neo resistance cassette was done by electroporation of a Cre recombinase expression vector (pBS185, Invitrogen) into ES cells and selection of ES cell colonies resistant to 2 μM gancyclovir.
Fig 1.
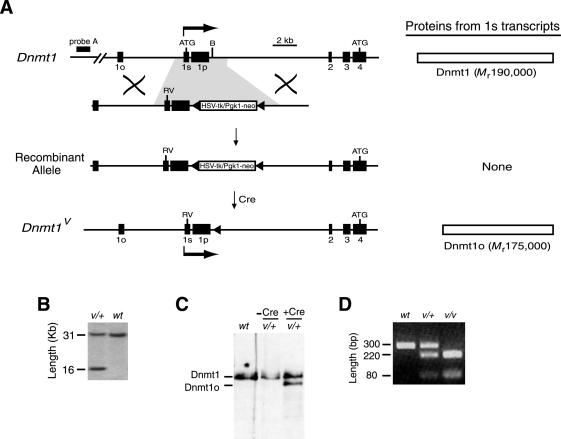
Expression of the Mr 175,000 Dnmt1o protein from the Dnmt1 1s promoter by mutating the translation initiation codon in exon 1s. (A) The transcripts for Dnmt1o and Dnmt1 (called Dnmt1o and Dnmt1, respectively) are normally produced from different promoters associated with exon 1o and exon 1s, respectively. The Dnmt1o first exon (exon 1o) and the Dnmt1 first exon (exon 1s) are both spliced into the common exon 2 (4). Exon 1p is an alternative first exon whose associated promoter is active in spermatocytes (4). The linear targeting construct contains a mutated ATG initiation codon in exon 1s and a loxP-flanked cassette of HSV-tk and Pgk1-neor genes at a BamHI site in the intron between exon 1p and exon 2. The loxP sites are filled triangles. After homologous recombination between the targeting construct and a WT Dnmt1 allele, and removal of the antibiotic-resistance genes by Cre-mediated excision, the mutant Dnmt1V allele is produced. The Dnmt1 protein is synthesized from 1s transcripts of the Dnmt1 allele, and the Dnmt1o protein is synthesized from 1s transcripts of the Dnmt1V allele. B, BamHI; RV, EcoRV. (B) Southern blot confirmation of homologous recombination at the Dnmt1 locus. DNA was digested with EcoRV, and hybridization was performed with Probe A (A). v = Dnmt1V allele; + or wt = WT Dnmt1 allele or locus. (C) Effect of Cre-mediated deletion of HSV-tk/Pgk1-neor from targeted allele on protein expression in ES cells. Dnmt1o and Dnmt1 proteins were detected with an immunoblot assay by using the UPTC21 Ab (9). −Cre, before Cre-mediated excision; +Cre, after Cre-mediated excision. (D) Genotyping of Dnmt1 and Dnmt1V alleles by using primers flanking exon 1s followed by EcoRV digestion of PCR product. EcoRV digests Dnmt1V allele to yield 220- and 80-bp products.
Immunoblot Assay.
Affinity-purified chicken anti-Dnmt1 Ab UPTC21 (9) was used at 1:1,000 dilution. The anti-chicken IgY-HRP secondary Ab (Jackson ImmunoResearch) was used at 1:50,000 dilution. The enhanced chemiluminescence detection kit was from Amersham Pharmacia.
PCR.
For genotyping, oligonucleotides MTE1S (GCCCTTCCCAATTGGTTTCCG) and MTI1S (AAGCACAGCCCGCGCCAAACG) were used, and PCR was performed by using Taq polymerase (Invitrogen) with annealing temperature of 58°C. Oligonucleotides MTE1S and MT28 (TTCTGCTCTCCAGGTTGGCCG) were used for RT-PCR measurements of gene expression; High Fidelity PCR system (Roche) was used with an annealing temperature of 60°C.
Methyltransferase Activity Assay.
The assay was adapted from Adams et al. (11) and Issa et al. (12) with the following modifications. The reaction mix was extracted with phenol:chloroform by using phase-lock gels from Eppendorf. The filters were washed eight times with 5% cold trichloroacetic acid and twice with 70% ethanol before counting.
