Abstract
We report a generally applicable strategy for transferring zygotic lethal mutations through the zebrafish germ line. By using a morpholino oligonucleotide that blocks primordial germ cell (PGC) development, we generate embryos devoid of endogenous PGCs to serve as hosts for the transplantation of germ cells derived from homozygous mutant donors. Successful transfers are identified by the localization of specifically labeled donor PGCs to the region of the developing gonad in chimeric embryos. This strategy, which results in the complete replacement of the host germ line with donor PGCs, was validated by the generation of maternal and maternal-zygotic mutants for the miles apart locus. This germ-line replacement technique provides a powerful tool for studying the maternal effects of zygotic lethal mutations. Furthermore, the ability to generate large clutches of purely mutant embryos will greatly facilitate embryological, genetic, genomic, and biochemical studies.
Large-scale mutational screens in the fruit fly, Drosophila melanogaster, and in the zebrafish, Danio rerio, have identified numerous genetic loci the are required zygotically for normal embryogenesis (1–3). In most animal species, the genomes of fertilized embryos are not transcriptionally active at early developmental stages; therefore, maternally derived products, which are synthesized and accumulated during oogenesis, act to regulate early embryonic development. In Drosophila, many genes originally identified in zygotic mutational screens are also expressed maternally. Studies have shown that this maternal contribution is often required for early embryonic patterning (reviewed in ref. 4) and can partially compensate for the loss of zygotic gene function (5, 6). The production of germ-line clones has allowed for systematic analysis of the maternal effects of known zygotic lethal mutations in Drosophila (6, 7). Typically, genetically mosaic animals are generated by using dominant female-sterile mitotic recombination methods (8). In this case, only eggs from homozygous mutant germ cells are produced. In a second approach, cell transplantation techniques are used to transfer mutant pole cells into sterile wild-type (WT) host embryos (9). This approach results in the production of females carrying germ lines that have been completely repopulated by homozygous mutant cells. These strategies have allowed for the efficient production and analysis of maternal mutant progeny in Drosophila.
Studies in Xenopus and zebrafish have shown that in both of these organisms, maternal gene products also contribute to early embryonic development and patterning (10–13). Moreover, the production of maternal-zygotic mutants for one-eyed-pinhead (14) or lost-a-fin (15) have revealed previously unexpected roles for genes initially identified by zygotic mutant phenotype. These results have demonstrated that, as in Drosophila, maternally derived RNAs and proteins can partially mask the effects of zygotic mutations in zebrafish. In these studies homozygous mutant females were generated by injecting WT mRNA into mutant embryos, thereby rescuing early embryonic phenotypes. However, rescue by mRNA injection is not a generally applicable tool for studying maternal phenotypes as it requires (i) that a mutation be cloned, (ii) that ectopic/over-expression of the mRNA does not result in a lethal phenotype, and (iii) that the gene function is dispensable during later stages of development. The limitations of this approach underscore the need for improved methods for systematic analysis of possible maternal effects. A potential strategy that overcomes these drawbacks involves the generation of germ-line chimeras. Zebrafish chimeras have been generated through the transplantation of midblastula stage cells, including primordial germ cells (PGCs), from pigmented donor embryos to albino hosts (16). However, only 5 germ-line chimeras were produced from >400 midblastula transplants, and the extent of donor cell contribution to the chimeric germ line was generally <15% (16). Thus, whereas this study demonstrated the possibility of transferring PGCs between genotypically distinct embryos, the low efficiency of obtaining the chimeric animals and the high level of germ-line mosaicism make it extremely labor intensive and not practical for the analysis of a large number of genes.
In recent years, much has been learned about zebrafish PGC development. Identification of the zebrafish vasa homolog, which is expressed in the germ line (17, 18), and the detailed temporal and spatial characterization of its expression (19, 20), have provided insights into the development and migration of PGCs during embryogenesis. Furthermore, analyses of the posttranscriptional regulation of nanos and vasa RNA have yielded useful markers for studying PGC development in vivo (21–23). Moreover, genes whose function is essential for zebrafish PGC development have been isolated (ref. 21; G.W., B.T., C.T., and E.R., unpublished work). Here we exploit these recent advances to efficiently produce zebrafish chimeras in which host germ lines have been completely replaced by mutant donor cells. We describe fluorescent labeling techniques that permit efficient screening for donor PGC transfer in host embryos, and we report on a morpholino oligonucleotide (MO) antisense approach (24, 25) that effectively ablates the host germ line. As proof-of-principle, we have used this technology to generate maternal-zygotic mutants for the zygotic lethal miles apart (mil) mutation.
