Abstract
Gene-family evolution mostly relies on gene duplication coupled with functional diversification of gene products. However, other evolutionary mechanisms may also be important in generating protein diversity. The ubiquitous membrane intrinsic protein (MIP) gene family is an excellent model system to search for such alternative evolutionary mechanisms. MIPs are proteins that transport water, glycerol, and small solutes across cell membranes in all living organisms. We reconstructed the molecular phylogeny of MIPs based on amino acid sequence data by using neighbor-joining, maximum-likelihood, and Bayesian methods of phylogenetic inference. The recovered trees show an early and distinct separation of water and glycerol transporters, i.e., aquaporins (AQPs), and aquaglyceroporins. The latter are absent from plants. As expected, gene duplication and functional diversification account for most of the diversity of animal and plant members of the family. However, in contrast to this model, we find that the sister group of plant glycerol transporters are bacterial AQPs. This relationship suggests first that plant glycerol transporters may resulted from a single event of horizontal gene transfer from bacteria, which we have estimated to have occurred ≈1,200 million years ago, at the origin of plants, and second that bacterial AQPs were likely recruited to transport glycerol in plants because of their absence of aquaglyceroporins. This striking example of adaptive evolution at the molecular level was demonstrated further by finding convergent or parallel replacements at particular amino acid positions related to water- and glycerol-transporting specificity.
Keywords: MIP, aquaporins, aquaglyceroporins, functional convergence
All living organisms that have been examined contain membrane intrinsic proteins (MIPs) that facilitate the transport of water and small solutes across biological membranes (1, 2). Because of their importance to life, numerous studies have focused on MIPs, and a wealth of information is accumulating rapidly on their structure and physiological functions. MIPs are homotetramers, in which each monomer is organized into six transmembrane domains connected by five loops (A–E; ref. 1). Sequence analyses showed that the primary structure of a MIP can be divided into two similar halves with two highly conserved NPA (Asn-Pro-Ala) motifs that are located in the B (cytoplasmic) and E (extracellular) connecting loops, respectively (3).
Some MIPs are water-selective (aquaporins, AQPs), but others also transport small neutral molecules such as glycerol (aquaglyceroporins, GLPs). The crystal structures of the water-specific AQP1 (4) and glycerol-specific GlpF (5) have been determined recently. In both cases, a polar channel is formed in the center of the monomer. After a periplasmic wide vestibule, loops B and E interact with each other through their NPA motifs at the surface of the narrowest region of the pore (4, 5). Furthermore, the implication of several amino acid residues in the selectivity to water or glycerol was proposed on the basis of sequence (6, 7), mutagenesis (8), and topological (9) analyses.
More than 200 sequences of AQPs and GLPs from eubacteria, Archaea, fungi, plants, and metazoans have been described (2, 10). Eubacteria and fungi possess only one copy of the AQP gene and one of the GLP gene. In contrast, up to 10 different types of MIP genes have been found in vertebrates, AQP0–AQP9 (11). Of these, only three (AQP3, AQP7, and AQP9) transport glycerol (12). MIP genes are particularly abundant in plants, and for instance 35 different MIP genes have been identified in Arabidopsis (13). Plant MIPs are classified into four major groups: plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPs), NOD26-like intrinsic proteins (NIPs), and small basic intrinsic proteins (SIPs) (13). Of these, only NIPs transport glycerol (14).
Because of their widespread occurrence taxonomically, MIPs provide an excellent data set to broaden our understanding of the biological significance of molecular evolution by gene duplication coupled with structural and functional diversification. Several studies have focused on the evolutionary history of the gene family (2, 3, 10, 15, 16). As a result, a robust phylogenetic framework for the MIP family is emerging. Such a framework permits the establishment of homologous relationships within the family, an essential prerequisite to understanding the evolutionary mechanisms that generated the functional diversity of MIPs. Thus far, the evolution of the family has been explained by gene duplication followed by functional diversification and tissue specialization of the newly arisen proteins. Here we perform a detailed phylogenetic analysis of the MIP family that shows that plant glycerol transporters are an exception within the family, because they likely arose by horizontal gene transfer coupled with a functional recruitment.