TLC Assay for Total Methylation.
The TLC assay was adapted from Li et al. (5). Genomic DNA was digested with MspI and end-labeled with T4 polynucleotide kinase. It was then digested with nuclease P1 and run on thin layer chromatography plates in a mixture of ammonium hydroxide:ddH2O:isobutyric acid = 1:19.4:65.
Embryonic Fibroblasts.
Embryonic fibroblasts were isolated from E13.5 embryos and cultured in DMEM with 10% FBS. Cells were starved in DMEM without FBS. For inhibition of protein synthesis, 20 μM cycloheximide was added to DMEM containing 10% FBS.
Results
Dnmt1 and Dnmt1o transcripts are synthesized from alternative promoters associated with exon 1s and exon 1o, respectively (4). The initiation codon for protein synthesis from somatic Dnmt1 transcripts is in exon 1s, and the initiation codon for translation from oocyte Dnmt1o transcripts is in the common exon 4 (Fig. 1A). By targeted mutagenesis, we produced ES cells with the mutant Dnmt1V allele, in which the Dnmt1 translation initiation codon (AAGATGCCAG) was changed to an EcoRV site (tAGATatCAG) (Fig. 1). Because of these three point mutations, Dnmt1V transcripts, initiated from exon 1s, should be translated into the Dnmt1o protein. Indeed, Dnmt1o (from the Dnmt1V allele) and Dnmt1 (from the WT Dnmt1 allele) were found in equal amounts in heterozygous Dnmt1V/+ ES cells (Fig. 1C). Heterozygous and homozygous Dnmt1V mice produced from these ES cells were viable, fertile, and developed normally. Cohorts of 10 heterozygous and 10 homozygous Dnmt1V mice were followed for 18 mo from the time of birth. All 20 mice survived to at least 12 mo of age, and no tumors or gross behavioral abnormalities were observed in any of the mice.
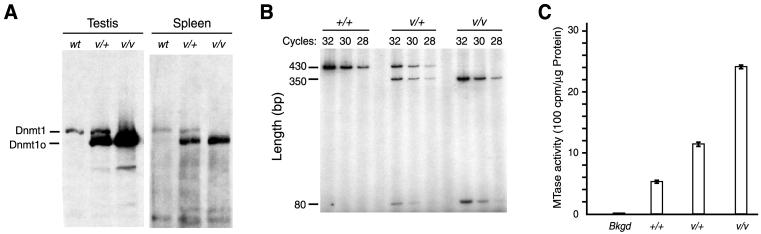
In testes and spleens of heterozygous Dnmt1V mice, the level of Dnmt1o was ≈5-fold higher than that of Dnmt1 (Fig. 2A). The Dnmt1V:Dnmt1 mRNA ratio in the Dnmt1V/+ spleen was 1:1, and nearly equal concentrations of total Dnmt1 (Dnmt1V+Dnmt1) mRNA were detected in WT, Dnmt1V/+, and Dnmt1V/Dnmt1V spleens (Fig. 2B). This means that the two mRNAs were transcribed at equal rates and were equally stable, as expected given the minor sequence difference between Dnmt1V and Dnmt1 mRNAs. Moreover, the high concentrations of Dnmt1o protein in spleens of Dnmt1V mutant mice corresponded to increased maintenance methyltransferase enzyme activity. A heterozygous Dnmt1V spleen had ≈2.7 times the activity, and a homozygous Dnmt1V spleen had ≈4.5 times the activity present in a WT spleen (Fig. 2C). Thus, the Dnmt1V allele produced ≈4.5 times as much activity as the WT Dnmt1 allele, even though the Dnmt1V and Dnmt1 alleles produced equal steady-state concentrations of mRNA (Fig. 2B).
Fig 2.