Materials and Methods
Microinjection and Cell Transplantation.
Fish and embryo maintenance (26) and cell transplantation techniques (27) have been described. Donor embryos for transplantation were obtained from intercrosses of milte273/+ fish (28); WT fish were used to generate host embryos. Donor and host embryos were dechorionated by Pronase treatment. Donor embryos were injected at the one- to two-cell stage with either 100 pl of 5% rhodamine-dextran (10 kDa; Molecular Probes), or 80 pg of GFP-nos1-3′UTR sense strand-capped mRNA that was synthesized by SP6 transcription from NotI-linearized plasmid (21) by using the mMESSAGE mMACHINE system (Ambion). Host embryos were injected at the one- to two-cell stage with 3 ng of a morpholino antisense oligonucleotide directed against the dead end mRNA (αPGC-MO, 5′-GCTGGGCATCCATGTCTCCGACCAT-3′; G.W., B.T., C.T., and E.R., unpublished work). RNA and MOs were diluted in 5 mg/ml phenol red in 0.2 M KCl; MO injection concentrations were calculated from manufacturer's specifications (Gene Tools, Philomath, OR). At mid-blastula stages, 50–100 cells were transplanted from the margin of donor embryos into either the margin or the animal pole of similarly staged hosts. Donor and hosts were cultured individually in 24-well plates on 2% agarose until 30 h post fertilization (hpf); host embryos were then screened for the transfer of fluorescently labeled donor-derived PGCs, and donor embryos were screened for the mil-mutant phenotype (28, 29).
PCR Genotyping.
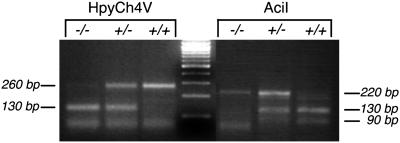
Embryos were genotyped for the mil locus by restriction enzyme digest of the 260-bp PCR product generated by using 5′-AGTGGTTCATACGAGAGGGG-3′ and 5′-TAGTATCGGGTGTTGAGGGG-3′ primer pairs. Typical amplification conditions consisted of 45 cycles of 30 sec at 94°C, 55°C, and 72°C. The amplified product encompasses the milte273 mutation (30), which results in loss of an AciI restriction site and creation of a HpyCH4V site in the amplicon. AciI digestion produces a 220-bp product in the presence of a milte273 allele, and a 130- + 90-bp doublet in the presence of the WT mil allele; conversely, HpyCH4V digestion yields a 260-bp product for the WT mil allele, and a 130-bp band for the milte273 allele. Products were resolved on a 3% agarose gel.
Results
Strategy for Germ-Line Replacement.
Our approach to generate germ-line replacement chimeras is outlined in Figs. 1 and 2. We reasoned that blocking host PGC development with specific antisense MOs would ensure the complete repopulation of the germ line by PGCs transplanted from donor embryos. Furthermore, fluorescent labels introduced into donor cells should allow the efficient in vivo screening of host embryos for potential PGC transfer. This approach would eliminate the need to generate and screen large numbers of adult fish, because only those chimeras that specifically had PGCs transferred from homozygous mutant donor embryos need be raised to adulthood.
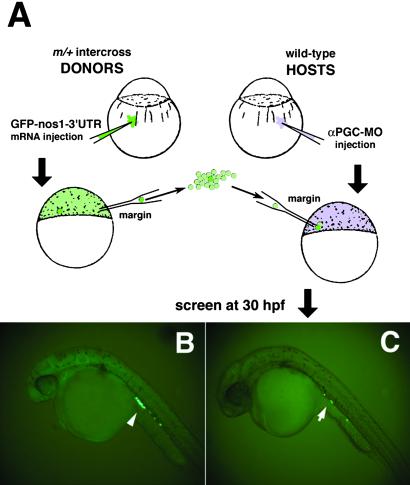
Fig 1.
Germ-line replacement strategy using a GFP-nos1-3′UTR labeling technique. (A) Overview of transplantation strategy showing the transfer of cells from the margin of GFP-nos1-3′UTR-labeled mutant donor embryos into the margin of αPGC-MO-injected WT hosts. “m” refers to any zygotic mutation, in this case milte273. (B) At 30 hpf, PGCs from injected control milix embryos are specifically labeled with GFP (arrowhead). (C) Host embryos were screened at 30 hpf for the presence of GFP-labeled donor PGCs and their normal migration into the gonadal mesoderm at the anterior region of the yolk extension (arrow).
