Abstract
RNA interference is an evolutionarily conserved process in which recognition of double-stranded RNA ultimately leads to posttranscriptional suppression of gene expression. This suppression is mediated by short (21- to 22-nt) small interfering RNAs (siRNAs), which induce degradation of mRNA based on complementary base pairing. The silencing of gene expression by siRNAs is emerging rapidly as a powerful method for genetic analysis. Recently, several groups have reported systems designed to express siRNAs in mammalian cells through transfection of either oligonucleotides or plasmids encoding siRNAs. Because these systems rely on transfection for delivery, the cell types available for study are restricted generally to transformed cell lines. Here, we describe a retroviral system for delivery of siRNA into cells. The use of retroviral vectors can greatly expand the types of cells available for RNA interference analysis. Furthermore, we demonstrate that this retroviral system allows for stable inactivation of genes in primary cells.
It has long been appreciated that detection of double-stranded RNA (dsRNA) within mammalian cells leads to an IFN response initiated by the sequence-independent recognition of dsRNA (1). More recently, it was discovered that dsRNA also could induce a sequence-specific degradation of mRNA, effectively silencing expression of a given gene (2). This process of RNA interference (RNAi) initially was discovered in the nematode Caenorhabditis elegans (3), but it is now clear that RNAi is an ancient, evolutionarily conserved process. Within invertebrate organisms, which lack the sequence-independent dsRNA response, RNAi has become a powerful method to quickly perform genetic analysis of a given gene.
The complete molecular mechanism responsible for RNAi is not yet known, although it has been shown that silencing does involve cleavage of both the initiating dsRNA and the target mRNA into 21- to 23-nt fragments (4). Furthermore, 21- to 23-nt dsRNAs, or small interfering RNAs (siRNAs), are capable of inducing RNAi in mammalian cells without initiating the sequence-independent dsRNA response (5). Consequently, the challenge to mammalian biologists has been to develop systems that deliver siRNAs efficiently into mammalian cells. Recently, a number of groups reported a solution based on transcription of short hairpin RNAs by RNA polymerase III (pol III) (6–10). The hairpins of these short RNAs are processed to generate siRNAs and induce gene silencing. Transfection of plasmids with pol III promoters driving hairpin RNAs can eliminate expression of a target gene.
The limitation of any plasmid- or oligonucleotide-based system is the dependence on transfection. Only certain cell lines can be transfected, and efficient transfection of primary cells is virtually impossible. In contrast, retroviral gene delivery is effective in most cell lines and many primary cell types. We have developed a retroviral system for delivery of siRNA into mammalian cells. This system can be used to silence gene expression in any cell line or primary cell type that can be infected by a retrovirus.
Materials and Methods
Constructs.
The RVH1 and LTRH1 vectors are based on the pQCXIH vector (CLONTECH). The hygromycin resistance gene was replaced with the human CD4 gene, which was cloned from splenic cDNA. To eliminate any potential signaling, a premature stop codon was introduced after amino acid 425, just after the transmembrane domain. The human H1 promoter was cloned from genomic DNA and inserted either upstream of the cytomegalovirus (CMV) promoter (RVH1) by using previously introduced XhoI and EcoRI sites or within the 3′ LTR by using SalI. The oligonucleotides encoding the human p53 siRNA, described by Brummelkamp et al. (6), were 5′-GATCCCCGACTCCAGTGGTAATCTACTTCAAGAGAGTAGATTACCACTGGAGTCTTTTTGGAAC-3′ and 5′-TCGAGTTCCAAAAAGACTCCAGTGGTAATCTACTCTCTTGAAGTAGATTACCACTGGAGTCGGG-3′. These oligonucleotides were annealed and ligated downstream of the H1 promoter. All oligonucleotides were synthesized by the Keck Facility at Yale University.
Generation of Virus.
The packaging cell line GP2-293 (CLONTECH) was grown in DMEM with 10% FCS/10 mM Hepes/2 mM l-glutamine/1 mM MEM sodium pyruvate/100 units/ml penicillin/100 μg/ml streptomycin (all from Invitrogen). Cells in 10-cm dishes were transfected by calcium phosphate precipitation with 15 μg pVSV-G (CLONTECH) and 15 μg of RVH1 or LTRH1. Chloroquine (Sigma) was added to a final concentration of 25 μM 5 min before transfection. The medium was replaced 8 h posttransfection. Twenty-four hours posttransfection, cells were placed at 32°C to increase viral titer. Forty-eight hours posttransfection, the supernatant was collected, filtered through a 0.45-μm syringe filter, and spun at 50,000 × g for 1.5 h. Pelleted virus was resuspended in 0.l or 0.05 the original volume of medium at 4°C for several hours.
Infection of Target Cells.