Dnmt1o protein level and methyltransferase activity are increased in heterozygous and homozygous Dnmt1V mice. (A) Immunoblots of protein extracts from spleens and testes probed with the UPTC21 Ab that detects both Dnmt1o and Dnmt1 (9). Extracts were from spleens and testes of age-matched adult males, and equal amounts of total protein were probed with the Ab. The ratio of Dnmt1o:Dnmt1 protein concentration was estimated to be four- to fivefold by performing immunoblots with twofold serial dilutions of heterozygous Dnmt1V spleen extract (data not shown). Similar elevations of Dnmt1o levels relative to Dnmt1 levels were also seen in other adult tissues (kidney, brain, heart, lung, and liver) from heterozygous Dnmt1V mice (data not shown). (B) Comparison of Dnmt1V and Dnmt1 mRNA levels in WT, heterozygous, and homozygous Dnmt1V spleens. RT-PCR was performed on equal amounts of RNA isolated from adult spleens of the three different genotypes. Radioactive [α-32P]dCTP was incorporated into the amplified products. The number of PCR amplification cycles was 28, 30, or 32, and the PCR amplification products were digested with EcoRV before resolving them on a 5% polyacrylamide gel. The bands were detected by autoradiography. The 430-bp RT-PCR fragment is from WT Dnmt1 transcripts, and the 350- and 80-bp fragments are from Dnmt1V transcripts. (C) Elevation of maintenance methyltransferase activity in heterozygous and homozygous Dnmt1V spleens, as measured by the incorporation of tritium-labeled methyl groups from S-adenosyl methionine into poly(dI-dC). Activity was determined in three separate experiments, and the results are plotted as mean ± SD for each of the three genotypes. The background (Bkgd) level of tritium incorporation in the absence of poly(dI-dC) is also shown. Actual experimental values for 5 μg of protein are Bkgd (72 +/− 4); wt (2,658 +/− 146); v/+ (5,742 +/− 219); and v/v (12,090 +/− 178).
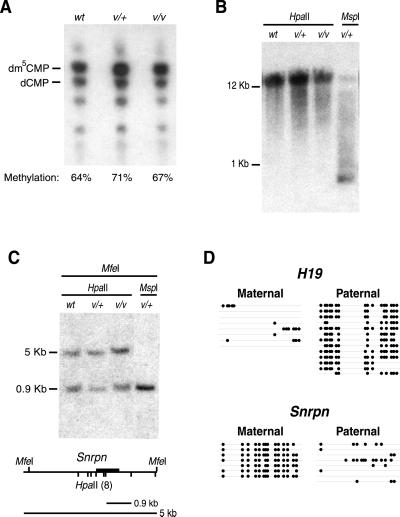
Because elevated levels of Dnmt1o methyltransferase activity in cell lines are associated with increases in genomic methylation (13), the extent of genomic methylation in Dnmt1V/+ and Dnmt1V/Dnmt1V spleens was determined (Fig. 3). DNA from Dnmt1V/Dnmt1V and Dnmt1V/+ spleens had normal levels of global genomic methylation (Fig. 3A), normal methylation patterns on intracisternal A particle proviral sequences (Fig. 3B), and normal parent-specific methylation patterns on the paternally imprinted H19 gene and the maternally imprinted Snrpn gene (Fig. 3 C and D). We conclude that increases in Dnmt1o protein concentration and the accompanying increases in methyltransferase activity in Dnmt1V mutant mice have little, if any, effect on the level of genomic methylation.
Fig 3.