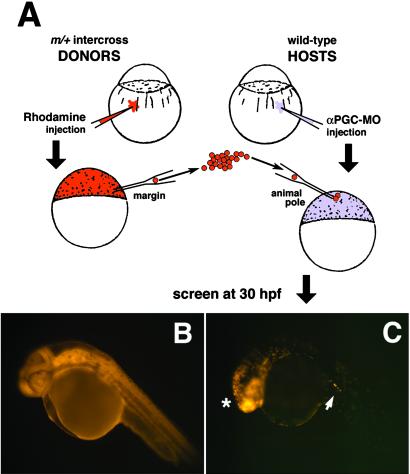
Fig 2.
Germ-line replacement strategy using the rhodamine-dextran labeling technique. (A) Overview of transplantation strategy showing the transfer of cells from the margin of rhodamine-dextran-labeled mutant donor embryos into the animal pole of αPGC-MO-injected WT hosts. (B) At 30 hpf, all cells within control donor embryos are efficiently labeled with rhodamine-dextran. (C) Chimeric host embryo at 30 hpf showing somatic contribution of rhodamine-dextran-labeled donor cells to anterior neuroectoderm lineages (*). Host embryos were screened for the presence of labeled donor PGCs that had migrated successfully into the gonadal mesoderm (arrow).
As proof-of-principle of the proposed strategy, we attempted to replace the germ line of WT host embryos with PGCs transplanted from mil-mutant donors. mil codes for a sphingosine-1-phosphate receptor that has been implicated in regulating embryonic cell migration (30). The milte273 allele is a fully penetrant recessive zygotic lethal mutation, and mil-mutant embryos develop with cardia bifida and epithelial tail blisters (28, 29). mil is maternally expressed, and previous attempts at rescuing the mil-mutant phenotype have failed because of dominant effects induced by mil mRNA injection (30). The mil locus is thus a typical candidate for the germ-line replacement approach to generate maternal mutants.
Transplantation and Preparation of Host Embryos.
Embryos were collected from intercrosses between milte273/+ fish (milix donors) as well as WT fish (host embryos), and transplants were performed between the oblong- and sphere-stages (31). PGC precursors have been shown to reside near the margin at these mid-blastula stages (17, 18). Therefore, ≈50–100 cells were transferred from the margin of milix donor embryos into WT hosts.
To ensure that the germ line of chimeric embryos is entirely replaced by milix donor cells, WT host embryos were injected at the one-cell stage with a MO that blocks translation of the dead end mRNA. dead end was identified in an in situ expression screen as a gene expressed in PGCs (C.T. and B.T., unpublished work). Recent studies have shown that inhibiting the translation of dead end by using antisense MOs inhibits PGC development and results in sterility without affecting viability (G.W., B.T., C.T., and E.R., unpublished work). We titrated the activity of this αPGC-MO at 1 ng, 3 ng, and 6 ng quantities, and found that 3 ng of MO blocked PGC development in all injected embryos (n > 30) as scored by the absence of vasa expression (17, 18) or GFP-nos1–3′UTR reporter expression (21) at 30 hpf (data not shown).
Labeling and Screening for Donor PGC Transfer.
We used two strategies to fluorescently label and transfer donor PGCs. In the first (Fig. 1A), milix embryos were injected at the one-cell stage with a GFP mRNA containing the nanos 3′UTR (GFP-nos1-3′UTR; ref. 21). Although GFP is expressed widely at the time of transplantation, the nos1-3′UTR directs the specific expression of GFP to PGCs by late gastrulation (Fig. 1B; ref. 21). Cells were transplanted from the margin of donor embryos into the margin of αPGC-MO-injected WT hosts. At 30 hpf, host embryos were screened for GFP-positive cells residing in the gonadal mesoderm near the anterior region of the yolk extension (arrow, Fig. 1C). These represent transplanted milix PGCs that have migrated to the normal embryonic domain of germcell development (19). In some instances, GFP reporter expression appeared variable among control milix embryos that were allowed to develop to 30 hpf, with GFP expression restricted to only one side of the embryo. Because transfer of unlabeled milix PGCs would go unnoticed, incomplete labeling of donor PGCs would decrease the efficiency with which successful germ-line chimeras are identified and selected. It was also observed that some chimeric embryos developed cardia bifida. Presumably, this was caused by somatic mil-mutant cell contribution to the host embryo as mil-mutant cells were transplanted to the margin of WT hosts, a region fated to give rise to the endoderm and mesodermal precursors of the future heart field (32, 33).