Materials and Methods
Sequence Alignment and Phylogenetic Reconstruction.
A total of 94 complete MIPs were retrieved from the NCBI database (www.ncbi.nlm.nih.gov) and analyzed at the amino acid level. Of these, 19 were GLPs, 10 were eubacterial AQPs, 23 were metazoan AQPs, 14 were plant TIPs, 15 were yeast and plant PIPs, 5 were plant SIPs, and 8 were plant NIPs. Sequences were aligned by using CLUSTALX (17) and refined by eye. Gaps resulting from the alignment were treated as missing data. Ambiguous alignments in highly variable regions were excluded from the phylogenetic analyses (aligned sequences and the exclusion set are available from the authors on request). A neighbor-joining (NJ) analysis (18) of the amino acid alignment was based on mean character distances. Robustness of the phylogenetic results was tested by bootstrap analyses with 500 pseudoreplications (19). NJ phylogenetic analysis was performed by using PAUP* 4.0b8 (20). Maximum-likelihood (ML) and Bayesian analyses were performed in a reduced data set (because of computational constraints) of 47 sequences (maintaining a proportional representation of the different paralogs) by using the JTT model (21). ML phylogenetic analyses were performed by using TREE-PUZZLE 5.0 (22) and 10,000 quartet-puzzling steps. Bayesian inference was performed by using MRBAYES 2.01 (23) and simulating a Markov chain for 100,000 generations.
Because MIPs do not show constant evolutionary rates across paralogs (10), we used the nonparametric rate-smoothing (NPRS) method (24) to estimate divergence times from the 47-sequence phylogeny. The NPRS-corrected mean character distances for the split of mammal and frog AQP1 and the split of the Homo and Xenopus AQP3 were averaged (davg = 0.1). The divergence of amniotes and amphibians ≈360 million years ago (25) was used as calibration point.
Character Evolution.
Character states for internal nodes of the NJ tree were reconstructed with parsimony by using the delayed transformation (DELTRAN) procedure as implemented in PAUP* 4.0b8 (20). The trace character option of MACCLADE 3.08 (26) was used to visualize character evolution on the NJ tree and to determine shared derived characters.
Results and Discussion
Distinct and Early Separation of Water- and Glycerol-Transporting Proteins.
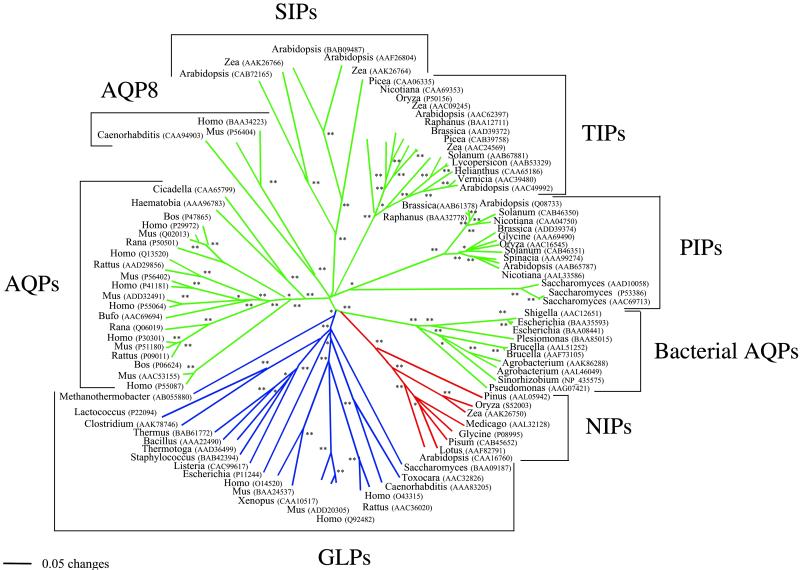
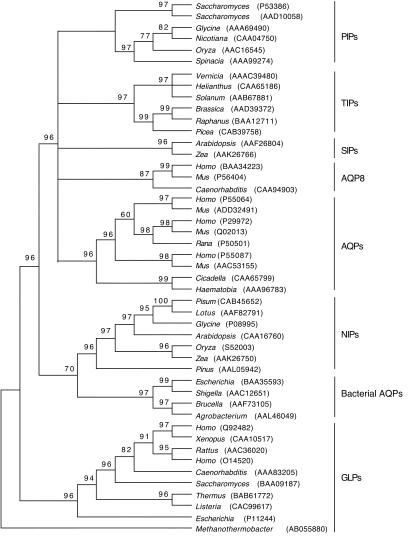
MIP amino acid sequences produced an alignment of 469 positions. As expected, positional identity was difficult to establish for most sites, and a total of 284 positions were excluded from the analyses because of ambiguity. Of the remaining, only 2 positions were invariant, and 180 were parsimony-informative. Amino acid composition was homogeneous across sequences according to a 5% χ2 test with the exception of Zea (GenBank accession no. AAK26766). We reconstructed a molecular phylogeny of 94 AQPs and related proteins based on their amino acid sequences by using the NJ phylogenetic method of inference and mean (uncorrected) character distances (Fig. 1). Phylogenetic inference with ML (not shown) and Bayesian methods based on a reduced data set of 47 sequences arrived at similar and congruent trees (Fig. 2). Several interesting evolutionary trends can be drawn from these trees. First, there is a distinct and early separation of water- and glycerol-transporting systems (57% NJ bootstrap support, 91% quartet-puzzling ML support, and 96% Bayesian posterior probability; refs. 2 and 3). This split is a clear example of how gene duplication and functional divergence can accompany the acquisition of new protein functions. Second, GLP homologs are present in all living organisms except plants. The most parsimonious interpretation of this evidence is that the common ancestor of plants already lacked a GLP protein. Third, plant AQP main paralogs (TIPs, PIPs, and SIPs) show greater divergences among themselves than do metazoan main paralogs (with the exception of AQP8). The greater diversity of AQPs in plants may reflect the importance of having a fine control of water uptake in these organisms (13). Fourth, AQP8 is an early divergent metazoan AQP that seems to be present as a single copy in animals. The remaining metazoan AQPs are clustered together, indicating diversification within the metazoan clade. In addition, according to the phylogenetic tree, gene duplications occurred after the divergence of vertebrates. The existence of three main groups of vertebrate AQPs (AQP4, AQP1, and the remaining) and GLPs (AQP3, AQP7, and AQP9) may reflect the two genomic duplication events proposed for vertebrates (27).
Fig 1.
Fifty percent majority-rule bootstrap consensus tree reconstructed with the NJ method based on a mean (uncorrected) character distance matrix. Nodes with 50–69% (*) or 70–100% (**) bootstrap support are indicated. Based on their function, AQPs are shown in green, GLPs are in blue, and NIPs are in red.
Fig 2.
Fifty percent majority-rule consensus tree reconstructed with the Bayesian method by using the JTT model. Numbers above branches are Bayesian posterior probabilities. A close relationship between plant NIPs and bacterial AQPs was recovered.
Plant NIPs Likely Were Acquired by Horizontal Gene Transfer.
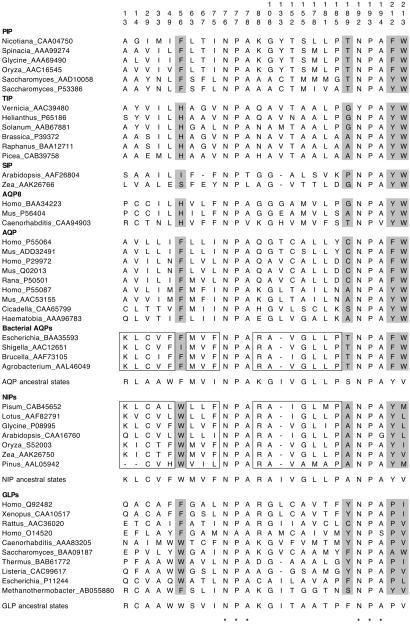
The most striking result from the phylogenetic analyses is the sister-group relationship of plant NIPs and bacterial AQPs (Figs. 1 and 2), which suggests that plant NIPs resulted from a single event of horizontal gene transfer from bacteria. The grouping is supported by a 70% bootstrap value in the NJ tree (Fig. 1), a 70% posterior probability value in the Bayesian tree (Fig. 2), and a 69% quartet-puzzling value in the ML tree (not shown). To find shared derived characters between plant NIPs and bacterial AQPs, amino acid ancestral character states were reconstructed. The close phylogenetic relationship of plant NIPs and bacterial AQPs is supported, at least, by 16 putative shared derived characters that are found in both groups but only spuriously in others (Fig. 3). Interestingly, the close phylogenetic relationship of NIPs and bacterial AQPs was recovered already in previous phylogenetic analyses (2, 3, 16) but apparently was overlooked. The unrooted phylogram shows an old divergence of both sets of genes from a common ancestor (Fig. 1). The age estimated for the most recent common ancestor of bacterial AQPs and NIPs based on the nonparametric rate-smoothing method (24) was 1,188 million years. This date agrees with the estimated origin of plants ≈1,200 million years ago (28). It will be interesting to search for NIPs in nonvascular plants and protozoan species to confirm that the lateral transfer occurred before the origin of plants.