Normal levels of genomic methylation in heterozygous and homozygous Dnmt1V mutant mice. (A) Similar levels of global genomic methylation among WT (wt), heterozygous Dnmt1V mutant mice (v/+), and homozygous Dnmt1V mutant mice (v/v). DNA was cleaved with the methylation-insensitive enzyme MspI before end-labeling with [γ-32]ATP, digestion to 5′-mononucleotides with nuclease P1, and separation of 5′-deoxynucleotides by TLC on cellulose plates (5). The positions of dm5CMP and dCMP are shown. The position of dCMP was determined by HpaII digestion of WT DNA (data not shown). Global genomic methylation levels are determined by the ratio of dm5CMP to total 5′-deoxycytidine (dm5CMP + dCMP). Actual experimental values are: wt = 64 ± 3%; v/+ = 71 ± 0.2%; v/v = 67 ± 3%. (B) Normal methylation of intracisternal A particle proviral sequences in genomic DNA from heterozygous and homozygous Dnmt1V mutant mice. HpaII and MspI digests of genomic DNA were hybridized with a probe to the LTR sequence of the agouti Aiapy allele (14). (C) Normal methylation of the differentially methylated domain of the imprinted Snrpn gene. Southern blot of genomic DNA digested with MfeI plus either HpaII or MspI. Restriction map of the 5′-end of the Snrpn gene is shown. Dark bar is the probe. (D) Normal methylation of the differentially methylated domains of the imprinted H19 and Snrpn genes (15, 16). H19 and Snrpn methylation patterns in a genomic DNA sample from the spleen of a heterozygous Dnmt1V/+ mouse (from a cross between a homozygous Dnmt1V female and a Mus musculus castaneus male) were analyzed by bisulfite genomic sequencing (17). Parental alleles were distinguished by single nucleotide polymorphisms between Dnmt1V mice and M. musculus castaneus mice (8, 18). Alleles are represented by horizontal lines. Positions of methylated CpG dinucleotides are indicated by filled circles. The Snrpn gene was analyzed between nucleotides 2,151 and 2,562 (GenBank accession no. AF081460) and the H19 gene was analyzed between nucleotides 1,301 and 1,732 (GenBank accession no. U19619). H19 is normally methylated at all CpG dinucleotides on the paternal allele; Snrpn is normally methylated at all CpG dinucleotides on maternal allele.
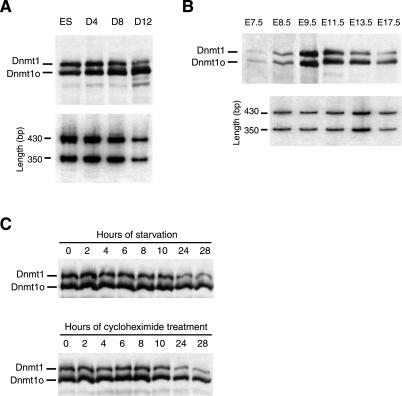
The 4.5:1 ratio of Dnmt1o:Dnmt1 in adult tissues from Dnmt1V/+ mice was not evident in Dnmt1V/+ ES cells, where the ratio was 1:1 (Figs. 1C and 2A). To investigate the possibility that Dnmt1o and Dnmt1 levels are developmentally regulated, they were measured during Dnmt1V/+ ES cell differentiation. Over a 12-day course of differentiation as embryoid bodies, Dnmt1V and Dnmt1 mRNA levels declined at roughly the same rate (Fig. 4A Lower). The level of Dnmt1 protein declined with a similar time course to that of Dnmt1 mRNA, whereas the Dnmt1o protein level remained elevated on day 12, despite a marked reduction in Dnmt1V mRNA concentration at this time (Fig. 4A Upper). A similar change in the Dnmt1o:Dnmt1 ratio was also observed during embryonic development. In E7.5–E9.5 Dnmt1V/+ embryos the Dnmt1o:Dnmt1 ratio was 1:1. This ratio steadily increased throughout later fetal stages (Fig. 4B Upper), even though the Dnmt1V:Dnmt1 mRNA ratio remained constant (Fig. 4B Lower). In E17.5 heterozygous embryos, the Dnmt1o:Dnmt1 protein ratio was 4:1, similar to the ratio in heterozygous Dnmt1V spleen (Fig. 2A). Equal amounts of protein extracts from E11.5–E17.5 embryos were loaded in the last three lanes of Fig. 4B. Hence, the increasing Dnmt1o:Dnmt1 ratio during the later stages of fetal development was due to the combination of a declining Dnmt1 concentration and a relatively constant Dnmt1o level.
Fig 4.