HEK293T cells were grown in DMEM with 10% FCS/10 mM Hepes/2 mM l-glutamine/1 mM MEM sodium pyruvate/100 units/ml penicillin/100 μg/ml streptomycin. Primary human fibroblasts (AG01522C cells; Coriell Cell Repositories, Camden, NJ) were grown in MEM α (Invitrogen) with 15% FCS/2 mM l-glutamine/100 units/ml penicillin/100 μg/ml streptomycin. The day before infection, 3 × 105 293T cells or 2 × 105 AG01522 cells were plated per well of six-well plates. The next day, virus supernatant was added with polybrene (5 μg/ml final concentration), and the cells were placed at 32°C overnight. The next day, the medium was replaced and the cells were returned to 37°C until analysis. Etoposide, used to treat AG01522 cells, was purchased from Calbiochem.
Western Blots.
Cells were harvested at the indicated times, washed twice with cold PBS, and lysed in TNT lysis buffer (20 mM Tris/200 mM NaCl/1% Triton X-100) plus protease inhibitors. After 20 min on ice, lysates were spun at 15,000 × g for 10 min to remove insoluble material. Protein concentrations were determined by BCA assay (Pierce). Thirty micrograms of lysate was separated on 10% or 12% SDS/PAGE gels, transferred to Immobilon-P membrane (Millipore), and incubated with anti-p53 or anti-p21 antisera (both from Santa Cruz Biotechnology), followed by incubation with donkey anti-rabbit secondary antibody conjugated to horseradish peroxidase (Amersham Pharmacia). Western blots were developed by using enhanced chemiluminescence (Amersham Pharmacia).
Flow Cytometry.
Infected cells were harvested, washed twice with FACS buffer (PBS/3% FCS/0.0025% sodium azide), and incubated for 30 min with anti-hCD4-PE antibody (PharMingen). Cell analysis was performed with a FACScan or FACSCalibur instrument by using cellquest software (PharMingen).
Southern Blots.
Genomic DNA (10 μg) was digested with EcoRI and NheI overnight, separated on a 1.2% agarose gel, and transferred to Hybond nylon membrane (Amersham Pharmacia) under alkaline conditions. A DNA probe consisting of an EcoRI/XbaI fragment from RVH1 (encoding the CMV promoter) was labeled with horseradish peroxidase by using the North2South Direct HRP Labeling and Detection Kit (Pierce). Hybridization and developing of the blot were performed according to the manufacturer's instructions.
Results and Discussion
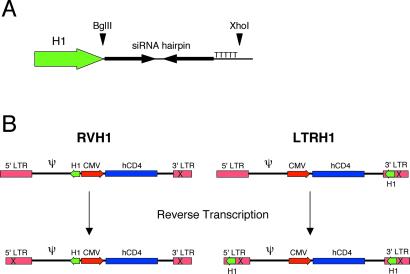
It has been demonstrated recently that pol III-dependent promoters can be used to drive expression of short hairpin RNAs, which then are processed into siRNAs (6–10). We sought to develop a retroviral vector that would drive expression of siRNAs from the pol III-dependent H1 promoter. Anticipating that expression of a pol III promoter from within a provirus would be hampered by interference from the strong LTR promoter (11), we constructed two viruses to circumvent this problem (Fig. 1). The first, RVH1, lacks the U3 portion of the 3′ LTR. Through the course of reverse transcription, this deletion is copied into the 5′ LTR. Consequently, the 5′ LTR of the integrated provirus is transcriptionally inactive. The second vector, LTRH1, contains the H1 cassette within the 3′ LTR such that the entire cassette is copied into the 5′ LTR, upstream of the promoter elements, during reverse transcription. This design has the added benefit of duplicating the H1 cassette such that the integrated provirus has two copies. Unique restriction endonuclease sites allow the hairpin-encoding oligonucleotides to be cloned directly into both retroviral vectors. In addition, both vectors express a version of human CD4 that lacks the intracellular domain, so infected cells can be identified by cell surface staining with anti-CD4 antibodies.
Fig 1.
(A) Schematic representation of the H1 promoter cassette. Unique restriction endonuclease sites allow cloning of hairpin-encoding oligonucleotides directly into the retroviral vectors. The five thymidines serve as a termination signal for RNA pol III. (B) Schematic representations of RVH1 and LTRH1 retroviral vectors. The vectors are shown above and the integrated proviruses are shown below. During reverse transcription, portions of the 3′ LTR serve as the template for generating the new 5′ LTR. The “X” in the 3′ LTR in the upper schematics represents deletion of the U3 region, which is copied into the 5′ LTR.