Fig 3.
Conserved positions of the MIP alignment and character evolution. Positions are numbered according to human AQP1 (GenBank accession no. P29972). NPA motifs are denoted by asterisks. Putative shared derived characters between NIPs and bacterial AQPs are boxed. Amino acid residues that have been proposed to be involved in selectivity to water or glycerol are in gray. Ancestral amino acid states at these positions were reconstructed on the NJ tree for AQPs, GLPs, and NIPs by using parsimony (DELTRAN procedure). NIPs show a distinct combination of residues to both AQPs and GLPs at those positions involved in substrate selectivity.
It is well known that gene horizontal transfer was key in the origin of the three main kingdoms (Bacteria, Archaea, and Eukarya), and that at present it is a crucial means of transferring adaptive traits across species in bacterial evolution (29). Our results suggest that horizontal gene transfer also may have been essential in plant evolution. Indeed, there is evidence that other plant genes such as Arabidopsis enolase 2 and 3 most likely derived also from lateral gene transfer (30).
Glycerol Transporting in Plants Resulted from a Functional Shift.
Plant NIPs first were described in the peribacteroid membranes of symbiotic legume root nodules where they control metabolite flux between the plant cytosol and symbiotic nitrogen-fixing bacteria (31). However, these plant proteins are not restricted to root symbiosomes. Plant NIPs are found also in, for example, Pinus (32) and Arabidopsis (14) and exhibit glycerol permease activity. NIPs are considered the glycerol transporters of plants, because as mentioned above, there are no GLP homologs in plants.
However, from an evolutionary perspective and according to the phylogenetic tree, NIPs are members of the AQP clade. Because the horizontal gene transfer involved an AQP, plant NIPs must have acquired the capacity for glycerol transport at a later time by recruitment or exaptation (acquiring a function different from the one for which the protein was selected originally; ref. 33).
The recruitment of an AQP as glycerol transporter requires convergent or parallel replacements at specific amino acid positions. Ancestral amino acid states at these sites were reconstructed by using parsimony. It has been determined that two residues found in the sixth transmembrane helix of the MIP protein are important for selectivity to water or glycerol (8). These two residues are tyrosine (or phenylalanine) and tryptophan in AQPs but are proline and valine (or isoleucine) in GLPs. At the same positions, NIPs show tyrosine and leucine (or valine or methionine), respectively (Fig. 3). The ancestral character state reconstruction analysis shows that NIPs retain the ancestral character state at these positions (tyrosine and valine), whereas GLPs have changed the first residue to proline, and AQPs have changed the second residue to tryptophan.
Another two residues are important in specifying the physiological properties of the channel. A tryptophan located before the first NPA motif of the molecule seems to be involved in glycerol transport by Escherichia coli GLP (5). However, this tryptophan is not conserved in all GLPs, because some show phenylalanine at this position (Fig. 3). Trytophan is also found in all NIPs but not in AQPs, which possess phenylalanine or histidine at this position (Fig. 3). The ancestral character state reconstruction shows that there has been a convergence to tryptophan at this position in NIPs and GLPs. Similarly, a phenylalanine located before the second NPA motif of the molecule has been implicated also in glycerol transport by E. coli GLP (5). However, this residue is not conserved in other GLPs nor among other groups of MIPs (Fig. 3). The intermediate combination of amino acids at specific sites of NIPs with respect to AQPs and GLPs has been noted already (14). It is apparent that glycerol transport by NIPs and GLPs represents a functional convergence, because it is not mediated by the same combination of residues.