The relative stabilities of Dnmt1o and Dnmt1 proteins change with ES cell differentiation, fetal development, and cell cycle alterations. (A) Ratio of Dnmt1o:Dnmt1 in heterozygous Dnmt1V ES cells increases during embryoid body differentiation. Embryoid bodies were grown for 12 days in the absence of lymphocyte inhibitory factor in Petri dishes (19). (Upper) Immunoblot probed with the UPTC21 Ab (9). Equal amounts of protein were loaded in each lane. The faint bands of lower molecular weight than Dnmt1o and Dnmt1 are probably degradation products of Dnmt1o and/or Dnmt1. (Lower) RT-PCR amplification of Dnmt1V and Dnmt1 mRNAs from total RNA, as described in Fig. 2B. Titration of PCR cycles was performed to ensure the amplification was within the linear range of PCR, and the results of 30 PCR cycles are shown. The 430- and 350-bp bands are from the Dnmt1 and Dnmt1V mRNAs, respectively. (B) Ratio of Dnmt1o:Dnmt1 in heterozygous Dnmt1V embryos increases during embryonic development. Immunoblots of protein extracts from E7.5 to E17.5 embryos were probed with the UPTC21 Ab. For the E11.5, E13.5, and E17.5 lanes, equal amounts of protein were loaded. (Lower) RT-PCR amplification of heterozygous Dnmt1V/+ embryo RNA, as described above and in Fig. 2B. The results of 32 cycles of PCR are shown. (C) Ratio of Dnmt1o:Dnmt1 in E13.5 embryonic fibroblasts increases during starvation in medium without serum (Upper) and during protein synthesis inhibition by cycloheximide (Lower). The 2 × 106 cells growing in log phase in 100-mm dishes were either starved or treated with 20 μM cycloheximide for the indicated length of time and harvested for immunoblot analysis with the UPTC21 Ab.
The changes in the Dnmt1o:Dnmt1 ratio described above indicate that the Dnmt1o and Dnmt1 proteins are handled differently during embryonic development and cellular differentiation. To explore this issue in a more homogeneous cell population than differentiating ES cells, Dnmt1o and Dnmt1 proteins were measured in Dnmt1V/+ embryonic fibroblasts derived from an E13.5 embryo (Fig. 4C). At this stage, a difference in the levels of the two proteins is clearly evident (Fig. 4 B and C). When serum was withdrawn from the medium, the Dnmt1 protein level in Dnmt1V/+ embryonic fibroblasts fell rapidly over the course of 28 h, whereas the Dnmt1o protein level fell at a much slower rate (Fig. 4C Upper). To determine whether this difference was due to a difference in protein stability rather than a difference in translational regulation, we treated Dnmt1V/+ embryonic fibroblasts with 20 μM cycloheximide, which inhibits protein synthesis in mammalian cells, and followed the levels of the two proteins for 28 h. In the absence of protein synthesis, the steady-state level of Dnmt1 fell rapidly, whereas Dnmt1o showed a much slower decline (Fig. 4C Lower). Thus, the Dnmt1o protein is more stable than Dnmt1 in differentiated cells, suggesting that the unique 118-aa domain of Dnmt1 directly or indirectly mediates the degradation of Dnmt1.
Discussion
The marked increase in stability of the Dnmt1o protein does not have any obvious detrimental effect on the viability of Dnmt1V mice, and the Dnmt1o protein substitutes functionally for the Dnmt1 protein in somatic cells of Dnmt1V mice. The two proteins have, in effect, equivalent maintenance methyltransferase function, with methylation patterns being maintained equally well in Dnmt1-containing WT mice and in Dnmt1o-containing Dnmt1V mice. Consequently, WT mice and Dnmt1V mice have identical, or nearly identical, genomic DNA methylation patterns, despite the fivefold difference in enzyme activity between the two strains of mice (Figs. 2 and 3). We can conclude from these observations that the extent of genomic methylation in somatic cells is not governed strictly by the activity of Dnmt1 methyltransferase. A minimal activity of Dnmt1 (or Dnmt1o) is probably needed to efficiently maintain genomic methylation, and additional activity does not lead to any measurable increase in genomic methylation.