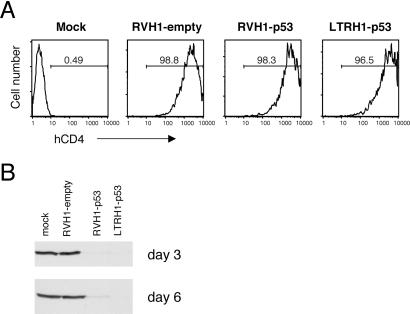
We initially targeted human p53 gene expression in HEK293T cells, using a 21-nt sequence previously shown to induce RNAi against human p53 (6). 293T cells have high endogenous levels of p53 because of expression of SV40 large T antigen, which stabilizes and inactivates p53. To produce high titer virus, we generated pseudotyped virus by using the vesicular stomatitis virus G protein in place of the normal retroviral envelope. The stability of vesicular stomatitis virus G protein permits concentration of virus by ultracentrifugation (12). Consequently, we were able to infect all cells in a given experiment, eliminating the need to select or sort for infected cells (Fig. 2A). Infection of 293T cells with high titer RVH1-p53 or LTRH1-p53 virus eliminated p53 expression completely when compared with mock-infected cells or cells infected with a virus lacking the p53 hairpin, RVH1-empty (Fig. 2B). Importantly, because these retroviral vectors integrate into the genomes of the target cells, the elimination of expression was stable over time, effectively creating a mutant cell line.
Fig 2.
Infection of 293T cells with RVH1-p53 or LTRH1-p53 eliminates expression of p53. (A) CD4 expression on 293T cells infected with RVH1 vector, RVH1-p53, or LTRH1-p53 or mock-infected, as measured by flow cytometry. The percentage of infected cells 3 days postinfection is indicated. (B) 293T cells infected as in A were analyzed for p53 expression by Western blotting. Western blots are shown 3 and 6 days postinfection.
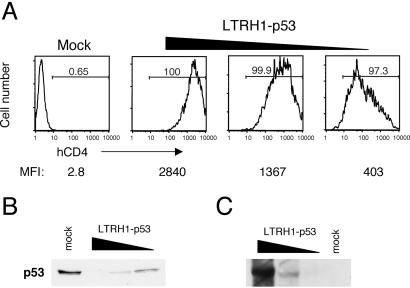
Through the course of our analysis, we noticed that the reduction in p53 expression in infected cells correlated with expression of CD4 (Fig. 3). The best silencing was achieved in superinfected cells, i.e., cells infected by multiple viruses. As shown in Fig. 3A, populations of 293T cells infected with serial dilutions of virus expressed decreasing levels of CD4 even though all of the cells within the population were infected. When we examined p53 expression in these cells, we found that the higher expression of CD4 correlated with more efficient silencing of p53 (Fig. 3B). In fact, in cases in which all of the cells expressed CD4, but at relatively low levels, silencing was not always complete. These data indicate that silencing is most efficient when multiple virions infect a given cell, leading to multiple copies of the H1 cassette integrated within the cellular genome. To demonstrate this directly, we performed Southern blot analysis on genomic DNA from infected cells by using a probe for the retroviral vector. As suggested by the analysis of CD4 expression, populations of infected cells with more complete silencing of p53 had more copies of the integrated retroviral vectors within their genomes (Fig. 3C). These data agree with the reports of plasmid-based pol III siRNA systems in which silencing was dose-dependent and may suggest that expression from pol III promoters is relatively weak (6), although the need for high expression may vary depending on which gene is targeted. Generation of vectors with multiple copies of the H1 cassette may overcome the need for high copy number or superinfection.
Fig 3.
Multiple copies of the H1 cassette are necessary for complete silencing. (A) 293T cells were infected with serial dilutions of LTRH1-p53, and p53 expression was measured 3 days postinfection. The percentage of infected cells was determined by measuring CD4 expression by flow cytometry. The infection percentage and mean fluorescence intensity corresponding to each dilution are indicated. (B) 293T cells infected as described in A were analyzed for p53 expression by Western blot. (C) 293T cells infected as in A were analyzed for integration of the retroviral vector by Southern blot. A probe corresponding to the CMV promoter was used to detect the integrated viruses within genomic DNA from infected cells.
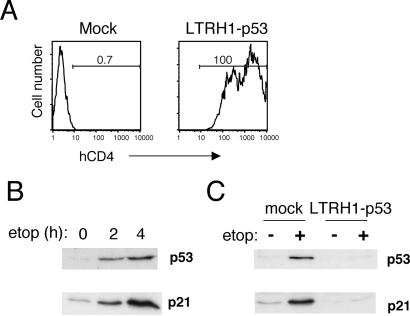
One of the powerful aspects of retroviral gene delivery is the ability to introduce genes into otherwise intractable cell types. In particular, primary cells often can be transduced efficiently by using retroviral vectors. Using high-titer virus, we were able to infect primary human fibroblasts at efficiencies of greater than 99% (Fig. 4A). We infected AG01522 cells (primary human fibroblasts) with the LTRH1-p53 vector and analyzed p53 induction after treatment with the topoisomerase inhibitor, etoposide, which induces DNA damage (Fig. 4). Etoposide treatment of AG01522 cells led to up-regulation of p53 after 2 h (Fig. 4B), demonstrating that these cells have an intact DNA damage response. DNA damage also led to induction of the p53-dependent gene p21WAF1, which inhibits cell cycle progression. Infection of primary fibroblasts with LTRH1-p53 virus before etoposide treatment blocked the up-regulation of p53 and p21WAF1, whereas mock-infected cells were still able to mount the appropriate DNA damage response (Fig. 4B). These data demonstrate that LTRH1-p53-infected cells are functionally p53-deficient. Not only is p53 expression abrogated, but induction of p53-dependent genes is also prevented.