Other cases of functional recruitment have been reported for developmental proteins (34) and enzymes (35). For example, Indian hedgehog was recruited to participate in muscle development in zebrafish, neural plate development in frogs, and cartilage development in mouse (34). In another striking example, lysozyme was recruited independently three times for stomach function in ruminants, some colobine monkeys, and the hoatzin (35) to digest fermentative bacteria. In this latter case, convergent or parallel replacements were observed and attributed to selective pressures. Our results on AQPs and GLPs provide another example of this apparently widespread method for acquiring new molecular functions.
Concluding Remarks.
The molecular phylogeny of MIPs supports that glycerol transporting in plants was acquired by horizontal gene transfer and functional recruitment of bacterial AQPs. It is likely that these events were triggered by the absence of a GLP homolog in the common ancestor of plants. We find that plant NIPs and GLPs share convergent or parallel amino replacements needed to transport glycerol and therefore represent a remarkable example of adaptive evolution at the molecular level. Our results emphasize the need of looking for NIPs in nonvascular plants and protozoans as well as the importance of a better characterization of NIP functions and cellular localizations.
Acknowledgments
We thank Peter Agre and Scott Edwards for insightful comments on the manuscript. This work was supported partly by Ministerio de Ciencia y Tecnología from Spain Project REN2001-1514 (to R.Z.) and in part by a grant from the National Research Initiative Competitive Grants Program/U.S. Department of Agriculture (to M.J.C.).
Abbreviations
MIP, membrane intrinsic protein
AQP, aquaporin
GLP, aquaglyceroporin
PIP, plasma membrane intrinsic protein
SIP, small basic intrinsic protein
TIP, tonoplast intrinsic protein
NIP, NOD26-like intrinsic protein
NJ, neighbor joining
ML, maximum likelihood
References
- 1.Chrispeels M. J. & Agre, P. (1994) Trends Biochem. Sci. 19 421-425. [DOI] [PubMed] [Google Scholar]
- 2.Heymann J. B. & Engel, A. (1999) News Physiol. Sci. 14 187-193. [DOI] [PubMed] [Google Scholar]
- 3.Park J. H. & Saier, M. H. (1996) J. Membr. Biol. 153 171-180. [DOI] [PubMed] [Google Scholar]
- 4.Murata K., Mitsuoka, K., Hiral, T., Walz, T., Agre, P., Heymann, J. B., Engel, A. & Fujiyoshi, Y. (2000) Nature 407 599-605. [DOI] [PubMed] [Google Scholar]
- 5.Fu D., Libson, A., Miercke, L. J. W., Weitzman, C., Nollert, P., Krucinski, J. & Stroud, R. M. (2000) Science 290 481-486. [DOI] [PubMed] [Google Scholar]
- 6.Froger A., Tallur, B., Thomas, D. & Delamarche, C. (1998) Protein Sci. 7 1458-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heymann J. B. & Engel, A. (2000) J. Mol. Biol. 295 1039-1053. [DOI] [PubMed] [Google Scholar]
- 8.Lagrée V., Froger, A., Deschamps, S., Hubert, J. F., Delamarche, C., Bonnec, G., Thomas, D., Gouranton, J. & Pellerin, I. (1999) J. Biol. Chem. 274 6817-6819. [DOI] [PubMed] [Google Scholar]
- 9.Tajkhorshid E., Nollert, P., Jense, M. Ø., Miercke, L. J. W., O'Connell, J., Stroud, R. M. & Schulten, K. (2002) Science 296 525-530. [DOI] [PubMed] [Google Scholar]