Because of the equivalent maintenance methylation function of Dnmt1 and Dnmt1o, we would predict that the Dnmt1 protein, if expressed in the preimplantation embryo, would maintain the same patterns of DNA methylation as does the Dnmt1o protein. Any observed difference between the two proteins during preimplantation development would probably reside in a function other than the intrinsic methylating properties of the two proteins. We propose that the highly stable nature of Dnmt1o allows ooplasmic stores of Dnmt1o protein to persist in high concentrations during the first three embryonic cleavage divisions. This persistence of Dnmt1o, in turn, would allow Dnmt1o to traffick into the nuclei of eight-cell stage blastomeres, where it most likely maintains methylation patterns on alleles of imprinted genes (8). The Dnmt1 protein would be a much less stable protein during preimplantation development than the Dnmt1o protein and therefore would be less efficient at maintaining DNA methylation patterns.
Although the molecular basis for the differential stability of Dnmt1o and Dnmt1 is not known, the amino terminal domain of human DNMT1 is known to interact with the transcriptional repressor protein DMAP1 (10). Dnmt1 presumably interacts with the mouse Dmap1 protein, and an additional function of Dmap1 might be to control the degradation of Dnmt1. Dnmt1o would be excluded from this Dmap1 regulation and consequently would be a more stable protein than Dnmt1. An intriguing possibility is that Dnmt1 degradation is regulated through a three-way interaction involving Dnmt1, Dmap1 and the product of the tumor suppressor gene tsg101. DMAP1 is known to interact with the human TSG101 protein (10). tsg101, which is similar to inactive ubiquitin conjugase homologs, controls p53 protein levels by interacting with Mdm2, a negative regulator of p53, and blocking its ubiquitination and degradation (20). If tsg101 regulates the level of Dmap1 by a similar mechanism, it would indirectly regulate a Dmap1-controlled degradation of Dnmt1. Alternatively, other processes, acting independently of Dmap1 binding, may mediate the degradation of Dnmt1.
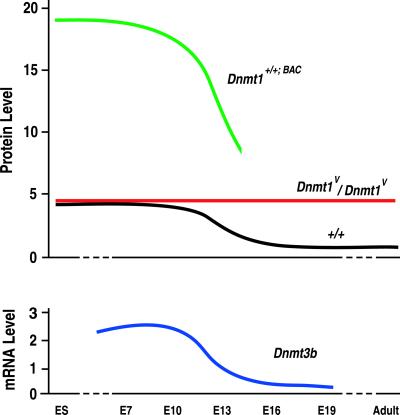
Homozygous Dnmt1V mice survived to adulthood, with 4.5 times the methyltransferase activity as WT mice. In contrast, overexpressing Dnmt1 from a bacterial artificial chromosome (BAC) transgene containing the complete Dnmt1 gene led to four times as much methyltransferase activity as in WT mice and caused genomic hypermethylation and death before E14.5 (21). This difference in outcomes is perplexing, particularly given the roughly equivalent activities of the Dnmt1o and Dnmt1 proteins (Fig. 2; refs. 3, 13, and 22). Perhaps the explanation lies in the dynamics of Dnmt1 activity during embryogenesis (Fig. 5). If the average level of methyltransferase activity in a WT adult cell is defined as 1.0 unit, the level of activity in Dnmt1V/Dnmt1V cells of adults and late-gestation embryos is 4.5 units. Because of the absence of a Dnmt1-specific degradation mechanism before E10 (as indicated by equal levels of Dnmt1o and Dnmt1 protein in Dnmt1V/+ embryos in Fig. 4), both WT and Dnmt1V/Dnmt1V cells have 4.5 units of activity, whereas Dnmt1+/+;BAC embryos would be expected to have 18 units of Dnmt1 activity. We theorize that this elevated Dnmt1 activity in early fetal development is the cause of the embryonic lethality in Dnmt1+/+;BAC transgenic mice.
Fig 5.
Time courses of protein and mRNA concentrations during development in Dnmt1+/+;BAC transgenic, WT, and Dnmt1V mice. (Upper) The relative concentrations of the Dnmt1o and Dnmt1 proteins are plotted in ES cells, embryos, and adults. The estimated time course of Dnmt1 protein concentration in Dnmt1+/+;BAC transgenic mice (21) was determined by assuming a continuous fourfold excess of Dnmt1 protein compared to WT levels. Dnmt1+/+;BAC embryos died by E14. The relative time courses of Dnmt1o and Dnmt1 concentrations in homozygous Dnmt1V/Dnmt1V and WT mice, respectively, are taken from Figs. 2A and 4 A and B. (Lower) The relative time course of Dnmt3b mRNA concentration was determined from the data presented in Okano et al. (23).