Fig 4.
Retroviral delivery of siRNA into primary fibroblasts eliminates p53 expression. (A) FACS analysis of CD4 expression on primary human fibroblasts infected with LTRH1-p53 or mock-infected. (B) Primary human fibroblasts (AG01522 cells) were treated with 100 μM etoposide for the indicated times, after which the cells were harvested and p53 and p21 expression levels were determined by Western blot analysis. (C) Primary fibroblasts were infected with LTRH1-p53 virus as in A. Three days postinfection, cells were treated with 100 μM etoposide (etop). After 4 h of etoposide treatment, cells were harvested and expression levels of p53 and p21 were measured by Western blot analysis.
In conclusion, we have developed a retroviral system capable of initiating RNAi against target genes in infected cells. This system can be used to assess rapidly the effect of specific genetic deficiencies in a wide range of cell lines and primary cell types. Furthermore, the ability to stably inactivate target genes in primary cells, in many cases, will save researchers the considerable expense and time required for generation of knockout mice. The approaches we have taken to increase the efficiency of retroviral-mediated RNAi can be applied easily to other retroviral vectors, most notably lentiviral systems with even broader host ranges. The development of appropriate siRNA delivery vectors eventually may have important clinical applications, especially in combating viruses that prove resistant to treatment, such as HIV (10, 13–15).
Acknowledgments
We thank A. Unni for thoughtful advice and discussions, C. Pasare for helpful comments on the manuscript, and T. Horng for providing the H1 promoter. This work was supported by the National Institutes of Health and The Howard Hughes Medical Institute.
Abbreviations
dsRNA, double-stranded RNA
RNAi, RNA interference
siRNA, small interfering RNA
pol III, RNA polymerase III
CMV, cytomegalovirus
References
- 1.Williams B. R. (1999) Oncogene 18, 6112-6120. [DOI] [PubMed] [Google Scholar]
- 2.Sharp P. A. (2001) Genes Dev. 15, 485-490. [DOI] [PubMed] [Google Scholar]
- 3.Fire A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E. & Mello, C. C. (1998) Nature 391, 806-811. [DOI] [PubMed] [Google Scholar]
- 4.Elbashir S. M., Lendeckel, W. & Tuschl, T. (2001) Genes Dev. 15, 188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elbashir S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- 6.Brummelkamp T. R., Bernards, R. & Agami, R. (2002) Science 296, 550-553. [DOI] [PubMed] [Google Scholar]
- 7.Paddison P. J., Caudy, A. A., Bernstein, E., Hannon, G. J. & Conklin, D. S. (2002) Genes Dev. 16, 948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paul C. P., Good, P. D., Winer, I. & Engelke, D. R. (2002) Nat. Biotechnol. 20, 505-508. [DOI] [PubMed] [Google Scholar]
- 9.Miyagishi M. & Taira, K. (2002) Nat. Biotechnol. 20, 497-500. [DOI] [PubMed] [Google Scholar]
- 10.Lee N. S., Dohjima, T., Bauer, G., Li, H., Li, M. J., Ehsani, A., Salvaterra, P. & Rossi, J. (2002) Nat. Biotechnol. 20, 500-505. [DOI] [PubMed] [Google Scholar]
- 11.Emerman M. & Temin, H. M. (1984) Cell 39, 449-467. [PubMed] [Google Scholar]
- 12.Burns J. C., Friedmann, T., Driever, W., Burrascano, M. & Yee, J. K. (1993) Proc. Natl. Acad. Sci. USA 90, 8033-8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gitlin L., Karelsky, S. & Andino, R. (2002) Nature 418, 430-434. [DOI] [PubMed] [Google Scholar]
- 14.Jacque J. M., Triques, K. & Stevenson, M. (2002) Nature 418, 435-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novina C. D., Murray, M. F., Dykxhoorn, D. M., Beresford, P. J., Riess, J., Lee, S. K., Collman, R. G., Lieberman, J., Shankar, P. & Sharp, P. A. (2002) Nat. Med. 8, 681-686. [DOI] [PubMed] [Google Scholar]