The second strategy used to label and screen for PGC transfer is depicted in Fig. 2A. milix donor embryos were injected at the one-cell stage with rhodamine-dextran, which effectively labels all cells of the host embryo (Fig. 2B). In these experiments, cells were transplanted from the margin of milix donors into the animal pole of WT hosts, a region fated to become anterior neural tissue (32, 34). Recent studies have suggested that PGCs are attracted to migrate into the gonadal mesoderm (35). We therefore reasoned that somatic cells transplanted to the animal region should contribute to anterior neuroectoderm (36), and that PGCs would actively migrate from this region toward the presumptive gonad. Host embryos were screened at 30 hpf for the distribution of fluorescently labeled milix donor cells. As expected, somatic milix cell contribution was largely confined to the head of chimeric embryos (Fig. 2C). Successful transfer of donor PGCs was scored by the presence of rhodamine-dextran-labeled cells in the gonadal mesoderm near the anterior region of the yolk extension (arrow, Fig. 2C). Rhodamine-dextran-labeled PGCs appeared larger and brighter than surrounding donor-derived somatic cells, presumably because of the lower mitotic index of PGCs (17). This finding made it possible to screen for milix PGC transfer even when donor cell contribution was not restricted wholly to anterior neural tissue (data not shown).
Table 1 summarizes the efficiency of PGC transplantation in four of our most recent experiments, using rhodamine-dextran or GFP-nos1–3′UTR labeling strategies. In total, the overall efficiency of both techniques was nearly 12%.
Table 1.
Efficiencies of PGC transfer by using alternative screening and labeling strategies
| Donor PGC label | Exp. | Total no. of transplants performed | No. of successful PGC transfers | Efficiency, % |
|---|---|---|---|---|
| GFP-nos1-3′ UTR mRNA | 1 | 160 | 18 | |
| 2 | 180 | 22 | ||
| Total | 340 | 40 | 11.8 | |
| Rhodamine-dextran | 3 | 144 | 12 | |
| 4 | 144 | 22 | ||
| Total | 288 | 34 | 11.8 |
Dead chimeras were included in the presented totals. Although survival rates were not recorded for all individual transplant experiments, we approximate a 20% mortality rate; with improved technique, PGC transfer efficiencies of 15% may therefore be obtainable.
Fertile Chimeras with mil-Mutant-Derived Germ Lines.
On identification of PGC transfer into host embryos, corresponding donor embryos were screened for the mil-mutant phenotype. All chimeric embryos that had received mil-mutant PGCs were selected to be raised to adulthood; some chimeras that had received PGCs transferred from WT donors were also kept as controls. To date, 26 of a total of 39 selected chimeras have survived until adulthood: 12 of these chimeras were derived from mil-mutant donors, and 14 were derived from milte273/+ or +/+ donors. Of the 26 chimeras raised, 12 have proven fertile (4 females and 8 males).
To test whether donor-derived cells had completely replaced the germ lines of the hosts, chimeric fish were mated with milte273/+ stocks and the progeny were screened for WT or mil-mutant phenotypes (see Table 2). If the germ line of host embryos has been completely replaced by milte273/milte273 PGCs, then crosses to milte273/+ fish should produce WT and mil-mutant progeny at a 1:1 ratio. However, if the germ line of the +/+ host was not completely ablated, then a greater number of WT embryos should be observed. Of the 628 progeny from such mil-mutant chimera × milte273/+ intercrosses, 320 mil-mutant and 308 WT embryos were identified; the phenotype of maternal-zygotic mil-mutants was not appreciably stronger than zygotic mutants. χ2 analyses indicate no significant deviation from the expected 1:1 mutant to WT progeny ratio for all milte273/milte273 chimeras tested (Table 2).
Table 2.
Phenotypes and genotypes of progeny obtained from matings between milte273/+ fish and adult chimeras
| Chimera | Total no. of progeny obtained
|
Phenotype | χ2 value
|
Total no. of WT genotyped
|
Genotype | ||||
|---|---|---|---|---|---|---|---|---|---|
| Donor genotype | ID | Sex | mil-mutant | WT | +/+ | milte273/+ | |||
| milte273/milte273 | m22 | F | 322 | 166 | 156 | 0.31 | 83 | 0 | 83 |
| m3 | F | 164 | 83 | 81 | 0.02 | 40 | 0 | 40 | |
| m17 | M | 66 | 37 | 29 | 0.97 | 27 | 0 | 27 | |
| m1 | M | 76 | 34 | 42 | 0.84 | 34 | 0 | 34 | |
| Total | 320 | 308 | Total | 0 | 184 | ||||
| milte273/+ | m26 | F | 169 | 42 | 127 | 0.002 | |||
| m15 | F | 70 | 18 | 52 | 0.02 | ||||
| m20 | M | 82 | 25 | 57 | 1.32 | ||||
| m9 | M | 73 | 18 | 55 | 0.004 | ||||
| Total | 103 | 291 | |||||||
For 1 degree of freedom, a χ2 value of <3.84 indicates no statistically significant deviation from expected mutant:WT progeny ratios (see text).