Why might high levels of Dnmt1 adversely affect early embryos? The genome of the implanted embryo gradually gains CpG methylation through a combination of de novo and maintenance methyltransferase activities. Dnmt1 probably accounts for the maintenance methyltransferase activity, and the likely source of the de novo methytransferase activity is the Dnmt3b protein (23). We speculate that some genomic methylation patterns are set during early embryogenesis by a combination of Dnmt3b and Dnmt1 activities and that an elevation in either Dnmt1 or Dnmt3b activity leads to DNA hypermethylation. The elevation of Dnmt1 activity in Dnmt1+/+;BAC embryos would lead to hypermethylation of the genome, disruptions in gene regulation and death of the embryos. In support of this notion, there is evidence that Dnmt1 and Dnmt3b cooperate to silence genes in human cancer cells by generating aberrant methylation, including hypermethylation of certain sequences (24). A similar hypermethylation would not occur in Dnmt1V embryos because an increase in Dnmt1o methyltransferase activity does not occur until the later half of gestation, after Dnmt3b levels fall from their peak embryonic levels (Fig. 5).
Lower than normal Dnmt1 protein levels are associated with a reduced incidence of certain tumors (25). By inference, we would expect a higher incidence of these tumors in Dnmt1V mice. However, if cooperation between Dnmt1 (or Dnmt1o) and Dnmt3b is the significant feature that relates methyltransferase activity to tumorigenesis, then we would not necessarily expect more tumors in Dnmt1V mice, as the increase in maintenance methyltransferase level in Dnmt1V mice occurs after the embryonic peak of Dnmt3b mRNA (Fig. 5). Also, tumor incidence may be related to the level of Dnmt1 protein per se, rather than to the level of maintenance methyltransferase activity. The reason for this is that the level of Dnmt1 protein may affect the level of the tsg101 tumor suppressor protein because of an interaction of Dmap1 with both Dnmt1 and tsg101 (see above). In turn, the level of tsg101 protein may affect the incidence of certain tumors. If low Dnmt1 levels lead to higher than normal levels of tsg101, then we would predict a reduction in tumor incidence in homozygous Dnmt1V mice, which have no Dnmt1 protein. Only the evaluation of the effect of the Dnmt1V allele on the incidence of tumors, particularly in tumor-prone models such as the Apcmin mouse (25), will lead to a resolution of this issue.
Acknowledgments
We thank Colin Stewart for the gift of W9.5 ES cells, and Jeff Yoder, Michael Rountree, and Jeff Brodsky for helpful discussions. This work was supported by a grant from the National Institutes of Health (to J.R.C.).