- 10.Zardoya R. & Villalba, S. (2001) J. Mol. Evol. 52 391-404. [DOI] [PubMed] [Google Scholar]
- 11.Agre P. (1997) Biol. Cell 89 255-257. [PubMed] [Google Scholar]
- 12.Ishibashi K., Sasaki, S., Fushimi, K., Uchida, S., Kuwahara, M., Saito, H., Furukawa, T., Nakajima, K., Yamaguchi, Y., Gojobori, T. & Marumo, F. (1994) Proc. Natl. Acad. Sci. USA 91 6269-6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johanson U., Karlsson, M., Johansson, I., Gustavsson, S., Sjövall, S., Fraysse, L., Weig, A. R. & Kjellbom, P. (2001) Plant Physiol. 126 1358-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weig A. R. & Jakob, C. (2000) FEBS Lett. 481 293-298. [DOI] [PubMed] [Google Scholar]
- 15.Pao G. M., Wu, L. F., Johnson, K. D., Höfte, H., Chrispeels, M. J., Sweet, G., Sandal, N. N. & Saier, M. H. (1991) Mol. Microbiol. 5 33-37. [DOI] [PubMed] [Google Scholar]
- 16.Johansson I., Karlsson, M., Larsson, C. & Kjellbom, P. (2000) Biochim. Biophys. Acta 1465 324-342. [DOI] [PubMed] [Google Scholar]
- 17.Thompson J. D., Gibson, T. J., Plewniak, F., Jeanmougin, J. & Higgins, D. G. (1997) Nucleic Acids Res. 25 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saitou N. & Nei, M. (1987) Mol. Biol. Evol. 4 406-425. [DOI] [PubMed] [Google Scholar]
- 19.Felsenstein J. (1985) Evolution (Lawrence, Kans.) 39 783-791. [DOI] [PubMed] [Google Scholar]
- 20.Swofford D. L., (1998) PAUP*, Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA), Version 4.0.
- 21.Jones D. T., Taylor, W. R. & Thornton, J. M. (1992) Comput. Appl. Biosci. 8 275-282. [DOI] [PubMed] [Google Scholar]
- 22.Strimmer K. & von Haeseler, A. (1996) Mol. Biol. Evol. 13 964-969. [Google Scholar]
- 23.Huelsenbeck J. P. & Ronquist, F. R. (2001) Bioinformatics 17 754-755. [DOI] [PubMed] [Google Scholar]
- 24.Sanderson M. J. (1997) Mol. Biol. Evol. 14 1218-1231. [Google Scholar]
- 25.Kumar S. & Hedges, S. B. (1998) Nature 392 917-920. [DOI] [PubMed] [Google Scholar]
- 26.Maddison W. P. & Maddison, D. R., (1992) MACCLADE, Analysis of Phylogeny and Character Evolution (Sinauer, Sunderland, MA). [DOI] [PubMed]
- 27.Meyer A. & Schartl, M. (1999) Curr. Opin. Cell Biol. 11 699-704. [DOI] [PubMed] [Google Scholar]
- 28.Feng D. F., Cho, G. & Doolittle, R. F. (1997) Proc. Natl. Acad. Sci. USA 94 13028-13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doolittle R. F. (2002) Nature 416 697-700. [DOI] [PubMed] [Google Scholar]
- 30.Keeling P. J. & Palmer, J. D. (2001) Proc. Natl. Acad. Sci. USA 98 10745-10750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rivers R. L., Dean, R. M., Chandy, G., Hall, J. E., Roberts, D. M. & Zeidel, M. L. (1997) J. Biol. Chem. 272 16256-16261. [DOI] [PubMed] [Google Scholar]
- 32.Ciavatta V. T., Morillon, R., Pullman, G. S., Chrispeels, M. J. & Cairney, J. (2001) Plant Physiol. 127 1556-1567. [PMC free article] [PubMed] [Google Scholar]
- 33.Gould S. J. & Vrba, E. S. (1982) Paleobiology 8 4-15. [Google Scholar]
- 34.Zardoya R., Abouheif, E. & Meyer, A. (1997) Trends Genet. 12 496-497. [DOI] [PubMed] [Google Scholar]
- 35.Kornegay J. R., Schilling, J. W. & Wilson, A. C. (1994) Mol. Biol. Evol. 11 921-928. [DOI] [PubMed] [Google Scholar]