If the host germ line had been completely ablated and reconstituted only with milte273/milte273 donor cells, then all offspring would inherit the milte273 allele from a chimeric parent. To test this hypothesis, the WT offspring of crosses between chimeras and milte273/+ partners were PCR genotyped to distinguish milte273/+ from +/+ embryos (Fig. 3). To date, 0/184 WT embryos have genotyped as +/+, indicating that the germ lines of chimeric embryos have been completely repopulated by milte273/milte273 PGCs (Table 2). Furthermore, intercrosses between mil-mutant chimeras have yielded only mil-mutant embryos (n = 48/48). These results indicate that αPGC-MO treatment can efficiently block host PGC development and that transferred cells can completely replace the host germ line to give rise to functional sperm and eggs.
Fig 3.
PCR genotyping of mil-mutant and WT embryos. Shown is 3% agarose gel resolution of HpyCh4V and AciI digestion products of PCR fragments amplified from genomic DNA prepared from milte273/milte273, milte273/+, and +/+ embryos.
Similar analyses were conducted with WT control chimeras that had been derived from milte273/+ donor embryos (Table 2). If the germ line of host embryos has been completely replaced by milte273/+ PGCs, then matings with milte273/+ fish should generate mil-mutant and WT progeny at a ratio of 1:3. Of the 394 progeny obtained from these crosses, 103 mil-mutant and 291 WT embryos were identified. χ2 analysis indicates no significant deviation from the expected 1:3 mutant to WT progeny ratio for all milte273/+ chimeras tested. This result underscores the efficiency of αPGC-MO mediated germ-line ablation.
Discussion
To our knowledge, this is the first report, in vertebrates, of a method that allows for the complete and functional replacement of a host germ line by PGCs transplanted from homozygous mutant donors. While we have specifically demonstrated the propagation of milte273/milte273 germ cells through chimeric animals, the protocol should be widely applicable for most zygotic mutants, provided the mutation does not interfere in a cell autonomous manner with PGC survival. As such, it should provide an invaluable tool for screening both the maternal effects and less common paternal effects (37) of known zygotic lethal mutations. We have also shown that prudent choices for the host transplantation site (e.g., animal pole instead of margin in the case of mil) can help avoid potential negative consequences of mutant somatic cell contribution to the chimera, and further ensure the success and general applicability of the procedure.
Efficiency of Method.
Two methods were used to label and screen for donor-PGC transfer into host embryos: (i) GFP-nos1–3′UTR mRNA injection, which directed GFP expression specifically to PGCs by late gastrulation (21), and (ii) rhodamine-dextran injection, which labeled all cells and therefore required an ability to discern between labeled PGCs and soma within chimeric embryos. Interestingly, we observed that PGCs could migrate to their normal target tissue at the anterior region of the yolk extension, even when ectopically transplanted to the animal pole of host blastulas. This result provided the separation of rhodamine-dextran labeled soma and PGCs that was required for screening purposes. These observations support a model for PGC migration that favors the activity of long-range positional cues over site-dependent, localized factors in directing PGC movement (19, 35).
The efficiencies of PGC transfer for labeling and screening techniques were similar. Although GFP-nos1–3′UTR injection did not reliably label all PGCs, screening for the presence of GFP-positive PGCs in host embryos was simple and efficient. Conversely, rhodamine-dextran injection consistently labeled all donor PGCs; a higher PGC transfer rate was therefore expected. It is possible, however, that not all transferred PGCs migrated normally from their ectopic transplantation site at the animal pole. Regardless of labeling strategy, the ability to screen for mutant PGC transfer in living host embryos eliminated the need to raise and screen large numbers of adult fish for donor PGC contribution to the germ line.