Abbreviations
BAC, bacterial artificial chromosome
E, embryonic day
ES, embryonic stem
References
- 1.Trasler J. M., Trasler, D. G., Bestor, T. H., Li, E. & Ghibu, F. (1996) Dev. Dyn. 206, 239-247. [DOI] [PubMed] [Google Scholar]
- 2.Yoder J. A., Soman, N. S., Verdine, G. L. & Bestor, T. H. (1997) J. Mol. Biol. 270, 385-395. [DOI] [PubMed] [Google Scholar]
- 3.Carlson L. L., Page, A. W. & Bestor, T. H. (1992) Genes Dev. 6, 2536-2541. [DOI] [PubMed] [Google Scholar]
- 4.Mertineit C., Yoder, J. A., Taketo, T., Laird, D. W., Trasler, J. M. & Bestor, T. H. (1998) Development (Cambridge, U.K.) 125, 889-897. [DOI] [PubMed] [Google Scholar]
- 5.Li E., Bestor, T. H. & Jaenisch, R. (1992) Cell 69, 915-926. [DOI] [PubMed] [Google Scholar]
- 6.Li E., Beard, C. & Jaenisch, R. (1993) Nature 366, 362-365. [DOI] [PubMed] [Google Scholar]
- 7.Lei H., Oh, S. P., Okano, M., Jutterman, R., Goss, K. A., Jaenisch, R. & Li, E. (1996) Development (Cambridge, U.K.) 122, 3195-3205. [DOI] [PubMed] [Google Scholar]
- 8.Howell C. Y., Bestor, T. H., Ding, F., Latham, K. E., Mertineit, C., Trasler, J. M. & Chaillet, J. R. (2001) Cell 104, 829-838. [DOI] [PubMed] [Google Scholar]
- 9.Ratnam S., Mertineit, C., Ding, F., Howell, C. Y., Clarke, H. J., Bestor, T. H., Chaillet, J. R. & Trasler, J. M. (2002) Dev. Biol. 245, 304-314. [DOI] [PubMed] [Google Scholar]
- 10.Rountree M. R., Bachman, K. E. & Baylin, S. B. (2000) Nat. Genet. 25, 269-277. [DOI] [PubMed] [Google Scholar]
- 11.Adams R. P. L., Rinaldi, A. & Seivwright, C. (1991) J. Biochem. Biophys. Methods 22, 19-22. [DOI] [PubMed] [Google Scholar]
- 12.Issa J. P., Vertino, P. M., Wu, J., Sazawal, S., Celano, P., Nelkin, B. D., Hamilton, S. R. & Baylin, S. B. (1993) J. Natl. Cancer Inst. 85, 1235-1240. [DOI] [PubMed] [Google Scholar]
- 13.Vertino P. M., Yen, R.-W. C., Gao, J. & Baylin, S. B. (1996) Mol. Cell. Biol. 16, 4555-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michaud E. J., van Vugt, M. J., Bultman, S. J., Sweet, H. O., Davisson, M. T. & Woychik, R. P. (1994) Genes Dev. 8, 1463-1472. [DOI] [PubMed] [Google Scholar]
- 15.Gabriel J. M., Gray, T. A., Stubbs, L., Saitoh, S., Ohta, T. & Nicholls, R. D. (1998) Mamm. Genome 9, 788-793. [DOI] [PubMed] [Google Scholar]
- 16.Tremblay K. D., Duran, K. L. & Bartolomei, M. S. (1997) Mol. Cell. Biol. 17, 4322-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark S. J., Harrison, J., Paul, C. L. & Frommer, M. (1994) Nucleic Acids Res. 22, 2990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szabo P. E. & Mann, J. R. (1995) Genes Dev. 9, 3097-3108. [DOI] [PubMed] [Google Scholar]
- 19.Fan Y., Melhem, M. F. & Chaillet, J. R. (1999) Dev. Biol. 210, 481-496. [DOI] [PubMed] [Google Scholar]
- 20.Li L., Liao, J., Ruland, J., Mak, T. W. & Cohen, S. N. (2001) Proc. Natl. Acad. Sci. USA 98, 1619-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biniszkiewicz D., Gribnau, J., Ramsahoye, B., Gaudet, F., Eggan, K., Humpherys, D., Mastrangelo, M. A., Jun, Z., Walter, J. & Jaenisch, R. (2002) Mol. Cell. Biol. 22, 2124-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaudet F., Talbot, D., Leonhardt, H. & Jaenisch, R. (1998) J. Biol. Chem. 273, 32725-32729. [DOI] [PubMed] [Google Scholar]
- 23.Okano M., Xie, S. & Li, E. (1998) Nat. Genet. 19, 219-220. [DOI] [PubMed] [Google Scholar]
- 24.Rhee I., Bachman, K. E., Park, B. H., Jair, K. W., Yen, R. W., Schuebel, K. E., Cui, H., Feinberg, A. P., Lengauer, C., Kinzler, K. W., et al. (2002) Nature 416, 552-556. [DOI] [PubMed] [Google Scholar]
- 25.Laird P. W., Jackson-Grusby, L., Fazeli, A., Dickinson, S. L., Jung, W. E., Li, E., Weinberg, R. A. & Jaenisch, R. (1995) Cell 81, 197-205. [DOI] [PubMed] [Google Scholar]