Of the 26 chimeric fish raised to adulthood, 12 have proven fertile. Moreover, all 8 of the 12 fertile chimeras chosen for further analysis appear to have germ lines that have been completely reconstituted by donor PGCs. Although the efficiency of “complete” germ-line replacement will be better judged when larger numbers of such chimeric animals have been analyzed, our results indicate that αPGC-MO treatment can effectively inhibit PGC development. This conclusion is corroborated by the large number of sterile chimeras obtained. In these cases, rhodamine-dextran- or GFP-labeled PGCs that were observed in the vicinity of the anterior yolk extension may not have migrated to their actual targets and might thus have been excluded from the developing gonad. We expect that proficiency in identifying functional PGC transplantation has increased with experience, and that fertility rates in future experiments will prove higher. Alternatively, too few PGCs may have been transferred in some chimeric embryos to allow for efficient reconstitution of the host germ line. Although studies in Drosophila have demonstrated that a single germ cell has the ability to repopulate the germ line (R. Lehmann, personal communication), little is known about the absolute number of PGCs required for normal zebrafish germ line development. The PGC labeling and transfer techniques outlined in this report should allow the empirical resolution of this issue.
In summary, we have reported a 12% efficiency for PGC transplantation at midblastula stages. Because only 25% of PGC transfers are expected to come from donor embryos homozygous for a given recessive zygotic lethal mutation, the efficiency of homozygous mutant PGC transfer is reduced to 3%. In our facility, 67% of selected chimeras survived until adulthood, of which ≈50% have proven fertile. Given that we have routinely performed 150 transfers per day, one can reasonably expect to generate 1 or 2 fertile adult germ-line replacement chimeras per transplantation experiment.
Potential Applications.
The germ-line replacement strategy should provide a powerful means to analyze the maternal effect of known zygotic mutations. It is free from the inherent disadvantages associated with current MO antisense approaches, which do not eliminate maternally loaded protein and are susceptible to inconsistent and nonspecific phenotypes depending on the timing and concentration of MO injection (38, 39). Furthermore, the germ-line replacement strategy can complement forward genetic analyses and allow investigation into the maternal effects of zygotic mutations that have not yet been cloned.
We have reported the ability to produce both male and female zebrafish chimeras that have germ lines completely reconstituted by donor PGCs. Once the lack of mosaicism in parental germ lines is established, large clutches of embryos that consist entirely of homozygous mutants can be generated; these can subsequently be used as donor embryos, allowing for more efficient production of future generations of germ-line replacement chimeras.
The phenotype of maternal-zygotic mil mutants is not appreciably stronger than zygotic mil mutants. Accordingly, the maternal expression of a gene does not necessarily indicate significant redundancy between the maternal and zygotic gene functions, and germ-line replacement chimeras will not always reveal maternal effects for previously identified zygotic lethal mutations. However, the ability to generate large clutches of entirely homozygous mutants by using germ-line replacement strategies will prove invaluable in embryological and genetic analyses that require positive identification of mutant embryos before the onset of a zygotic phenotype. For example, embryonic fate mapping within different mutant backgrounds (40), as well as mosaic analyses into the cell autonomy of a given mutation (27), can be performed much more efficiently. Large amounts of mutant tissue can be collected for biochemical or microarray experiments without the need for elaborate embryo sorting technologies (41). Finally, homozygous mutant clutches provide abundant material for performing complementation or modifier screens, or MO gene knock-down analyses in any given mutant background. Although we have illustrated the germ-line replacement strategy by using the single mil mutation, the efficiency of the protocol should also allow for the propagation of double and even triple mutations.
The production of zebrafish germ-line chimeras from cultured embryonic cells has been reported (42). This finding raises the possibility that in vitro transgenesis and gene targeting technologies will become feasible in the fish. Germ-line replacement will provide a powerful tool in these strategies: the ability to ablate the PGCs of host embryos by using MOs against dead end should allow for the efficient transmission of genetic alterations into the germ line of chimeric embryos, and the complete replacement of the host germ line with genetically altered material. The phenotype of any genetic manipulation could therefore be studied in the next generation. This result would rival in both speed and efficiency the current transgenic and reverse genetic approaches used to manipulate the mouse genome (43, 44).
Acknowledgments
We thank D. Yelon, R. Lehmann, R. G. Martinho, and members of the Schier and Yelon laboratories for discussions, and S. Zimmerman, T. Bruno, and N. Dillon for fish care. H.K. thanks Christiane Nüsslein-Volhard for support. B.C. received support from the Canadian Institutes of Health Research and the Human Frontier Science Program. H.K. was supported by a predoctoral scholarship from the Boehringer-Ingelheim Fonds. B.T. and C.T. are supported by funds from the Institut National de la Santé et de la Recherche Médicale, the Centre National de la Recherche Scientifique, the Hôpital Universitaire de Strasbourg, the Association pour la Recherche sur le Cancer, the Ligue Nationale Contre le Cancer, and the National Institutes of Health. E.R. is supported by grants from the Deutsche Forschungsgemeinschaft and the Volkswagen-Stiftung. A.F.S. is a Scholar of the McKnight Endowment Fund for Neuroscience, an Irma T. Hirschl Trust Career Scientist, and an Established Investigator of the American Heart Association, and is supported by grants from the National Institutes of Health.
Abbreviations
PGC, primordial germ cell
MO, morpholino oligonucleotide
hpf, hours post fertilization
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Nüsslein-Volhard C. & Wieschaus, E. (1980) Nature 287, 795-801. [DOI] [PubMed] [Google Scholar]
- 2.Haffter P., Granato, M., Brand, M., Mullins, M. C., Hammerschmidt, M., Kane, D. A., Odenthal, J., van Eeden, F. J., Jiang, Y. J., Heisenberg, C. P., et al. (1996) Development (Cambridge, U.K.) 123, 1-36. [DOI] [PubMed] [Google Scholar]
- 3.Driever W., Solnica-Krezel, L., Schier, A. F., Neuhauss, S. C., Malicki, J., Stemple, D. L., Stainier, D. Y., Zwartkruis, F., Abdelilah, S., Rangini, Z., et al. (1996) Development (Cambridge, U.K.) 123, 37-46. [DOI] [PubMed] [Google Scholar]
- 4.St. Johnston D. & Nüsslein-Volhard, C. (1992) Cell 68, 201-219. [DOI] [PubMed] [Google Scholar]
- 5.Jimenez F. & Campos-Ortega, J. A. (1982) Wilhelm Roux's Arch. Entwickslungsmech. Org. 191, 191-201. [DOI] [PubMed] [Google Scholar]
- 6.Perrimon N., Engstrom, L. & Mahowald, A. P. (1984) Dev. Biol. 105, 404-414. [DOI] [PubMed] [Google Scholar]
- 7.Perrimon N., Engstrom, L. & Mahowald, A. P. (1989) Genetics 121, 333-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chou T. B., Noll, E. & Perrimon, N. (1993) Development (Cambridge, U.K.) 119, 1359-1369. [DOI] [PubMed] [Google Scholar]
- 9.Lehmann R. & Nüsslein-Volhard, C. (1986) Cell 47, 141-152. [DOI] [PubMed] [Google Scholar]
- 10.Heasman J. (1997) Development (Cambridge, U.K.) 124, 4179-4191. [DOI] [PubMed] [Google Scholar]
- 11.Abdelilah S., Solnica-Krezel, L., Stainier, D. Y. & Driever, W. (1994) Nature 370, 468-471. [DOI] [PubMed] [Google Scholar]
- 12.Pelegri F., Knaut, H., Maischein, H. M., Schulte-Merker, S. & Nüsslein-Volhard, C. (1999) Curr. Biol. 9, 1431-1440. [DOI] [PubMed] [Google Scholar]
- 13.Kelly C., Chin, A. J., Leatherman, J. L., Kozlowski, D. J. & Weinberg, E. S. (2000) Development (Cambridge, U.K.) 127, 3899-3911. [DOI] [PubMed] [Google Scholar]
- 14.Gritsman K., Zhang, J., Cheng, S., Heckscher, E., Talbot, W. S. & Schier, A. F. (1999) Cell 97, 121-132. [DOI] [PubMed] [Google Scholar]
- 15.Mintzer K. A., Lee, M. A., Runke, G., Trout, J., Whitman, M. & Mullins, M. C. (2001) Development (Cambridge, U.K.) 128, 859-869. [DOI] [PubMed] [Google Scholar]
- 16.Lin S., Long, W., Chen, J. & Hopkins, N. (1992) Proc. Natl. Acad. Sci. USA 89, 4519-4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoon C., Kawakami, K. & Hopkins, N. (1997) Development (Cambridge, U.K.) 124, 3157-3165. [DOI] [PubMed] [Google Scholar]
- 18.Olsen L. C., Aasland, R. & Fjose, A. (1997) Mech. Dev. 66, 95-105. [DOI] [PubMed] [Google Scholar]
- 19.Weidinger G., Wolke, U., Koprunner, M., Klinger, M. & Raz, E. (1999) Development (Cambridge, U.K.) 126, 5295-5307. [DOI] [PubMed] [Google Scholar]
- 20.Knaut H., Pelegri, F., Bohmann, K., Schwarz, H. & Nüsslein-Volhard, C. (2000) J. Cell Biol. 149, 875-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koprunner M., Thisse, C., Thisse, B. & Raz, E. (2001) Genes Dev. 15, 2877-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolke U., Weidinger, G., Koprunner, M. & Raz, E. (2002) Curr. Biol. 12, 289-294. [DOI] [PubMed] [Google Scholar]
- 23.Knaut H., Steinbeisser, H., Schwarz, H. & Nüsslein-Volhard, C. (2002) Curr. Biol. 12, 454-466. [DOI] [PubMed] [Google Scholar]
- 24.Summerton J. & Weller, D. (1997) Antisense Nucleic Acid Drug Dev. 7, 187-195. [DOI] [PubMed] [Google Scholar]
- 25.Nasevicius A. & Ekker, S. C. (2000) Nat. Genet. 26, 216-220. [DOI] [PubMed] [Google Scholar]
- 26.Solnica-Krezel L., Schier, A. F. & Driever, W. (1994) Genetics 136, 1401-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho R. K. & Kane, D. A. (1990) Nature 348, 728-730. [DOI] [PubMed] [Google Scholar]
- 28.Chen J. N., Haffter, P., Odenthal, J., Vogelsang, E., Brand, M., van Eeden, F. J., Furutani-Seiki, M., Granato, M., Hammerschmidt, M., Heisenberg, C. P., et al. (1996) Development (Cambridge, U.K.) 123, 293-302. [DOI] [PubMed] [Google Scholar]
- 29.Stainier D. Y., Fouquet, B., Chen, J. N., Warren, K. S., Weinstein, B. M., Meiler, S. E., Mohideen, M. A., Neuhauss, S. C., Solnica-Krezel, L., Schier, A. F., et al. (1996) Development (Cambridge, U.K.) 123, 285-292. [DOI] [PubMed] [Google Scholar]
- 30.Kupperman E., An, S., Osborne, N., Waldron, S. & Stainier, D. Y. (2000) Nature 406, 192-195. [DOI] [PubMed] [Google Scholar]
- 31.Kimmel C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B. & Schilling, T. F. (1995) Dev. Dyn. 203, 253-310. [DOI] [PubMed] [Google Scholar]
- 32.Kimmel C. B., Warga, R. M. & Schilling, T. F. (1990) Development (Cambridge, U.K.) 108, 581-594. [DOI] [PubMed] [Google Scholar]
- 33.Warga R. M. & Nüsslein-Volhard, C. (1999) Development (Cambridge, U.K.) 126, 827-838. [DOI] [PubMed] [Google Scholar]
- 34.Woo K. & Fraser, S. E. (1995) Development (Cambridge, U.K.) 121, 2595-2609. [DOI] [PubMed] [Google Scholar]
- 35.Weidinger G., Wolke, U., Koprunner, M., Thisse, C., Thisse, B. & Raz, E. (2002) Development (Cambridge, U.K.) 129, 25-36. [DOI] [PubMed] [Google Scholar]
- 36.Ho R. K. & Kimmel, C. B. (1993) Science 261, 109-111. [DOI] [PubMed] [Google Scholar]
- 37.Fitch K. R., Yasuda, G. K., Owens, K. N. & Wakimoto, B. T. (1998) Curr. Top. Dev. Biol. 38, 1-34. [DOI] [PubMed] [Google Scholar]
- 38.Ekker S. C. & Larson, J. D. (2001) Genesis 30, 89-93. [DOI] [PubMed] [Google Scholar]
- 39.Heasman J. (2002) Dev. Biol. 243, 209-214. [DOI] [PubMed] [Google Scholar]
- 40.Melby A. E., Warga, R. M. & Kimmel, C. B. (1996) Development (Cambridge, U.K.) 122, 2225-2237. [DOI] [PubMed] [Google Scholar]
- 41.Furlong E. E., Andersen, E. C., Null, B., White, K. P. & Scott, M. P. (2001) Science 293, 1629-1633. [DOI] [PubMed] [Google Scholar]
- 42.Ma C., Fan, L., Ganassin, R., Bols, N. & Collodi, P. (2001) Proc. Natl. Acad. Sci. USA 98, 2461-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Capecchi M. R. (1989) Science 244, 1288-1292. [DOI] [PubMed] [Google Scholar]
- 44.Rossant J. & Nagy, A. (1995) Nat. Med. 1, 592-594. [DOI] [PubMed] [Google Scholar]